Abstract
We present molecular dating analyses for land plants that incorporate 33 fossil calibrations, permit rates of molecular evolution to be uncorrelated across the tree, and take into account uncertainties in phylogenetic relationships and the fossil record. We attached a prior probability to each fossil-based minimum age, and explored the effects of relying on the first appearance of tricolpate pollen grains as a lower bound for the age of eudicots. Many of our divergence-time estimates for major clades coincide well with both the known fossil record and with previous estimates. However, our estimates for the origin of crown-clade angiosperms, which center on the Late Triassic, are considerably older than the unequivocal fossil record of flowering plants or than the molecular dates presented in recent studies. Nevertheless, we argue that our older estimates should be taken into account in studying the causes and consequences of the angiosperm radiation in relation to other major events, including the diversification of holometabolous insects. Although the methods used here do help to correct for lineage-specific heterogeneity in rates of molecular evolution (associated, for example, with evolutionary shifts in life history), we remain concerned that some such effects (e.g., the early radiation of herbaceous clades within angiosperms) may still be biasing our inferences.
Keywords: divergence times, rates of molecular evolution, angiosperms, eudicots, land plants
Our understanding of the history of life depends critically on knowledge of the ages of major clades. The timing of land plant evolution is fundamental to the interpretation of earth history and macroevolution throughout the Phanerozoic. Age estimates bear directly on our interpretation of the tempo and mode of morphological and molecular evolution of plants themselves, but also on our interpretation of the evolution of many other groups. For example, the age of origin of the angiosperms has variously been related to the evolution of other plant lineages (e.g., ferns) (1) and biomes (e.g., tropical forests) (2), as well as to the major insect clades and their feeding habits (3–7), and even to the evolution of fungi (8, 9) and dinosaurs (10).
In plants, as in other major groups (e.g., mammals) (11), the ages of clades estimated from molecular phylogenetic analyses have not always corresponded well with the accepted fossil record. In particular, the application of molecular-clock methods has tended to yield older dates (12), in some cases much older than has seemed credible based on the stratigraphic record (13–15). The approaches used in molecular dating have been problematical for several reasons, and in some cases the results have been too easy to dismiss. Early attempts used a strict molecular clock (e.g., ref. 16). Recently, so-called “relaxed clock” methods have been used, which variously allow departures from clock-like behavior (17–20), but here too there are difficulties. First, molecular dating analyses have tended to treat the tree topology as complete and fixed, as opposed to taking into account phylogenetic uncertainties (21–25). For angiosperms this is problematical, considering the limited confidence we still have in the order of branching early in their diversification (24, 26).
Second, the information provided by fossils is often treated as fixed. Uncertainties associated with fossil calibrations are inherent, given the nature of fossilization (27), the difficulty of dating fossil localities, and the standard logic used in placing fossils into phylogenies to obtain minimum ages for lineages (28). One concern is the default practice of assigning fossils to the stem of the most inclusive crown clade to which they probably belong, thereby possibly biasing estimated ages (possibly throughout the tree) to be younger (29). In many cases, estimates have relied on lower bounds, based on what is assumed to be a tightly constrained fossil record. In angiosperms, the origin of eudicots, marked by the appearance of tricolpate pollen in the Late Barremian-Early Aptian (∼125 Myr), has widely been used as a hard constraint (i.e., maximum age) to either calibrate or assess angiosperm molecular divergence times (21–25). However, using the first appearance of tricolpate pollen as a fixed calibration may underestimate the origin of eudicots and, by extension, other age estimates that have relied on this constraint. Tricoplate grains first appear in separated geographical areas and the grains themselves are not uniform in morphology (30–33), both observations implying that the tricoplate clade originated some time before its appearance in the fossil record. Although there may be value for some purposes in using the same fossil constraints in different studies, the reliance on the eudicot maximum-age constraint in multiple studies ties them all to the same underlying assumption, thereby compromising their independence.
Third, previous studies have relied on methods that have probably not accommodated sufficiently for heterogeneity in rates of molecular evolution (34). Rarely are datasets found to conform to a molecular clock, and broadly sampled plant phylogenies are no exception (22, 35, 36). Rates of evolution can vary among genes within a lineage, or among lineages. Incorporating data from multiple genes can help to compensate for rate heterogeneity across genes (23), but lineage-specific rate heterogeneity has not been adequately addressed and remains a potentially large source of error. Several analyses have demonstrated striking differences in molecular rates across large plant clades (36–39). These differences have been associated with traits such as growth habit, generation time, and population size, all of which are labile and might change multiple times along a branch, which will be especially difficult to detect when taxonomic sampling is sparse. When not accounted for, such rate heterogeneity can systematically bias slow-rate branches to appear younger and fast-rate branches to appear older (38, 40). Some methods for accommodating nonclock-like rates were designed to smooth differences across branches, under the assumption that rates are autocorrelated (17, 18): that is, they assume the inheritance of rate from parent node to child node. However, factors such as the evolution of life-history characteristics are expected to result in large differences between adjacent nodes, in which case autocorrelated methods are problematical (34). With the development of models of molecular evolution that are uncorrelated over a phylogeny (41, 42), we can at least begin to accommodate such rate shifts.
Here, we present a hypothesis of divergence times for land plants, with special emphasis on the timing of the origin of crown-clade angiosperms. To address the issues noted above, we have used the uncorrelated lognormal (UCLN) relaxed-clock model of Drummond et al. (41), which permits the rate of molecular substitution to be uncorrelated across the tree while incorporating uncertainty in both tree topology and multiple fossil calibrations. We also allow fossil calibrations to act as probabilistic priors rather than as point estimates, and explore the possibility of dramatic rate differences associated with life-history evolution. Our results bear on the ages now often assumed in broad comparative analyses in plants (43, 44) and on the possible link between the radiation of plants and other organisms. However, as we discuss below, we remain concerned about the possible effects of rate heterogeneity (especially related to shifts in life history) and highlight the need to develop methods that more explicitly take this into account.
Results and Discussion
Phylogenetic Results.
We conducted Bayesian and maximum-likelihood (ML) phylogenetic analyses on 154 species of land plants using previously published sequences of 18S, atpB, and rbcL (Table S1). These genes were chosen because of their utility in identifying major plant clades (45, 46). Our taxonomic sampling within Angiospermae (flowering plants) represents most large clades of Mesangiospermae (mesangiosperms, or “core angiosperms”), including Magnoliidae (magnoliids), Monocotyledoneae (monocots), and Eudicotyledoneae (eudicots), all in the sense of Cantino et al. (47), as well as species sampled in previous large-scale tracheophyte analyses (36, 46). To obtain preliminary estimates for the divergence times of crown clades within angiosperms, at least two species were sampled to represent very large clades. Both our ML and Bayesian analyses confirmed previous phylogenetic inferences within angiosperms, with the few exceptions likely because of our full partitioning of the data into gene regions and our somewhat different sample of taxa (see Materials and Methods for details). Likewise, relationships among the major clades of land plants largely confirmed previous analyses based on ML nonpartitioned analyses (36, 46). Monophyletic Spermatophyta (seed plants), Acrogymnospermae (containing the four major lineages of extant “gymnosperms”), and Angiospermae are well supported in both our ML and Bayesian analyses. Monocots and eudicots are well supported as monophyletic. In agreement with recent analyses, Ceratophyllum is placed sister to eudicots (24), and we see an accelerated rate of molecular evolution in Gnetales (Fig. 1A and Figs. S1, S2, S3 and S4) (48).
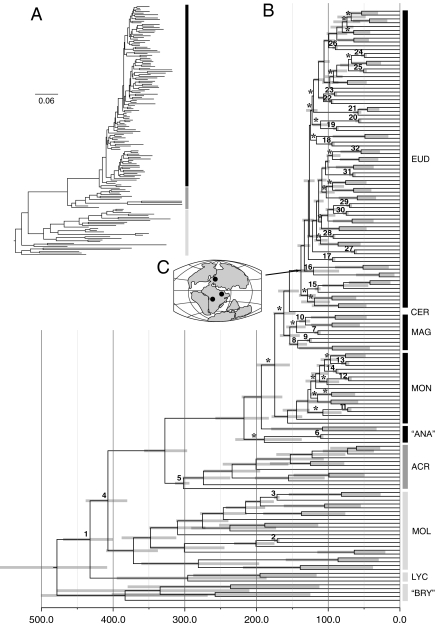
Fig. 1.
Phylogenetic tree and divergence time estimates for land plants. (A) MrBayes consensus tree from a three-gene (atpB, rbcL, and 18S) analysis (see Fig S1 for taxon names). Branch lengths represent average substitutions per site. Shaded bars represent Angiospermae (black), Acrogymnospermae (dark gray), and the rest of the land plants (light gray). (B) The maximum clade credibility tree from the divergence time analysis of the same three-gene dataset as in A. Studies focused on the root of the land plants (e.g., ref. 81), including outgroups, place the root along the liverwort branch (“bryophytes” paraphyletic). Nodes marked by an asterisk (*) are supported by <0.95 posterior probability. The 95% highest posterior density (HPD) estimates for each well-supported clade are represented by bars. Numbers at nodes correspond to the fossil calibrations in Table S2. (C) Map with localities for the first tricolpate pollen records. Clade names follow Cantino et al. (47): ACR, Acrogymnospermae; ANA, ANITA grade; BRY, bryophytes; CER, Ceratophyllum; EUD, Eudicotyledonae; LYC, Lycopodiophyta; MAG, Magnoliidae; MOL, Monilophyta; MON, Monocotyledonae.
Previous phylogenetic analyses have differed with respect to the relationships of Amborella and Nymphaeales to the rest of angiosperms (49) and among the magnoliids, monocots, and eudicots (24, 26). The most comprehensive analysis to date, based on 61 chloroplast genes, supported Amborella as sister to the rest of angiosperms but was uncertain regarding magnoliids, eudicots, and monocots (24). We conducted ML analyses on individual genes to help identify differential support for problematical relationships (Figs. S2, S3, and S4). The 18S sequences support Nymphaeales as sister to the rest of angiosperms, and weakly (51% BS) support the nonmonophyly of Acrogymnospermae, with cycads sister to angiosperms. atpB sequences support the monophyly of Acrogymnospermae and place magnoliids as sister to eudicots, but uncertainty remains as to the placement of Amborella and Nymphaeales in relation to the rest of the angiosperms. rbcL sequences do not clearly resolve the placement of magnoliids or monocots, and weakly support Amborella as sister to Nymphaeales (79% BS) and a monophyletic Acrogymnospermae. Our divergence time analyses take into account this uncertainty in the underlying tree topology.
Dating Analyses and Results.
Our age estimates for divergences within land plants were based on 33 fossil calibrations (Table S2). The origin of land plants centered on 477 Myr (95% HPD: 407–557 Myr) (Table 1), which corresponds well with the earliest known occurrence of microfossils assigned to land plants from the middle Ordovician (∼470 Myr) (50). The origin of tracheophytes was estimated at 432 Myr (95% HPD: 399–469 Mya), which corresponds to the middle Silurian. The first fossil fragments widely assigned to early Tracheophyta are also of Silurian age (∼419) (51). We estimated the origin of Spermatophyta (crown seed plants) in the Middle Carboniferous (327 Myr; 95% HPD: 296–356 Myr), which broadly corresponds with the fossil record (52). A Middle Carboniferous age for crown seed plants follows the Devonian (>360 Myr) evidence of the “progymnosperm” lineages, Archaeopteridales and Aneurophytales (53, 54), and Early Carboniferous evidence of Paleozoic seed ferns (55). Acrogymnospermae (301 Myr, 95% HPD: 293–313) are estimated to have originated some 30 million years after crown seed plants.
Table 1.
Divergence-time estimates (in Myr) for major clades of land plants as estimated using 33 fossil calibrations (including the eudicot pollen calibration) and 32 fossil calibrations (excluding the eudicot pollen calibration)
| Clade | With eudicot calibration | Without eudicot calibration |
| Land plants | 477 (407–557) | 474 (419–577) |
| Tracheophyta | 432 (399–469) | 434 (404–490) |
| Spermatophyta | 327 (296–356) | 330 (301–366) |
| Acrogymnospermae | 301 (293–313) | 302 (294–316) |
| Gnetophyta | 155 (104–202) | 158 (110–208) |
| Angiospermae | 217 (182–257) | 228 (193–270) |
| Mesangiospermae | 174 (153–200) | 184 (160–210) |
| Magnoliidae | 155 (136–181) | 163 (138–198) |
| Monocotyledoneae | 156 (139–167) | 164 (141–191) |
| Eudicotyledoneae | 137 (128–147) | 153 (138–172) |
Dates within parentheses denote the 95% HPD.
We estimated the origin of crown angiosperms to be 217 Myr (95% HPD: 182–257 Myr), in the Late Triassic. This result was robust to the inclusion or exclusion of a 125 Myr minimum-age calibration on the node corresponding to crown eudicots (Table 1). A Late Triassic origin for crown angiosperms is typically not estimated with molecular methods. Of the several analyses carried out by Sanderson and Doyle (35), a few estimated a Late Triassic origin of crown angiosperms. However, this was sensitive to the underlying tree topology, codon position, and taxon sampling, with a majority of estimates falling instead between 140 and 190 Myr. Most molecular divergence-time analyses for crown angiosperms have reported dates within this range of 140 to 190 Myr (reviewed by 34). For example, Bell et al. (22) and Magallón and Sanderson (23) estimated crown angiosperms to be 140 to 180 Myr (with Bayesian relaxed-clock methods) and 163 to 189 Myr (using penalized likelihood), respectively. More recently, Moore et al. (24) estimated the origin of crown angiosperms to be ∼170 Myr using a data set of 61 plastid genes. Magallón and Castillo (25) reported an age of 130 Myr; however, this depended on a maximum age constraint applied to the crown corresponding to the oldest putative fossil angiosperm pollen from the Valanginian to Hauterivian (∼130–140 Myr) (33).
Our age estimates for nodes within the Mesangiospermae suggest that crown magnoliids, monocots, and eudicots had all originated by the Late Jurassic (Table 1). Although the relationships among these three lineages remains uncertain, the short time interval between the origin of the corresponding crown groups suggests a rapid succession in the origin of the major angiosperms lineages. Although a Late Jurassic origin for magnoliids, monocots, and eudicots is much older than previous analyses have reported, it is important to note that we cannot reject an Early Cretaceous origin (the lower 95% HPD <144 Myr) (Table 1). When we removed the minimum age calibration for crown eudicots at 125 Myr, the estimates for the origin of crown magnoliids and monocots remained centered on the Late Jurassic, again with the lower 95% credibility interval encompassing the Early Cretaceous (Table 1). Taken together, our age estimates for the origin of major crown groups within mesangiosperms clearly predate the first putative angiosperm fossils (∼130–140 Myr) (33).
Rate Heterogeneity.
One possible explanation for the lack of correspondence between our molecular divergence estimate and the accepted fossil record is that we have failed to properly account for rate heterogeneity. Two metrics were used to evaluate the appropriateness of assuming a model of uncorrelated rates of molecular variation and to assess overall rate heterogeneity across the tree. First, the degree of autocorrelation of molecular rates from parent to child throughout the phylogeny was estimated through the covariance parameter. Across land plants, we estimated a rate covariance of ρ = 0.074, which was not significantly different from zero (95% HPD: −0.038 to 0.176). Although this result does not support the autocorrelation of rates, we note that the UCLN may be inadequate in detecting significant rate autocorrelation even when it exists (56). Second, we examined the coefficient of variation to assess the overall degree of rate heterogeneity across our tree. Assuming uncorrelated substitution rates, the coefficient of variation is the variance of rates scaled by the associated mean. A significant portion of the posterior density centered near zero is evidence that the data are “clock-like,” whereas a posterior density that does not encompass zero provides evidence for significant rate heterogeneity. The estimated coefficient of variation indicated that the rate of molecular substitution varied by 69.7% (95% HPD: 63.2–76.3%) across the entire tree. This high degree of rate variation suggests an influence of rate heterogeneity.
It has been noted that nonsynonymous sites may show less rate heterogeneity than synonymous sites (37, 39, 57, 58). Different rates of evolution associated with different life histories may be more evident in synonymous sites. To examine whether the UCLN model had sufficiently accommodated for rate heterogeneity, we estimated divergence times using only the first and second codon positions for atpB and rbcL, which focused rate and date estimates on nonsynonymous sites. In this analysis, only the divergence time for land plants changed significantly, with the coding data supporting a slightly younger date of 442 Myr (95% HPD: 401–514 Myr) (Table S3). Importantly, the coefficient of variation indicated that significant rate heterogeneity remained, as the substitution rate varied 67.6% from the mean (95% HPD: 59.5–75.8%). Thus, although using only first and second sites may remove some bias caused by rate heterogeneity, it certainly does not eliminate it (39), and this is presumably true not just for the UCLN method but for methods that assume autocorrelated rates.
To explore whether the evolution of different life histories might help to explain shifts in the rate of molecular evolution (38, 39, 58), and specifically to test whether a shift to the herbaceous habit at the base of the angiosperms (59) could explain the long branch subtending crown angiosperms, we reconstructed growth habit over the posterior distribution of dated trees (Fig. 1B). We found an 88.6% and a 97.4% posterior probability of “woody” being the ancestral state for crown angiosperms and crown seed plants, respectively. This finding supports the results of Feild et al. (60, 61), who also argued on the basis of such reconstructions coupled with physiological data that the first angiosperms were woody plants living in “dark and disturbed” environments. Here it is relevant that fossil lineages inferred to be along the stem subtending the angiosperms, such as the glossopterids, Caytoniales, and Bennittitales (62, 63; but see ref. 64), are considered to be woody. These observations work against a simple argument that a shift to herbaceous habit along the line leading to angiosperms resulted in a faster rate of molecular evolution and, hence, to the inference that crown angiosperms are much older than they actually are. However, it is important to appreciate that the length of the branch leading to crown angiosperms spans ∼100 Myr, which would allow many potentially confounding and undetected changes in life history and population size. Of course, the long branch subtending the angiosperms may not entirely reflect a faster rate of molecular evolution; it may simply indicate high extinction along that branch. We return to these concerns below.
Use of First Occurrence of Tricolpate Pollen.
Tricolpate pollen grains (and derivative conditions) characterize modern eudicots, and the appearance of such grains in the fossil record is taken as evidence of the existence of the eudicot lineage, if not the crown clade. The first appearance of tricolpate pollen grains at the Barremian-Aptian boundary (∼125 Myr) has commonly been used as a fossil calibration in divergence-time analyses. Whether placed along the stem or at the crown of the eudicots, this tricolpate pollen calibration has primarily been treated as a maximum-age constraint (22–25, 36, 65). This treatment could be justified based on the observation that the pollen record is substantial and that tricolpate grains, which are easy to identify, have not been recovered from any earlier sediments. It may be possible in this case to infer a likely maximum age using the statistical methods proposed by Marshall (66). In the meantime, several aspects of the appearance of tricolpate grains in the fossil record suggest that 125 Myr may not be an appropriate maximum age for eudicots. The first tricolpate grains have the same aperture configuration (tricolpate, not tricolporoidate, or triporate), but show “considerable structural variety” (33) in the sculpturing of the exine. In addition, the Barremian-Aptian tricolpate pollen localities are geographically widespread, first at several Gondwanan sites (present-day northern and equatorial Africa), with specimens becoming more common in Laurasia during the Aptian-Albian (Fig. 1C) (30, 31). Based on these observations, it is possible that the appearance of tricolpate grains reflects the rise to dominance of the eudicot lineage as opposed to the origin of the group. Finally, it is not clear whether these pollen grains represent the emergence of the tricolpate apomorphy along the branch leading to crown eudicots or whether they represent the appearance of modern lineages of eudicots (i.e., within the crown). There are too few characters to place them with any certainty within the eudicot phylogeny (33). In view of these caveats, we favor the use of 125 Myr as a minimum age for the origin of the eudicot crown clade, with an associated probabilistic prior (67).
Reexamining Biological Patterns.
Of particular interest for botanists and entomologists is the possible correlation of the early evolution of angiosperms with the rise of the major lineages of holometabolous insects (Coleoptera, Hymenoptera, Diptera, and Lepidoptera). Labandeira and Sepkoski (3) noted that a number of major insect radiations date to the late Permian (∼254 Myr), with trophic diversity proliferating dramatically during this period. Based on the apparent incongruity with the angiosperm fossil record, they concluded that angiosperms had little impact on the early evolution of holometabolous insects.
Our results, taken at face value, push the origin of the angiosperm crown clade much closer in time to the diversification of the major lineages of holometabolous insects. Molecular estimates for the origin of Coleoptera (285 Myr) (5) predate crown angiosperms, but the origin of the most diverse herbivorous lineages of Coleoptera (i.e., Chrysomeloidea, Curculionoidea) are estimated to be ∼230 Myr, which is in the range of our age estimate for crown angiosperms (68). Also congruent are Triassic fossils of Diptera and Hymenoptera (68, 69). Molecular age estimates for the origin of Lepidoptera, as well as for the ant and bee clades nested within Hymnoptera, correspond well with our age estimates for the major crown clades within Mesangiospermae (7, 70, 71), as does the fossil record of long-proboscid Mecoptera (72). However, it is important to note that even if our inferred dates were correct, the absence of clear-cut angiosperm fossils during the Triassic and Jurassic may signify that the first angiosperms were not abundant, widespread, or ecologically very significant, in which case it would be difficult to argue that the appearance of the angiosperms dramatically increased insect diversity during that time period.
Conclusions
Regarding the tempo of plant evolution, our results show generally good correspondence with the fossil record (e.g., for crown tracheophytes and seed plants). However, they also imply that crown angiosperms originated in the Triassic (or possibly in the Jurassic), well before the Cretaceous radiations that were responsible for the dramatic rise of the angiosperms. That is, they suggest that crown angiosperms were in existence for some 50 Myr (or more) before the radiation of the mesangiosperms, and some 60 Myr (or more) before the diversification of monocots and eudicots. The only living remnants of the lineages that existed in this inferred interval are Amborella, Nymphaeales/Hydatellales, and Austrobaileyales. Today, these lineages are species-poor, but they exhibit tremendous morphological and ecological disparity. One possibility is that these groups were once much more diverse, and that we are left today with only a few survivors. In this case, it may be that most of these plants lived in environments that were not conducive to fossilization. However, it is also possible that angiosperms were simply not diverse or ecologically dominant plants during the Late Triassic and Jurassic. For example, as Feild et al. (60, 61) have argued, the physiology and ecological preferences of the early angiosperms (living in dark, wet, and disturbed understory habitats, probably with low population sizes) may have restricted their abundance, geographical spread, and diversification. These same factors might also account for the lack of fossils during this interval.
Regarding eudicots, our results suggest that the first appearance of tricolpate grains at ca. 125 Myr underestimates the origin of the tricolpate clade by perhaps 3 to 22 Myr. This finding is problematic because the record of fossil pollen is judged to be very good through this time period (33). If our inferences are correct, the appearance of tricolpate grains may not signal the origin of the crown group, but rather the rise in abundance and geographic expansion of the tricolpate lineage.
Dismissing our angiosperm date as an artifact will be tempting. However, as the date reflects the current state of knowledge of fossils and phylogeny, as well as the current state of development of relevant analytical tools, we believe that these dates should not be set aside lightly. Yet, we hasten to acknowledge that our analysis is unlikely to be the final word on the subject and, moreover, there are several reasons to proceed cautiously. Perhaps most importantly, we remain concerned about the impacts of lineage-specific rate heterogeneity on molecular age estimates, despite having tried to accommodate this. It is increasingly clear that there may be extreme differences in molecular rate depending upon life history and other factors (39), and current methods may be unable to cope. It is possible that the effects of lineage-specific rate heterogeneity can “trickle-down” to nodes at some distance from the inferred shift in life history and molecular rate. For this reason we are concerned that, although a shift to herbaceousness may not have marked the origin on the angiosperms, multiple shifts to the herbaceous habit not far within angiosperm, followed by several rapid radiations, might result in an older age estimate for angiosperms as a whole. This possibility needs to be explored further using simulations and also suggests the need to develop methods (akin, perhaps, to so-called local-clock methods) (20) that allow shifts to different rate categories as a function of evolutionary shifts in an underlying parameter that might drive rate changes, such as life history or population size.
It is interesting to reflect, however, that as older fossils are discovered and incorporated into various lineages, this will tend to shift the angiosperm date back further in time. Furthermore, as our taxonomic sampling improves, and as knowledge of fossils increases to the point of allowing us to place them within clades with greater precision, there may be a general tendency for these to be placed further up within the clades with which they are associated. This process will also tend to push the age of crown angiosperms further back in time. It is possible that a closer match between molecular inferences and the stratigraphic record will eventually be obtained, as dating methods are improved to cope with extreme rate heterogeneity and as older fossils are discovered. However, it is also possible that a significant gap will remain and, if so, this might tell us something important about the rise of flowering plants.
Materials and Methods
Phylogenetic Analyses.
Sequences for each gene region were aligned separately with MUSCLE (73) within three partitions: angiosperms, acrogymnosperms, and the remaining green plants. These separate alignments were then combined using profile alignment techniques (73; see also ref. 74), and the aligned gene regions were concatenated using Phyutility (75).
Maximum-likelihood analyses were conducted with RAxML (Ver. 7.0.1) (76). Runs were partitioned into gene regions with parameters unlinked. We used the GTRMIX substitution and rate heterogeneity model. ML analyses were conducted by first running 100 rapid bootstrap analyses; every tenth bootstrap tree was used as a start tree for a full ML search. The best tree from those searches was considered to be the ML tree. Bootstraps were summarized with Phyutility (75). We conducted these analyses on the concatenated dataset as well as on the individual gene regions (Figs S1, S2, S3, and S4).
Bayesian phylogenetic analyses were conducted with MrBayes (Ver. 3.1.2) (77, 78) using the Metropolis coupled Markov Chain Monte Carlo (MCMC) algorithm. Two analyses, each consisting of four incrementally heated chains, were run for 10 million generations, sampling every 1,000th tree. The posterior distribution of trees was summarized after removing 1 million generations as burn-in. A GTR+Γ model was applied to each gene region and the associated parameters were unlinked. Posterior distributions for parameter estimates and likelihood scores were visualized in Tracer (Ver. 1.4) to approximate convergence.
Divergence-Time Estimation.
Simultaneous divergence-time and phylogenetic analyses were conducted using MCMC methods implemented in BEAST (Ver. 1.4.7; 42). BEAST employs an uncorrelated relaxed-clock (UCLN) model to estimate divergence times and allows topologies to be considered “fixed” or estimated to accommodate for phylogenetic uncertainty (41, 42). Here, we allowed BEAST to estimate the topology. For each branch, the UCLN independently draws substitution rates from a lognormal distribution, allowing substitution rates to be uncorrelated across the phylogeny. The absolute estimates of divergence times are then calculated from fossil calibrations, each with an associated probabilistic prior. We attached a lognormal prior probability (67) to the minimum-age estimates obtained from 33 fossil calibrations (using the International Commission on Stratigraphy 2007), 27 of these for crown clades within angiosperms (Table S2). The mean and standard distributions of these calibrations were chosen to acknowledge that, although the fossil age represents the minimum age of the lineage, there remains a probability that the true age extends (in most cases ∼10–15 Myr) further back in time.
Our divergence-time analyses were carried out using two partitioning strategies. The first partitioned the data by gene region (atpB, rbcL, and 18S), with the rate parameters unlinked and assuming a GTR+Γ substitution model. The second partitioned the first and second codon positions of atpB and rbcL only. Again, the parameters were unlinked and we assumed a GTR+Γ substitution model. For each partitioning strategy, we initiated five separate MCMC chains, each consisting of 10 to 50 million generations with convergence monitored by Tracer (Ver. 1.4). We determined the number of runs to conduct based on the effective samples sizes of each estimated parameter, where we required the posterior, prior, and likelihood to be at least 200. We heuristically removed a percentage of each run as burn-in and the resulting trees for each replicate were combined. Trees were summarized with TreeAnnotator and represent the maximum clade credibility tree. Ninty-five percent HPD were estimated using the R package (79) Bayesm (Ver. 2.2–1).
Ancestral Life-History Reconstructions.
We used the Bayesian implementation of the program MultiState in BayesTraits (80) to reconstruct the probable life-history of crown seed plants and crown angiosperms across the posterior distribution of dated trees. Multistate implements a reversible-jump MCMC procedure for single multistate characters. Predominantly herbaceous clades were scored as 0, and predominantly woody clades as 1; any clade for which ancestral life form was judged to be equivocal was scored as missing data. We used an exponential hyperprior on the rate coefficients and sampled every 1,000th point from 10 million total generations. We discarded the first 2.5 million iterations as burn-in. The probability distributions obtained from the reversible-jump MCMC were examined using Tracer (Ver. 1.4).
Supplementary Material
Acknowledgments
We thank J. Oliver and D. Tank for helpful suggestions on the manuscript, J. Doyle for discussions about fossils, P. Crane and S. Mathews for helpful reviews, and S. Magallón, C. Bell, D. Soltis, and P. Soltis, for kindly sharing the results of their parallel studies of angiosperm diversification. S.A.S. was partially supported by the National Evolutionary Synthesis Center (NSF EF-0905606 and NSF EF-0423641). M.J.D. and J.M.B. have been supported through a National Science Foundation Angiosperm “Tree of Life” (ATOL) award (NSF EF-0431258).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001225107/DCSupplemental.
References
- 1.Schneider H, et al. Ferns diversified in the shadow of angiosperms. Nature. 2004;428:553–557. doi: 10.1038/nature02361. [DOI] [PubMed] [Google Scholar]
- 2.Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat. 2005;165:E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- 3.Labandeira CC, Sepkoski JJ., Jr Insect diversity in the fossil record. Science. 1993;261:310–315. doi: 10.1126/science.11536548. [DOI] [PubMed] [Google Scholar]
- 4.Grimaldi D. The co-radiations of pollinating insects and angiosperms in the Cretaceous. Ann Miss Bot Gard. 1999;86:373–406. [Google Scholar]
- 5.Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 6.McKenna DD, Farrell BD. Tropical forests are both evolutionary cradles and museums of leaf beetle diversity. Proc Natl Acad Sci USA. 2006;103:10947–10951. doi: 10.1073/pnas.0602712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the ants: diversification in the age of angiosperms. Science. 2006;312:101–104. doi: 10.1126/science.1124891. [DOI] [PubMed] [Google Scholar]
- 8.Matheny PB, et al. Out of the Palaeotropics? Historical biogeography and diversification of the cosmopolitan ectomycoorhizal mushroom family Inocybaceae. J Biogeogr. 2009;36:577–592. [Google Scholar]
- 9.Hibbett DS, Matheny PB. The relative ages of ectomycorrhizal mushrooms and their plant hosts estimated using Bayesian relaxed molecular clock analyses. BMC Biol. 2009;7 doi: 10.1186/1741-7007-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett PM, Willis KJ. Did dinosaurs invent flowers? Dinosaur-angiosperm coevolution revisited. Biol Rev Camb Philos Soc. 2001;76:411–447. doi: 10.1017/s1464793101005735. [DOI] [PubMed] [Google Scholar]
- 11.Tavaré S, Marshall CR, Will O, Soligo C, Martin RD. Using the fossil record to estimate the age of the last common ancestor of extant primates. Nature. 2002;416:726–729. doi: 10.1038/416726a. [DOI] [PubMed] [Google Scholar]
- 12.Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. doi: 10.1038/nrg1020. [DOI] [PubMed] [Google Scholar]
- 13.Crane PR, Friis EM, Pedersen KR. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- 14.Sanderson MJ. Molecular data from 27 proteins do not support a Precambrian origin of land plants. Am J Bot. 2003;90:954–956. doi: 10.3732/ajb.90.6.954. [DOI] [PubMed] [Google Scholar]
- 15.Foote M, Hunter JP, Janis CM, Sepkoski JJ., Jr Evolutionary and preservational constraints on origins of biologic groups: divergence times of eutherian mammals. Science. 1999;283:1310–1314. doi: 10.1126/science.283.5406.1310. [DOI] [PubMed] [Google Scholar]
- 16.Heckman DS, et al. Molecular evidence for the early colonization of land by fungi and plants. Science. 2001;293:1129–1133. doi: 10.1126/science.1061457. [DOI] [PubMed] [Google Scholar]
- 17.Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 18.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 19.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 21.Anderson CL, Bremer K, Friis EM. Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am J Bot. 2005;92:1737–1748. doi: 10.3732/ajb.92.10.1737. [DOI] [PubMed] [Google Scholar]
- 22.Bell CD, Soltis DE, Soltis PS. The age of the angiosperms: a molecular timescale without a clock. Evolution. 2005;59:1245–1258. [PubMed] [Google Scholar]
- 23.Magallón SA, Sanderson MJ. Angiosperm divergence times: the effect of genes, codon positions, and time constraints. Evolution. 2005;59:1653–1670. doi: 10.1554/04-565.1. [DOI] [PubMed] [Google Scholar]
- 24.Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci USA. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magallón SA, Castillo A. Angiosperm diversification through time. Am J Bot. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- 26.Soltis DE, Gitzendanner MA, Soltis PS. A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int J Plant Sci. 2007;168:137–157. [Google Scholar]
- 27.Marshall CR. In: The Adequacy of the Fossil Record. Donovan SK, Paul CRC, editors. New York: Wiley; 1998. pp. 23–53. [Google Scholar]
- 28.Doyle JA, Donoghue MJ. Phylogenies and angiosperm diversification. Paleobiology. 1993;19:141–167. [Google Scholar]
- 29.Bell CD, Donoghue MJ. Dating the Dipsacales: comparing models, genes, and evolutionary implications. Am J Bot. 2005;92:284–296. doi: 10.3732/ajb.92.2.284. [DOI] [PubMed] [Google Scholar]
- 30.Hughes NF, McDougall AB. Barremian-Aptian angiospermid pollen records from southern England. Rev Palaeobot Palynol. 1990;65:145–151. [Google Scholar]
- 31.Doyle JA. Revised palynological correlations of the lower Potomac Group (USA) and the Cocobeach sequence of Gabon (Barremian-Aptian) Cretaceous Res. 1992;13:337–349. [Google Scholar]
- 32.Brenner GJ. In: Flowering Plant Origin, Evolution, and Phylogeny. Taylor DW, Hickey LJ, editors. New York: Chapman & Hall; 1996. pp. 91–115. [Google Scholar]
- 33.Friis EM, Pedersen KR, Crane PR. Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;232:251–293. [Google Scholar]
- 34.Sanderson MJ, Thorne JL, Wikstrom N, Bremer K. Molecular evidence on plant divergence times. Am J Bot. 2004;91:1656–1665. doi: 10.3732/ajb.91.10.1656. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson MJ, Doyle JA. Sources of error and confidence intervals in estimating the age of angiosperms from rbcL and 18s rDNA data. Am J Bot. 2001;88:1499–1516. [PubMed] [Google Scholar]
- 36.Soltis PS, Soltis DE, Savolainen V, Crane PR, Barraclough TG. Rate heterogeneity among lineages of tracheophytes: integration of molecular and fossil data and evidence for molecular living fossils. Proc Natl Acad Sci USA. 2002;99:4430–4435. doi: 10.1073/pnas.032087199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaut BS, Muse SV, Clark WD, Clegg MT. Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J Mol Evol. 1992;35:292–303. doi: 10.1007/BF00161167. [DOI] [PubMed] [Google Scholar]
- 38.Kay KM, Whittall JB, Hodges SA. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evol Biol. 2006;6:36. doi: 10.1186/1471-2148-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- 40.Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K. Estimating divergence times in large phylogenetic trees. Syst Biol. 2007;56:741–752. doi: 10.1080/10635150701613783. [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moles AT, et al. Factors that shape seed mass evolution. Proc Natl Acad Sci USA. 2005;102:10540–10544. doi: 10.1073/pnas.0501473102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beaulieu JM, et al. Correlated evolution of genome size and seed mass. New Phytol. 2007;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- 45.Soltis PS, Soltis DE, Chase MW. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature. 1999;402:402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- 46.Pryer KM, et al. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature. 2001;409:618–622. doi: 10.1038/35054555. [DOI] [PubMed] [Google Scholar]
- 47.Cantino PD, et al. Towards a phylogenetic nomenclature of Tracheophyta. Taxon. 2007;56:822–846. [Google Scholar]
- 48.Burleigh JG, Mathews S. Phylogenetic signal in nucleotide data from seed plants: implications for resolving the seed plant tree of life. Am J Bot. 2004;91:1599–1613. doi: 10.3732/ajb.91.10.1599. [DOI] [PubMed] [Google Scholar]
- 49.Zanis MJ, Soltis DE, Soltis PS, Mathews S, Donoghue MJ. The root of the angiosperms revisited. Proc Natl Acad Sci USA. 2002;99:6848–6853. doi: 10.1073/pnas.092136399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wellman CH, Gray J. The microfossil record of early land plants. Philos Trans R Soc Lond B Biol Sci. 2000;355:717–731. doi: 10.1098/rstb.2000.0612. discussion 731–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenrick PK, Crane PR. The Origin and Early Diversification of Land Plants. Washington, London: Smithsonian Institution Press; 1997. [Google Scholar]
- 52.Doyle JA. Molecules, morphology, fossils, and the relationship of angiosperms and Gnetales. Mol Phylogenet Evol. 1998;9:448–462. doi: 10.1006/mpev.1998.0506. [DOI] [PubMed] [Google Scholar]
- 53.Rothwell GW, Scheckler SE, Gillespie WH. Elkinsia gen. nov., a late Devonian gymnosperm with cupulate ovules. Bot Gaz. 1989;150:170–189. [Google Scholar]
- 54.Gerrienne P, Meyer-Berthaud B, Fairon-Demaret M, Streel M, Steemans P. Runcaria, a middle devonian seed plant precursor. Science. 2004;306:856–858. doi: 10.1126/science.1102491. [DOI] [PubMed] [Google Scholar]
- 55.Dunn MT. The Fayetteville flora I: Upper Mississippian (middle Chesterian/lower Namurian A) plant assemblages of permineralized and compression remains from Arkansas, USA. Rev Palaeobot Palynol. 2004;132:79–102. [Google Scholar]
- 56.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am Nat. 2007;170(Suppl 2):S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 57.Bousquet J, Strauss SH, Doerksen AH, Price RA. Extensive variation in evolutionary rates of rbcL gene sequence among seed plants. Proc Natl Acad Sci USA. 1992;89:7844–7848. doi: 10.1073/pnas.89.16.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor DW, Hickey LJ. Phylogenetic evidence for the herbaceous origin of angiosperms. Plant Syst Evol. 1992;180:137–156. [Google Scholar]
- 60.Feild TS, Arens NC, Doyle JA, Dawson TE, Donoghue MJ. Dark and disturbed: a new image of early angiosperm ecology. Paleobiology. 2004;30:82–107. [Google Scholar]
- 61.Feild TS, Chatelet DS, Brodribb TJ. Ancestral xerophobia: a hypothesis on the whole plant ecophysiology of early angiosperms. Geobiology. 2009;7:237–264. doi: 10.1111/j.1472-4669.2009.00189.x. [DOI] [PubMed] [Google Scholar]
- 62.Doyle JA. Seed ferns and the origin of angiosperms. J Torrey Bot Soc. 2006;133:169–209. [Google Scholar]
- 63.Doyle JA. Integrating molecular phylogenetic and paleobotanical evidence on origin of the flower. Int J Plant Sci. 2008;169:816–843. [Google Scholar]
- 64.Friis EM, et al. Phase-contrast X-ray microtomography links Cretaceous seeds with Gnetales and Bennettitales. Nature. 2007;450:549–552. doi: 10.1038/nature06278. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA. 2009;106:3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall CR. Confidence intervals on stratigraphic ranges with nonrandom distributions of fossil horizons. Paleobiology. 1997;23:165–173. [Google Scholar]
- 67.Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- 68.Grimaldi D, Engel MS. Evolution of the Insects. New York: Cambridge University Press; 2005. [Google Scholar]
- 69.Reik EF. Fossil insects from the Triassic beds at Mt. Crosby, Queensland. Aust J Zool. 1955;3:654–691. [Google Scholar]
- 70.Poinar GO, Jr, Danforth BN. A fossil bee from Early Cretaceous Burmese amber. Science. 2006;314:614. doi: 10.1126/science.1134103. [DOI] [PubMed] [Google Scholar]
- 71.Danforth BN, Brady SG, Sipes SD, Pearson A. Single-copy nuclear genes recover cretaceous-age divergences in bees. Syst Biol. 2004;53:309–326. doi: 10.1080/10635150490423737. [DOI] [PubMed] [Google Scholar]
- 72.Ren D, et al. A probable pollination mode before angiosperms: Eurasian, long-proboscid scorpionflies. Science. 2009;326:840–847. doi: 10.1126/science.1178338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith SA, Beaulieu JM, Donoghue MJ. Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol Biol. 2009;9 doi: 10.1186/1471-2148-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith SA, Dunn CW. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24:715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- 76.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 77.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 78.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 79.R Development Core Team . Vienna, Austria: R Foundation for Statistical Computing; 2009. R: A language and environment for statistical computing. http://www.R-project.org. Accessed March 2, 2010. [Google Scholar]
- 80.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 81.Qiu Y, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.