Abstract
Endocytosis performs a wide range of functions in animals and plants. Clathrin-coated vesicle (CCV) formation is an initial step of endocytosis, and in animal cells is largely achieved by dynamins. However, little is known of its molecular mechanisms in plant cells. To identify dynamin-related proteins (DRPs) involved in endocytic CCV formation in plant cells, we compared the behaviors of two structurally different Arabidopsis DRPs, DRP2B and DRP1A, with those of the clathrin light chain (CLC), a marker of CCVs, at the plasma membrane by variable incidence angle fluorescent microscopy (VIAFM). DRP2B shares domain organization with animal dynamins whereas DRP1A is plant-specific. We show that green fluorescent protein (GFP)-tagged DRP2B and DRP1A colocalized with CLC tagged with monomeric Kusabira Orange (mKO) in Arabidopsis cultured cells. Time-lapse VIAFM observations suggested that both GFP-DRP2B and GFP-DRP1A appeared and accumulated on the existing mKO-CLC foci and disappeared at the same time as or immediately after the disappearance of mKO-CLC. Moreover, DRP2B and DRP1A colocalized and assembled/disassembled together at the plasma membrane in Arabidopsis cells. A yeast two-hybrid assay showed that DRP2B and DRP1A interacted with each other. An inhibitor of clathrin-mediated endocytosis, tyrphostin A23, disturbed the localization of DRP1A, but had little effect on the localization of DRP2B, indicating that DRP1A and DRP2B have different molecular properties. These results suggest that DRP2B and DRP1A participate together in endocytic CCV formation in Arabidopsis cells despite the difference of their molecular properties.
Keywords:
Endocytosis is a fundamental process in eukaryotic cells for engulfing external materials and for regulating the abundance and distribution of plasma membrane proteins and lipids (1). Endocytosis requires the participation of diverse molecular machineries. Compared with molecular mechanisms of endocytosis in animal and yeast cells, those in plant cells are poorly understood (2). Although plant genomes have many genes that are similar to endocytosis-related genes in animals and yeasts, few of the plant genes have been examined.
Clathrin is a major coat protein involved in endocytic vesicle formation. The clathrin coat is made up of a three-legged structure, called the triskelion, in which each leg consists of a clathrin heavy chain (CHC) and a clathrin light chain (CLC) (3). Triskelia are assembled at the vesicle formation sites on the plasma membrane and form a lattice surrounding the invaginating membrane. In animal and yeast cells during the formation of a vesicle from the plasma membrane, the behavior of clathrin is thought to correspond to that of CLC, as shown by total internal reflection fluorescence microscopy (TIRFM) of fluorescently labeled clathrin light chain (CLC) (4, 5). TIRFM is an optical technique for observing the fluorescence in a thin (∼400 nm) layer near the cover glass.
The Arabidopsis genome has a gene encoding CHC and three genes encoding CLC. Clathrin-dependent endocytosis was recently shown to be a predominant internalization pathway in plant cells (6). The localization and behavior of Arabidopsis CLC at the plasma membrane have been observed by a TIRFM-like technique, called variable angle epifluorescence microscopy (7–9) or variable incidence angle fluorescence microscopy (VIAFM) (10). This TIRFM-like technique, by using oblique incident light at the appropriate angle, makes it possible to observe fluorescence just beneath the plasma membrane in plant cells surrounded by the cell wall. The illumination depths in Arabidopsis root epidermal cells and in tobacco cultured cells were estimated to be 700 nm (8) and 550 nm (10), respectively.
Dynamin and dynamin-related proteins (DRPs) are large GTPases that tubulate or pinch off membranes (11). Numerous studies have revealed that these proteins are involved in diverse cellular membrane-remodeling events including vesicular transport, fissions of organelles, and cytokinesis (12). Dynamin is required for clathrin-mediated trafficking, including endocytosis in animals (13). During clathrin-coated vesicle (CCV) formation in endocytosis, dynamin assembles into helical structures at the neck of clathrin-coated buds (14) and constricts and pinches off the bud neck membrane (15). The dynamin helix undergoes a conformational change upon its hydrolysis of GTP (15). In TIRFM, the fluorescent fusions of dynamin accumulate at the clathrin assembly sites (4, 16). The Arabidopsis genome has 16 DRPs grouped into 6 subfamilies (DRP1–DRP6) on the basis of their amino acid sequence and predicted domains (17). The DRP1 and DRP2 subfamilies are predicted to have roles in CCV formation during endocytosis (18).
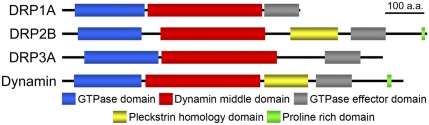
Dynamin family proteins contain three distinctive domains: an N-terminal GTPase domain that binds and hydrolyses GTP, a middle domain that is involved in oligomerization, and a GTPase-effector domain that regulates GTPase activity (11, 12). Dynamin contains two additional domains, a pleckstrin homology domain (PH) and a proline-rich domain (PRD), which are important for lipid binding (19) and movement to vesicle formation sites (20) (Fig. 1). Among Arabidopsis DRPs, only the DRP2 subfamily members contain both PH and PRD and have structural features similar to those of dynamin (17) (Fig. 1). The DRP1 subfamily members lack PH and PRD (17) (Fig. 1).
Fig. 1.
Domain structures of Arabidopsis DRPs analyzed in our study and animal dynamin. Each domain of DRP1A, DRP2B, DRP3A, and animal dynamin is identified and depicted by using the pfam program (http://pfam.sanger.ac.uk/).
The DRP1 subfamily consists of five members (DRP1A–DRP1E) (17) and have roles in cell expansion and cytokinesis (21, 22). DRP1A, DRP1C, and DRP1E accumulate in the cell plate during cytokinesis and localize to the cytoplasmic surface of the plasma membrane (21, 23). A null mutation of DRP1A reduced the internalization of FM4-64 (a styryl dye for endo-membrane) from the plasma membrane, which indicates that DRP1A is related to endocytosis (24). GFP-tagged DRP1A and DRP1C form dot-like foci that overlap the foci of orange fluorescently labeled CLC at the plasma membrane in Arabidopsis root epidermal cells (7, 9).
The DRP2 subfamily has two members, DRP2A and DRP2B (17). DRP2A, which is involved in trans-Golgi network trafficking (25), was fractionated with CCV and interacted with the clathrin adaptor γ-adaptin (26). Proteomic analysis of the plasma membrane suggested that DRP2B is associated with the plasma membrane (27). GFP-tagged DRP2B was observed as 200- to 500-nm diameter dot-like foci in the plasma membrane, similar to DRP1A foci in tobacco cultured cells (10). Moreover, during cytokinesis, DRP2B is colocalized with DRP1A on the leading edge of the forming cell plate.
However, it is not known whether DRP2 subfamily proteins are related to CCV formation in endocytosis. If they are, do DRP2 subfamily proteins function in CCV formation together with DRP1 subfamily proteins? To understand the molecular mechanism of endocytic CCV formation in plant cells, we compared the localizations and temporal behaviors of DRP2B and DRP1A with those of CLC at the plasma membrane by VIAFM. We also compared the localizations and temporal behaviors of DRP2B with those of DRP1A at the plasma membrane and investigated whether DRP2B interacts with DRP1A.
Results
DRP2B Accumulates to Clathrin Assembly Sites at the Plasma Membrane.
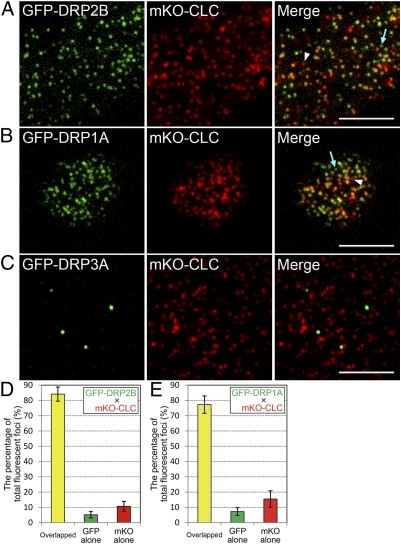
To examine whether DRP2B colocalizes with clathrin at the plasma membrane, we compared the fluorescent signals of DRP2B and CLC by variable incidence angle fluorescent microscopy (VIAFM). In VIAFM images of Arabidopsis cultured cells expressing GFP-DRP2B and mKO-CLC under the control of Cauliflower mosaic virus 35S promoter (CaMV35S), the fluorescent signals of these proteins were observed as discrete foci with diameters of 200–500 nm (Fig. 2A, Left and Center). Although foci of almost pure GFP and almost pure mKO were sometimes observed (Fig. 2A, Right, blue arrow and white arrowhead), most of the GFP-DRP2B fluorescent foci were colocalized with mKO-CLC foci (Fig. 2A, Right). In ten 100-μm2 VIAFM images containing a total of 1385 fluorescent foci (GFP and mKO overlapping foci, GFP-only foci, and mKO-only foci), 84.1 ± 4.6% (mean ± SD) of the foci were GFP-DRP2B and mKO-CLC overlapping foci (Fig. 2D). Like the DRP2B foci, most of the GFP-DRP1A foci also colocalized with mKO-CLC foci (Fig. 2B, Right). As for DRP1A, in ten 100-μm2 VIAFM images containing a total of 1261 fluorescent foci, 77.3 ± 5.7% (mean ± SD) of the foci were GFP-DRP1A and mKO-CLC overlapping foci (Fig. 2E). These results suggest that DRP2B, like DRP1A, is almost entirely colocalized with clathrin at the plasma membrane.
Fig. 2.
Localizations of DRP2B, DRP1A, and DRP3A with CLC at the plasma membrane. (A–C) VIAFM images of Arabidopsis cultured cells expressing GFP-tagged DRP2B (A), DRP1A (B), DRP3A (C), and mKO-tagged CLC. Left, Center, and Right in A–C represent GFP, mKO, and merged images, respectively. Blue arrows and white arrowheads in the merged images of A and B indicate GFP- and mKO-only foci, respectively. (Scale bars: 5 μm.) (D and E) Bar graphs representing the percentage of GFP and mKO overlapped foci (yellow bar), pure GFP foci (green bar), and pure mKO foci (red bar) in VIAFM images of Arabidopsis cultured cells expressing GFP-DRP2B and mKO-CLC (D) and GFP-DRP1A and mKO-CLC (E).
To check whether Arabidopsis DRPs generally colocalize with clathrin at the plasma membrane, we compared the fluorescent signals of CLC and DRP3A, which is involved in the fission of mitochondria and peroxisomes (28, 29) and which differs from DRP2B and DRP1A in its domain structure (Fig. 1). In VIAFM images of Arabidopsis cultured cells, GFP-tagged DRP3A also formed discrete foci like those of GFP-DRP2B and GFP-DRP1A (Fig. 2C, Left). However, GFP-DRP3A foci were much fewer in number than those of mKO-CLC and were not colocalized with mKO-CLC foci (Fig. 2C, Right). These results strengthen the possibility that the colocalization with clathrin is not a common feature of Arabidopsis DRPs, but a specific feature of the DRP1 and DRP2 subfamilies.
Temporal Behavior of DRP2B Is Similar to That of DRP1A on Clathrin Assembly Sites.
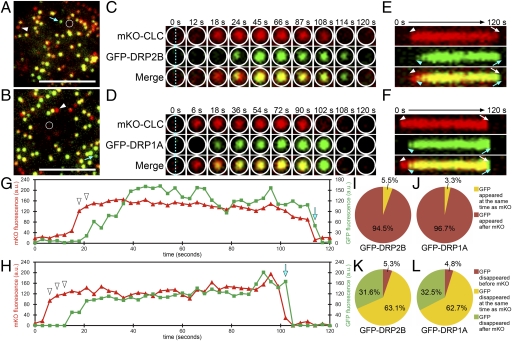
We compared the temporal behaviors of GFP-DRP2B and GFP-DRP1A foci to the temporal behavior of mKO-CLC foci. Fig. 3 A and B shows the VIAFM images of Arabidopsis cultured cells expressing GFP-DRP2B or GFP-DRP1A and mKO-CLC at the beginning (t = 0 s) of each time-lapse observation covering 120 s with 3-s intervals, respectively. Additional examples of appearing and disappearing behavior are shown in Movies S1 and S2. Fig. 3 C and D, respectively, show time-lapse images of the areas outlined by white lines in Fig. 3 A and B. In these images, interestingly, foci with only mKO-CLC fluorescence appeared in the center of the areas (Fig. 3 C and D: t = 18 and 6 s, respectively). Subsequently, GFP-DRP2B and GFP-DRP1A fluorescence also appeared on the mKO-CLC foci (Fig. 3 C and D: t = 24 and 18 s, respectively). At t = 114 and 102 s, respectively, in Fig. 3 C and D, mKO-CLC fluorescence disappeared and almost pure GFP-DRP2B and GFP-DRP1A foci remained. As time passed, the GFP-DRP2B and GFP-DRP1A fluorescence also disappeared in these areas (Fig. 3 C and D: t = 120 and 108 s, respectively).
Fig. 3.
Behaviors of DRP2B and DRP1A at CLC assembly sites at the plasma membrane. (A and B) VIAFM images of Arabidopsis cultured cells expressing mKO-CLC (in red) and GFP-DRP2B (A) or GFP-DRP1A (B) (in green) at the beginning of time-lapse observation covering 120 s. Blue arrows and white arrowheads in A and B indicate GFP- and mKO-only foci, respectively. (Scale bars: 5 μm.) (C and D) Ten frames of the region surrounded by white lines in A and B are from Movies S1 and S2. Top, Middle, and Bottom in each column represent mKO, GFP, and merged images, respectively. (E and F) Kymographs representing the fluorescence on the blue dotted lines of C and D. Top, Middle, and Bottom represent mKO, GFP, and merged images, respectively. Arrowheads and arrows in white and blue indicate the appearance and disappearance timing of mKO and GFP signals, respectively. (G and H) Time-intensity profiles of mKO-CLC fluorescence (red lines) and GFP-DRP2B (G) or GFP-DRP1A (H) fluorescence (green lines) within the areas surrounded by white lines in A and B. (I and J) Pie charts showing the percentage of the foci in which GFP-DRP2B (I) and GFP-DRP1A (J) appeared at the same time as (yellow) or after (red) mKO-CLC for 120 s. (K and L) Pie charts showing the percentage of the foci in which the fluorescence of GFP-DRP2B (K) and GFP-DRP1A (L) disappeared before (red), at the same time as (yellow), or after (green) mKO-CLC for 120 s.
Fig. 3 E and F show kymographs representing the time-course transition of mKO and GFP fluorescence intensities on the blue dotted line of the images at 0 s in Fig. 3 C and D. In these kymographs, GFP-DRP2B and GFP-DRP1A fluorescence (Fig. 3 E and F: blue arrowheads and arrows, respectively) appeared and disappeared later than mKO-CLC fluorescence (Fig. 3 E and F: white arrowheads and arrows).
A comparison of the profiles of mKO-CLC with those of GFP-DRP2B and GFP-DRP1A (Fig. 3 G and H: red and green lines, respectively) shows that the mKO-CLC fluorescence appeared and disappeared earlier than the GFP-DRP2B and GFP-DRP1A fluorescence (Fig. 3 G and H: white arrowheads and blue arrows, respectively). These results suggest that some almost pure mKO or almost pure GFP foci observed in single-time point VIAFM images (Fig. 2 A and B and Fig. 3 A and B: white arrowheads and blue arrows, respectively) were generated during the time between the appearance and disappearance of the overlapping CLC/DRP foci.
To determine what percentage of DRP2B and DRP1A foci appear or disappear later than CLC foci at the same position, we analyzed the appearing and disappearing behaviors of DRP2B foci (Movie S1) and DRP1A foci (Movie S2) on each CLC focus. In most of the appearing foci, GFP-DRP2B and GFP-DRP1A fluorescence appeared several frames later than mKO-CLC fluorescence (Fig. 3 I and J). On the other hand, in most of the disappearing foci, GFP-DRP2B and GFP-DRP1A fluorescence disappeared at the same frame as or a few frames later than the mKO-CLC fluorescence (Fig. 3 K and L). These results suggest that both DRP2B and DRP1A begin to accumulate at the vesicle formation sites of the plasma membrane after the clathrin assembly and detach from there at the same time as or immediately after the clathrin disassembly.
DRP2B and DRP1A Colocalize at the Plasma Membrane.
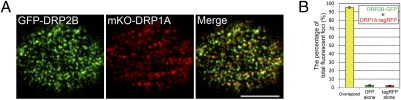
To investigate whether DRP2B colocalizes with DRP1A in the same foci at the plasma membrane, we compared the localizations of GFP-DRP2B and mKO-DRP1A in Arabidopsis cultured cells. In VIAFM images, almost all of the GFP and mKO signals also formed discrete foci (Fig. 4A, Left and Center) and colocalized with each other (Fig. 4A, Right). This result suggests that DRP2B colocalizes with DRP1A at the plasma membrane.
Fig. 4.
Colocalization of DRP2B and DRP1A at the plasma membrane. (A) VIAFM images of Arabidopsis cultured cells expressing GFP-DRP2B (in green) and mKO-DRP1A (in red). (Scale bar: 5 μm.) (B) Bar graphs representing the percentage of GFP and tagRFP overlapped foci (yellow bar), pure GFP foci (green bar), and pure tagRFP foci (red bar) in VIAFM images of Arabidopsis root epidermal cells expressing DRP2B-GFP and DRP1A-tagRFP under the control of their own promoters.
The VIAFM images showed only small numbers of GFP-DRP3A foci (Fig. 2C). To check whether DRP2B and DRP1A also colocalize with DRP3A, we compared the localization of GFP-DRP2B or GFP-DRP1A with that of mKO-DRP3A. In the VIAFM images, the signals of GFP and mKO did not overlap with each other (Fig. S1).
To confirm that DRP2B and DRP1A colocalize in Arabidopsis plants, we compared the localizations of GFP fused to the C terminus of DRP2B (DRP2B-GFP) and of tagRFP fused to the C terminus of DRP1A (DRP1A-tagRFP) in Arabidopsis root epidermal cells. These two fluorescent fusions were expressed under the control of ≈1.5-kb genomic sequences immediately upstream of DRP2B and DRP1A, respectively. In the VIAFM images of Arabidopsis root epidermal cells, the fluorescent signals of DRP2B-GFP and DRP1A-tagRFP were also observed as foci, and almost all of these foci overlapped with each other (Fig. S2). In five 100-μm2 VIAFM images containing a total of 694 fluorescent foci (GFP and tagRFP overlapping foci, GFP-only foci, and tagRFP-only foci), 95.3 ± 0.9% (mean ± SD) of the foci were DRP2B-GFP and DRP1A-tagRFP overlapping foci (Fig. 4B). These results indicate that DRP2B and DRP1A are expressed together and colocalize with each other at the plasma membrane in the cells (at least in the root epidermal cells) of Arabidopsis plants.
DRP2B and DRP1A Assemble and Disassemble Together at the Plasma Membrane.
To investigate whether DRP2B assembles and disassembles together with DRP1A, we compared the appearing and disappearing behaviors of DRP2B-GFP with those of DRP1A-tagRFP at the plasma membrane. Fig. 5A shows the VIAFM image of Arabidopsis root epidermal cells expressing DRP2B-GFP and DRP1A-tagRFP at the beginning (t = 0 s) of each time-lapse observation covering 5 s with 200-ms intervals, respectively. Additional examples of appearing and disappearing behavior are shown in Movie S3. Fig. 5 B and C show time-lapse images of the areas surrounded by the white and blue lines in Fig. 5A. In Fig. 5B, DRP2B-GFP and DRP1A-tagRFP foci (shown in green and red, respectively) seemed to appear at the same time (Fig. 5B, t = 3.8 s). As time passed, these GFP and tagRFP signals increased gradually. On the other hand, in Fig. 5C, GFP and tagRFP signals within the area surrounded by the blue lines gradually decreased, and these fluorescent foci seemed to disappear at the same time (Fig. 5C, t = 1.8 s).
Fig. 5.
Behaviors of DRP2B-GFP and DRP1A-tagRFP foci at the plasma membrane in an Arabidopsis root epidermal cell. (A) VIAFM image of a root epidermal cell expressing DRP2B-GFP (in green) and DRP1A-tagRFP (in red). (Scale bar: 5 μm.) (B and C) Six frames representing the area surrounded by the white (B) and blue (C) lines in A from Movie S3. (D and E) Time-intensity profiles of DRP2B-GFP fluorescence (green lines) and DRP1A-tagRFP fluorescence (red lines) within the areas surrounded by the white and blue lines in A.
Fig. 5 D and E show the profiles of DRP2B-GFP (green lines) and DRP1A-tagRFP (red lines) fluorescence within the encircled areas in Fig. 5 B and C. The fluorescence intensities at each time point were calculated by subtracting the background fluorescence (the fluorescence within the area surrounded by the white dotted line in Fig. 5A) from that within the areas surrounded by the white and blue lines in each frame of Movie S3. In Fig. 5 D and E, the DRP2B-GFP and DRP1A-tagRFP profiles roughly coincided during the increasing and decreasing of DRP2B-GFP and DRP1A-tagRFP fluorescence. These results suggest that DRP2B and DRP1A assemble and disassemble together at the plasma membrane.
DRP2B and DRP1A Interact.
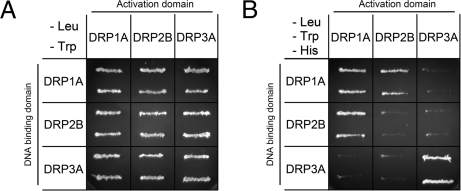
To examine whether DRP2B interacts with DRP1A, we conducted yeast two-hybrid assays. In this experiment, we examined the interactions among DRP1A, DRP2B, and DRP3A. Our previous report using yeast two-hybrid assays showed that (1) DRP1A interacts with itself, (2) DRP3A interacts with itself, and (3) DRP1A and DRP3A do not interact (30). Fig. 6 A and B show the growth of the yeast strain AH109 on a plate with synthetic defined medium lacking leucine and tryptophan (SD/−Leu/−Trp) for checking the transformation of both bait and prey constructs and on a plate with synthetic defined medium lacking leucine, tryptophan, and histidine (SD/−Leu/−Trp/−His) for analyzing the interactions between each combination of bait and prey. As shown in Fig. 6B, DRP1A-DRP1A interactions and DRP3A-DRP3A interactions were detected, whereas interactions between DRP1A and DRP3A were not detected, in agreement with our previous report. Interestingly, DRP2B did not interact with itself, but it interacted with DRP1A in both reciprocal combinations with bait and prey. These results indicate that DRP2B and DRP1A interact with each other.
Fig. 6.
Interactions among DRP1A, DRP2B, and DRP3A. (A and B) Yeast two-hybrid experiments. Yeast strain AH109 was transformed with the paired constructs for a fusion protein with the GAL4 activation domain (Left Column) and a fusion protein with the GAL4 DNA-binding domain (Top Row). (A) Transformants were streaked onto a SD/−Leu/−Trp plate (checking the introduction of two plasmids into the cells). (B) SD/−Leu/−Trp/−His plate (checking the interaction between two fusion proteins).
Tyrphostin A23 Has a Stronger Effect on DRP1A than on DRP2B.
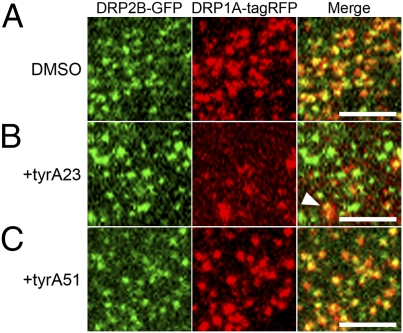
Tyrphostin A23 (tyrA23), an inhibitor of clathrin-mediated endocytosis in plant cells (6, 31), was found to disturb the dynamics and to increase the size of fluorescently tagged DRP1C foci at the plasma membrane in Arabidopsis root epidermal cells (9). To investigate whether tyrA23 has similar effects on DRP1A and DRP2B, we examined its effects on the localization of DRP1A-tagRFP and DRP2B-GFP in the same cell type. Thirty minutes of exposure to 50 μM tyrA23 caused DRP1A-tagRFP foci to increase in size and fluorescence intensity but decrease in number (Fig. 7B), whereas it had no such effects on the majority of DRP2B-GFP foci. However, we observed some overlapping immobile DRP1A-tagRFP and DRP2B-GFP foci of about 1 μm in diameter (Fig. 7B, Right, arrowhead). These effects were not observed in nontreated cells (0.1% DMSO, Fig. 7A) or in cells treated with a noneffective analog of tyrA23, tyrphostin A51 (tyrA51) (Fig. 7C). These results suggest that tyrA23 treatment strongly disturbs the localization of DRP1A at the plasma membrane but has little effect on DRP2B, which implies that the molecular properties of DRP1A and DRP2B are different.
Fig. 7.
Effects of tyrA23 on the localization of DRP2B-GFP and DRP1A-tagRFP foci at the plasma membrane. (A–C) VIAFM images of Arabidopsis root epidermal cells expressing DRP2B-GFP (in green) and DRP1A-tagRFP (in red) after a 30-min treatment with 0.1% DMSO (A), tyrA23 (B), and tyrA51 (C). Left, Center, and Right in A–C represent GFP, tagRFP, and merged images. An arrowhead in the merged image of B indicates an enlarged focus. (Scale bars: 3 μm.)
Discussion
Two Structurally Distinct Arabidopsis DRPs Accumulate on Clathrin Assembly Sites.
In our VIAFM images, fluorescent fusions of both DRP2B and DRP1A accumulated at the assembly sites of CLC with quite similar behaviors. This indicates that these two DRPs are related to endocytic CCV formation in Arabidopsis. Moreover, despite their differences in molecular structure and response to tyrA23, DRP2B and DRP1A colocalized at the plasma membrane and interacted with each other in the yeast two-hybrid assay, suggesting that they form a molecular complex on the CCV formation site. These results suggest that a DRP2 (DRP2B) and a DRP1 (DRP1A) work together to form endocytic CCVs but with distinct molecular properties in Arabidopsis cells. To our knowledge, this is previously undescribed evidence that two structurally distinct DRPs participate together in the same vesicle formation in any eukaryotic organism.
The different responses of DRP1A and DRP2B to tyrA23 provide evidence that these two DRPs have different molecular properties, possibly as a result of their different domain structures. The domain structures of animal dynamin are similar to those of DRP2s and distinct from those of DRP1s (17). This suggests that DRP2s, like animal dynamins, function alone at the plasma membrane. However, the GTPase domain of animal dynamin is much more similar to the GTPase domains of DRP1s (e.g., 66% identity to DRP1A) than to those of DRP2s (e.g., 27% identity to DRP2B). This raises the possibility that DRP2s and DRP1s each fulfill part of the function of animal dynamin. Moreover, a small number of the DRP1A and DRP2B foci were not colocalized (Fig S2, arrowhead and arrow), which raises an additional possibility that DRP1s and DRP2s also have functions on which they work alone.
In CCV formation, plant cells are thought to differ from animal cells not only in the numbers and types of dynamin family proteins that participate, but also in the binding partners of these proteins. One of the binding partners of animal dynamin is amphiphysin, which contains two distinctive domains, a Bin-Amphiphysin-Rvs (BAR) domain, which has membrane curvature-sensing properties, and a Src-homology 3 (SH3) domain, which binds the proline-rich domain (PRD) of dynamin (32). However, no apparent amphiphysin ortholog with both BAR and SH3 domains has been found in the Arabidopsis genome (33).
Temporal Behaviors of DRP2B and DRP1A During CCV Formation.
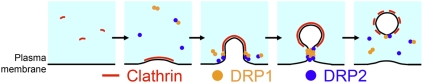
According to a recent review in yeast and animal cells (34), CCV formation can be divided into three distinct stages: (1) the assembly stage, in which a clathrin lattice assembles on the membrane; (2) the maturation stage, in which the clathrin coat and underlying membrane acquire curvature; and (3) the departure stage, in which the neck membrane of the clathrin-coated pit (CCP) is pinched off and the clathrin coat detaches from the membrane. In a TIRFM study using animal cells (35), the signal intensity of fluorescent fusions of CLC rose quickly in the first assembly stage, increased moderately or reached a plateau in the maturation stage, and fell suddenly in the departure stage. Assuming that these three stages also occur in Arabidopsis cells, in which stages do DRP2B and DRP1A assemble and disassemble? In frames from our VIAFM movies (Fig. 3 G and H), GFP-DRP2B and GFP-DRP1A fluorescence quickly increased at the beginning of the plateau phase of mKO-CLC fluorescence. On the other hand, GFP-DRP2B and GFP-DRP1A fluorescence quickly decreased at the same time as or immediately after the mKO-CLC fluorescence quickly decreased. These results suggest that DRP2B and DRP1A assemble together at the beginning of the maturation stage of the CCP (Fig. 8, Center and Center Right) and disassemble together at the departure stage of the CCP (Fig. 8, Right). These temporal behaviors of DRP2B and DRP1A raise the possibility that a complex of DRP2B and DRP1A regulates not only the fission of the neck of the CCP but also the maturation of the CCP in endocytosis.
Fig. 8.
Schematic of the proposed roles of DRP2B, DRP1A, and clathrin in endocytic vesicle formation in Arabidopsis. These schematics represent the putative process of endocytic vesicle formation in Arabidopsis. The light-blue background represents the cytosol. Black and red lines represent the plasma membrane and the clathrin coat, respectively. Orange and dark-blue dots represent DRP1 and DRP2 proteins, respectively. The behaviors of DRP2, DRP1, and clathrin are depicted on the basis of the results from our time-lapse VIAFM analysis.
Materials and Methods
VIAFM Observations.
Transformed Arabidopsis cultured cells and plants (A. thaliana ecotype Columbia-0) were observed with a fluorescence microscope (Nikon Eclipse TE2000-E and a CFI Apo TIRF 100 × H/1.49 numerical aperture objective) with a Nikon TIRF2 system (Nikon). The methods for growing and transforming Arabidopsis cultured cells and plants, for constructing Ti plasmids used in the transformation, and for drug treatment of Arabidopsis plants are described in SI Materials and Methods. Oligonucleotide primers used for Ti plasmid construction are shown in Table S1. Fifty microliters of culture medium and 7-day-old Arabidopsis seedlings were placed on a slide glass (76 × 26 mm; Matsunami) and covered with a 0.12–0.17-mm-thick cover glass (24 × 60 mm; Matsunami). GFP and mKO were excited, with 488 and 561 nm lasers, respectively, both simultaneously (Figs. 2, 4, 5, and 7, Figs. S1 and S2, and Movie S3) and sequentially (Fig. 3 and Movies S1 and S2). The fluorescence emission spectra were separated with a 565LP dichroic mirror and filtered through either a 515/30 (GFP) or 580LP (mKO and tagRFP) filter in a Dual View filter system (Photometrics). Images in Figs. 2, 4, and 5 and Figs. S1 and S2 were acquired with a Cool SNAP HQ2 CCD camera (Roper Scientific). The images in Fig. 7 were acquired with an iXonEM EMCCD camera (Andor Technology). These CCD cameras were controlled by NIS-Elements (Nikon). Each frame was exposed for 200 ms. In Movies S1 and S2, the frames were taken at 3-s intervals. In Movie S3, the frames were taken at 200-ms intervals. The acquired images were prepared and analyzed with Photoshop 7.0 (Adobe Systems), Image pro plus 4.0 (Media Cybernetics), and Image J (National Institutes of Health).
Yeast Two-Hybrid Assay.
Paired plasmids of pAD-GAL4-GWRFC and pBD-GAL4 GWRFC harboring DRP1A, DRP2B, and DRP3A were transformed into Saccharomyces cerevisiae strain AH109. The methods for constructing these plasmids are described in SI Materials and Methods. Oligonucleotide primers used for plasmid construction are shown in Table S1. AH109 was deficient in producing Leu, Trp, His, and uracil. The AH109 transformants were selected on SD/−Leu/−Thr plates (synthetic defined plates deficient in both Leu and Trp). The interactions were examined independently after 2 days of growth on SD/−Leu/−Trp/−His.
Supplementary Material
Acknowledgments
We thank Dr. M. Karimi (Ghent University, Belgium) and Dr. T. Nakagawa (Shimane University, Japan) for kindly donating Gateway destination vectors and Dr. T. Demura (Nara Institute of Science and Technology, Japan) for kindly donating pAD-GAL4-GWRFC and pBD-GAL4-GWRFC vectors. This research was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science, by grants-in-aid for Scientific Research for Plant Graduate Students from the Nara Institute of Science and Technology supported by the Ministry of Education, Culture, Sports, Science and Technology, Japan (M.F.), and by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants 18075005 and 18208002 to N.T., Grant 21248002 to A.N., and Grant 17780002 to S.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The GenBank accession numbers for DRP2B, DRP1A, DRP3A, and CLC are AV528687, BT001063, AV528354, and AY092995, respectively.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913562107/DCSupplemental.
References
- 1.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 2.Murphy AS, Bandyopadhyay A, Holstein SE, Peer WA. Endocytotic cycling of PM proteins. Annu Rev Plant Biol. 2005;56:221–251. doi: 10.1146/annurev.arplant.56.032604.144150. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky FM, Chen C-Y, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 4.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 5.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Dhonukshe P, et al. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- 7.Konopka CA, Bednarek SY. Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 2008;147:1590–1602. doi: 10.1104/pp.108.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 2008;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- 9.Konopka CA, Backues SK, Bednarek SY. Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell. 2008;20:1363–1380. doi: 10.1105/tpc.108.059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto M, Arimura S, Nakazono M, Tsutsumi N. Imaging of plant dynamin-related proteins and clathrin around the plasma membrane by variable incidence angle fluorescence microscopy. Plant Biotechnol. 2007;24:449–455. [Google Scholar]
- 11.Danino D, Hinshaw JE. Dynamin family of mechanoenzymes. Curr Opin Cell Biol. 2001;13:454–460. doi: 10.1016/s0955-0674(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 12.Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 13.Sever S. Dynamin and endocytosis. Curr Opin Cell Biol. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 14.Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 15.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent confor-mational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 16.Puthenveedu MA, von Zastrow M. Cargo regulates clathrin-coated pit dynamics. Cell. 2006;127:113–124. doi: 10.1016/j.cell.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 17.Hong Z, et al. A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol Biol. 2003;53:261–265. doi: 10.1023/b:plan.0000007000.29697.81. [DOI] [PubMed] [Google Scholar]
- 18.Samaj J, et al. Endocytosis, actin cytoskeleton, and signaling. Plant Physiol. 2004;135:1150–1161. doi: 10.1104/pp.104.040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 20.Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 21.Kang B-H, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003;15:899–913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang B-H, Busse JS, Dickey C, Rancour DM, Bednarek SY. The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang B-H, Rancour DM, Bednarek SY. The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J. 2003;35:1–15. doi: 10.1046/j.1365-313x.2003.01775.x. [DOI] [PubMed] [Google Scholar]
- 24.Collings DA, et al. Arabidopsis dynamin-like protein DRP1A: A null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J Exp Bot. 2008;59:361–376. doi: 10.1093/jxb/erm324. [DOI] [PubMed] [Google Scholar]
- 25.Jin JB, et al. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam BC-H, Sage TL, Bianchi F, Blumwald E. Regulation of ADL6 activity by its associated molecular network. Plant J. 2002;31:565–576. doi: 10.1046/j.1365-313x.2002.01377.x. [DOI] [PubMed] [Google Scholar]
- 27.Alexandersson E, Saalbach G, Larsson C, Kjellbom P. Arabidopsis plasma membrane proteomics identifies components of transport, signal transduction and membrane trafficking. Plant Cell Physiol. 2004;45:1543–1556. doi: 10.1093/pcp/pch209. [DOI] [PubMed] [Google Scholar]
- 28.Arimura S, Aida GP, Fujimoto M, Nakazono M, Tsutsumi N. Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 2004;45:236–242. doi: 10.1093/pcp/pch024. [DOI] [PubMed] [Google Scholar]
- 29.Mano S, Nakamori C, Kondo M, Hayashi M, Nishimura M. An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 2004;38:487–498. doi: 10.1111/j.1365-313X.2004.02063.x. [DOI] [PubMed] [Google Scholar]
- 30.Fujimoto M, et al. Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 2009;58:388–400. doi: 10.1111/j.1365-313X.2009.03786.x. [DOI] [PubMed] [Google Scholar]
- 31.Robinson DG, Jiang L, Schumacher K. The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 2008;147:1482–1492. doi: 10.1104/pp.108.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takei K, Yoshida Y, Yamada H. Regulatory mechanisms of dynamin-dependent endocytosis. J Biochem. 2005;137:243–247. doi: 10.1093/jb/mvi052. [DOI] [PubMed] [Google Scholar]
- 33.Holstein SEH. Clathrin and plant endocytosis. Traffic. 2002;3:614–620. doi: 10.1034/j.1600-0854.2002.30903.x. [DOI] [PubMed] [Google Scholar]
- 34.Pucadyil TJ, Schmid SL. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325:1217–1220. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loerke D, et al. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.