Abstract
A bioengineering method for self-assembly of multifunctional superstructures with in-advance programmable properties has been proposed. The method employs two unique proteins, barnase and barstar, to rapidly join the structural components together directly in water solutions. The properties of the superstructures can be designed on demand by linking different agents of various sizes and chemical nature, designated for specific goals. As a proof of concept, colloidally stable trifunctional structures have been assembled by binding together magnetic particles, quantum dots, and antibodies using barnase and barstar. The assembly has demonstrated that the bonds between these proteins are strong enough to hold macroscopic (5 nm–3 μm) particles together. Specific interaction of such superstructures with cancer cells resulted in fluorescent labeling of the cells and their responsiveness to magnetic field. The method can be used to join inorganic moieties, organic particles, and single biomolecules for synergistic use in different applications such as biosensors, photonics, and nanomedicine.
Keywords: superstructures, cancer cells, targeting, fusion proteins, magnetic nanoparticles
Recently, nanoparticles have become attractive objects for life science applications, in particular, in such rapidly growing areas as express diagnostics and advanced medical treatment. Encapsulation of nanoparticles with drug molecules (1, 2) or attaching them to viruses, bacteria, etc. are of special interest. Time-controlled release of the absorbed drugs would be advantageous for treatment of many diseases, e.g. diabetes, because of a decreased number of injections compared to that of molecular insulin. Furthermore, fluorescent or colored particles such as quantum dots (QD) (3), nanodiamonds (4), and gold nanoparticles (5) can be used for diagnostics as markers that provide visual information about the distribution of labeled agents in tissues and blood. Magnetic particles (MP) (6) can be also used as efficient labels for MRI diagnostics and can be precisely quantified even inside a living organism by an external induction probe (7, 8). At present, MP are widely studied for hyperthermia of tumors by heating in an AC magnetic field and for targeted delivery of drugs by magnetic field gradients, to avoid systemic intoxication of the organism (9, 10). Specific immunological targeting of nanoparticles by antibodies against pathogenic cells is another noteworthy application. Not only does it allow marking tumors for accurate dissection, but it also enhances drug delivery to the target cells.
The above-mentioned functional aspects of nanoparticles are brought into play in many life science applications. In certain cases, however, it would be beneficial to use multifunctional structures (11–13), which consist of several types of particles. Extensive studies were devoted to the synthesis of hybrid complexes of magnetic particles and different fluorophores (quantum dots or conventional chemical dyes) to allow visual MP tracking. Most approaches (14–16) employ sorption of dyes or QD on the MP surface, electro-static affinity between different components or complex chemical synthesis. Taking into account that these methods have a number of limitations such as instability in salt buffers or complexity of the synthesis procedures, new approaches are challenged.
Here we present a new generic method, named “protein-assisted nanoassembler,” for robust self-assembly of multifunctional superstructures consisting of different single-function particles: labels, carriers, recognition, and targeting agents, etc. The link between the particles is based on a very specific and strong noncovalent interaction between two proteins from Bacillus amyloliquefaciens—barnase and barstar (17–19).
Barnase and barstar noncovalent binding system (BBS) was chosen because of several remarkable properties. Firstly, the binding constant of barnase and barstar (17, 20, 21) is Ka ∼ 1014 M-1. That is only slightly smaller than the largest known binding constant of streptavidin and biotin (1015 M-1), but is much stronger than many other antibody–antigen constants, which normally range from 108 M-1 to 1011 M-1. One of the major advantages of BBS for the development of next-generation nanomedicine agents is its compatibility with in vivo applications. The barstar and barnase proteins are not displayed in mammals. On the contrary, (strept)avidin-biotin bridge could not be properly used in vivo because biotin, also known as vitamin H, is widely present in blood and tissues of mammals and may interfere with biotinylated agents during the drug administration. The closest homologue of barnase, binase (84% homology), does not induce any T-cell immune response (22), so one would accordingly expect the same for barnase. In addition, small size of barnase (12 kDa) and barstar (10 kDa) suggests lower immunogenicity than that of larger proteins like streptavidin. That makes further research of barnase–barstar system even more attractive. Moreover, because biotin is not a peptide, its attachment to proteins, e.g. antibodies, by gene engineering methods is not possible. Chemical linking is chaotic, usually nonspecific in terms of attachment geometry and often requires sophisticated postmodification separation of components. Although the technique for site-specific biotinylation of proteins is now developed, only cotransfection with a special plasmid in mammalian cells is possible in vitro or in vivo (23, 24). In contrast, barstar as well as barnase can be linked to antibodies by recombinant protein techniques and proteins of interest can be biosynthesized in different producents. This allows very attractive biotechnological production of fusion proteins (18, 25) and ensures standard composition and much higher reproducibility of the agent synthesis, activity of both proteins and the desired stoichiometry between antibody and one of the modular proteins. Another popular binding system based on DNA interaction (26, 27) also lacks the opportunity of nonchemical binding with antibodies. That results in less efficient cell targeting due to spatial hindrance of antigen-recognition sites of chaotically linked antibodies. BBS can be employed either alone or in combination with other techniques. For example, in traditional in vitro immunoassays, the multifunctional structures based on BBS can be used in combination with the (strept)avidin-biotin system, which is a commonly used analytical bridge between a variety of recognition antibodies and a generic label.
To describe the protein-assisted nanoassembler method, we would like to introduce the following glossary: Individual single-functional particles are conjugated with one of the BBS proteins (barnase or barstar) to form modules; modules are assembled together to form superstructures. Because the link between the modules is mediated by proteins and is very strong or quazicovalent, we say that the assembly is specific or protein-assisted to distinguish from ordered packing or ordinary agglomeration.
Results and Discussion
Assembly of Two-Module Superstructure with Magnetic and Fluorescent Functions.
As a first step, the protein-assisted nanoassembler principle was realized to join magnetic iron oxide particles with fluorescent nanoparticles. As a core module conjugated with barnase [MP-Bn], MP of different sizes and chemical composition were used: 500 nm magnetic particles (MagSense), 500 nm and 1 μm SiMAG magnetic beads (Chemicell) and 3 μm carboxyl magnetic beads (Spherotech). It should be emphasized that these particles, although not quite suitable for intravenous injections because of their big size, were chosen for the following reasons. First is the possibility of observation of such particles by an optical microscope, and second is our will to expand the range of the particles that can participate in the assembly process. As the fluorescent modules, we used Carboxyl Fluorescent Pink 53 nm polystyrene particles (Spherotech) and Qdot® 605 quantum dots (Invitrogen) conjugated with barstar ([Pink-Bs] and [QD-Bs], respectively). According to Spherotech, their particles are synthesized by polymerizing a fluorophore in styrene in the presence of polystyrene core particles. The surface conjugation chemistry for these particles is the same as for other polystyrene particles, which can, for example, absorb drug molecules. Binding of the proteins with particles was achieved using N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) (28) enhanced by N-Hydroxysulfosuccinimide sodium salt (Sulfo-NHS) according to the particle manufacturer recommendations. The assembly of magnetic and fluorescent modules into a single structure was performed by mixing suspensions of the modules in PBS buffer at room temperature for 30 min with excess of the fluorescent module in the presence of 0.5%–1% Tween 20. The unbound fluorescent modules were washed away while the assembled structures {[MP-Bn] + [Pink-Bs]} or {[MP-Bn] + [QD-Bs]} were kept by a magnetic field.
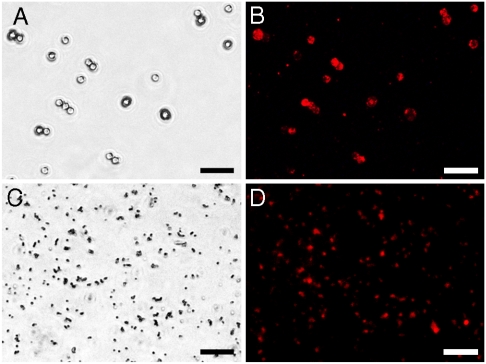
The results obtained for different combinations of the magnetic and fluorescent modules were similar. All the structures demonstrated efficient and highly specific assembly between the modules. An example of assembly between 500 nm magnetic particles (MagSense) or 3 μm carboxyl magnetic beads (Spherotech) and Carboxyl Fluorescent Pink 53 nm polystyrene particles (Spherotech) can be observed in Fig. 1. Quickly sedimenting 3 μm structures in the bright-field image (Fig. 1A) can be easily matched to their fluorescence in Fig. 1B. In case of 500 nm particles that are much smaller and constantly moving, the points in Fig. 1C and D match only partially, where the particles stick to the surface of the microscope glass. However, overall fluorescence of the structures is well detectable.
Fig. 1.
Photographs of the assembled bifunctional structures: (A and B) Structures assembled by mixing modules of 3-μm magnetic particles conjugated with barnase and Carboxyl Fluorescent Pink 53 nm polystyrene particles conjugated with barstar after magnetic washing of unbound fluorescent particles. (C and D) structures assembled by mixing modules of 500 nm magnetic particles conjugated with barnase and Carboxyl Fluorescent Pink 53 nm particles conjugated with barstar after magnetic washing of unbound fluorescent modules. (A and C) Bright-field pictures, (B and D) pictures made with excitation of fluorophore. (Scale bar: 10 μm.)
The crucial role of the barnase–barstar system for binding the components of the bifunctional structures together was proven by pretreatment of [MP-Bn] module with excess of free wild barstar before adding [Pink-Bs] or [QD-Bs] modules or by comparison of the assembly results when the magnetic particles were conjugated with BSA instead of barnase. The magnetically separated structures did not demonstrate any noticeable fluorescence at the same exposure (meaning no binding of fluorescent modules). The magnetic particles as well as [MP-Bn] modules not treated with [QD-Bs] also did not reveal any fluorescence at the same and longer exposures. Control experiments are illustrated in supporting information Fig. S1.
To accelerate the assembly of structures it is important to maximize the amount of the active BBS proteins on the surface of the particles. By active we imply a native protein capable of binding with the partner protein, i.e., having an accessible binding site. Because barnase is an RNAse, its enzymatic activity was used to estimate the surface density of the active protein on the particles to optimize the conjugation protocol. The evaluation was carried out by measuring an increase of yeast RNA absorbance at 260 nm during its hydrolysis by barnase (29) conjugated with MP (SI Text and Fig. S2). The estimated average density of active barnase on the magnetic and fluorescent polystyrene particles, which we used, was not < 1 molecule per 300 nm2. That means that the average distance between two active molecules was < 17 nm. Taking into account the diameters of the particles used, 50 and 500 nm, one could expect that the bond between two particles was realized by approximately one pair of the BBS proteins. The small size of the proteins (barnase -2 × 2.5 × 3.5 nm, barstar -1.5 × 2.5 × 2.5 nm) allows a close contact between the particles, whereas introduction of flexible spacers (18, 30) may permit the bond formation between the protein pairs located further from the contact point of particles for their stronger multipoint interlinking.
Assembly of Three-Module Superstructure with an Additional Antitumor-Targeted Function.
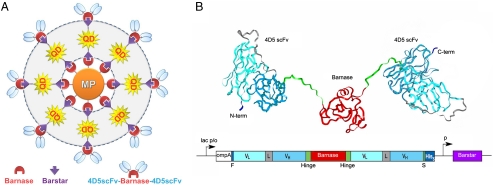
The same generic approach can be used for assembling of more complicated superstructures constructed from a larger number of different modules. For demonstration, we have assembled trifunctional superstructures by introducing an antitumor antibody module (Fig. 2A). Such structures could be used for simultaneous identification (by antibody), labeling (by quantum dot), and destruction of cancer cells (by magnetic particle after heating it in an AC magnetic field).
Fig. 2.
Conceptual design of trifunctional superstructures. (A) Layer-by-layer assembly: [MP-Bn] module as a core, then a layer of [QD-Bs] modules, and, finally, a layer of [4D5scFv-Bn] modules. (B) Construction of the recognition module: molecular model (ribbon representation, PDB 1fve and 1bnr) of the 4D5scFv-barnase-4D5scFv (above) and gene construct encoding the recombinant protein. The 4D5 scFv-barnase-4D5 scFv-His5 construct starts with N-terminal short FLAG tag (F, Dark Blue) followed by 4D5 scFv in VL-linker-VH orientation (VL, Turquoise; linker, Gray; VH, Cyan), 16-amino-acid hinge linker (Green); barnase (Red), hinge linker, and 4D5 scFv. The construct terminates in His5-tag (Dark Blue) attached via a short spacer (S) with the sequence Gly-Ala-Pro (Green) to the C-terminus of scFv-barnase-scFv fusion protein. The fusion gene is under control of the lac promoter and the ompA signal peptide is used to direct the secretion of the recombinant proteins to the E. coli periplasmic space. Barstar coexpression controlled by its own constitutive promoter (p) is required to suppress the cytotoxicity of barnase fusions. lac p/o, lac promoter/operator.
Single chain scFv antibody fragments are ideal for use as targeting moieties in selective delivery applications. They offer significant advantages over full-size antibodies due to a smaller size and rational design (19). Taking into account that conversion of scFv antibody fragments into dimeric format increases avidity to their antigens and decreases cell surface dissociation rates, we designed a bivalent scFv-barnase-scFv construct (Fig. 2B). The three-dimensional structure of the barnase–barstar complex shows that N and C termini of both proteins are localized away from the dimerization interface (17, 20). Therefore, all termini are accessible for fusions. The linking of scFv fragments to both termini of barnase was carried out via a flexible hinge peptide that allowed rotational and segmental flexibility preventing steric hindrance and enhancing the simultaneous binding of different receptor molecules on the cell surface. Thus, bifunctional fusion protein, scFv-barnase-scFv was constructed as the third antibody-targeted module. As an antibody, we chose anti-human epidermal growth factor receptor 2/neu (anti-HER2/neu) miniantibody (30–33), scFv 4D5 antibody fragment (below referred to as “4D5scFv”).
The trifunctional structures were assembled from the bifunctional particles (1-μm SiMAG magnetic beads from Chemicell and Qdot® 605 quantum dots from Invitrogen) described above by binding an additional layer of 4D5scFv-barnase-4D5scFv module ([4D5scFv-Bn]) as shown in Fig. 2A. The assembled complexes have the [MP-Bn] module as a core of 1 μm in diameter bound with the layer of [QD-Bs] modules approximately 10 nm, and, finally, with the layer of [4D5scFv-Bn] modules of approximately the same size.
Demonstration of Trifunctionality of the Assembled Superstructures.
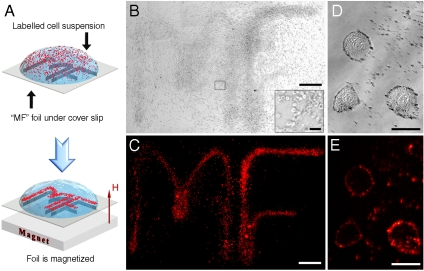
Designed multifunctionality of the superstructures was demonstrated with the following experimental procedure. A strip of ferromagnetic foil was cut and folded to form 3 mm high letters “MF” (MultiFunctional) as shown in Fig. 3A. The foil was placed above a NdFeB permanent magnet and was covered by a 0.1 mm glass slip. Such a setup produced strong magnetic field gradients on the glass surface to drag the magnetic particles toward the foil edges, i.e., along the contours of the letters MF.
Fig. 3.
Magnetic field-guided inscription of MF contour with cancer cells labeled by trifunctional superstructures: (A) Suspension of human ovarian cancer SKOV-3 cells labeled by the assembled trifunctional structures was dropped on a glass slip located above the ferromagnetic foil. The foil produced magnetic field gradients that dragged the cells labeled with the superstructures toward the contour of letters MF; (B and C) bright-field and fluorescent photos of the sample in the fluorescent microscope (multiple overlapping photos at 100x magnification were taken and then digitally combined into a single photograph to visualize individual cells over a large field of view). (Scale bar: -0.5 mm.) (Inset) Approximately 30 cells in the field of view. (Scale bar: -50 μm.) (D and E) Close-up bright-field and fluorescent photos showing three individual cells labeled with the trifunctional structures. (Scale bar: 17 μm.)
As a biological target, we chose human ovarian carcinoma SKOV-3 cells that overexpress the HER2/neu receptor on their surface. We added the assembled trifunctional superstructures in PBS buffer with 0.2% Tween 20 to these adhesive cells that were attached to the bottom of a culture plate, and incubated for 40 min at 4 °C to prevent antibody internalization. After three gentle washings of unbound superstructures with PBS buffer with 0.2% Tween 20, the cells linked with trifunctional complexes were suspended with 0.1 M EDTA (Versene) solution. A drop of the labeled cells suspension was placed on the glass slip located above the ferromagnetic foil, which was magnetized as shown in Fig. 3A. After 5 min, we examined the sample by fluorescence microscopy. The resulting images are displayed in Fig. 3B and C. It is clearly seen in the bright-field image that the cancer cells are mainly concentrated in the areas with strong magnetic field gradient displaying the letters MF (images of individual cells are shown in the magnified corner inset of Fig. 3B; a high resolution version of Fig. 3B is given in Fig. S3). This confirmed that MP of the superstructures are capable of moving the labeled cells to the permanent magnet poles. One can notice a few cells outside the MF contour. These are the cells that were located near the glass surface when the drop of cell suspension was placed on the glass. Once the cells settle on the glass surface, the magnetic forces cannot move the cells against friction. Fig. 3C also evidences that the superstructures have quantum dots, which form the fluorescence image of letters MF with high contrast. Hence, the conducted experiments demonstrate that the assembled superstructures possess all three designed functions: they are antitumor targeted, magnetic, and fluorescent.
Furthermore, the discussed interaction of the cells with the trifunctional superstructures due to the antibody module is also confirmed by the higher spatial resolution photographs shown in Fig. 3D and E. These images display both the clusterization of superstructures at the surface of cancer cells and individual superstructures. It is clearly seen that the locations of fluorescent dots in Fig. 3E are in good agreement with those of the structures (black dots in Fig. 3D).
To test the binding specificity, we used the magnetic particles conjugated with BSA, [MP-BSA] module, described above as a core of the structures. In the control experiments (Fig. S4) with particles {[MP-BSA] + [QD-Bs] + [4D5scFv-Bn]}, no binding with cells and no fluorescence were observed under the same conditions. This comprehensive investigation confirms a high specificity and multifunctionality of the assembled superstructures.
Variety of Superstructure Composition.
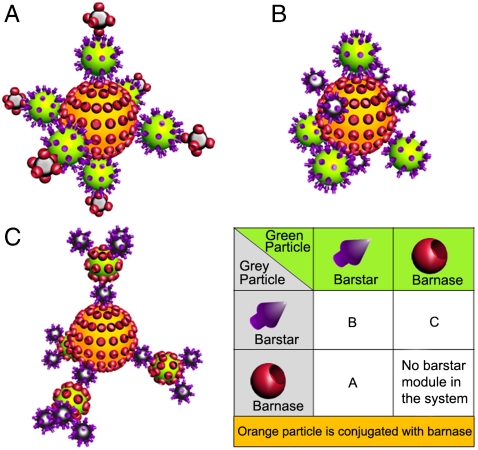
The superstructure composition synthesized with the protein-assisted nanoassembler method depends on the construction algorithm. By choosing various combinations of particles conjugated with the BBS proteins and the order of their incubation with each other, it is possible to get different sophisticatedly built complexes. For example, if we combine three different types of particles, we can get three different superstructures shown in Fig. 4A–C in 3D format or in Fig. S5 (2D version). The structures are drawn schematically for the concept demonstration. Let us assume that the core particle is conjugated with barnase. Then, depending on the combination of the proteins conjugated with the other two particles, it is possible to produce four combinations as shown in Fig. 4 (Table). However, the structure where all the proteins are barnase cannot be assembled: at least one barstar module should be involved.
Fig. 4.
Possible variants for assembling of three particles into a superstructure by the protein-assisted nanoassembler method. Correspondence of different combinations of BBS proteins conjugated with the green and gray particles and the structure number (A–C) is shown in the table. Barnase is chosen as the protein conjugated with the core orange particle. Two-dimensional representation can be found in Fig. S5.
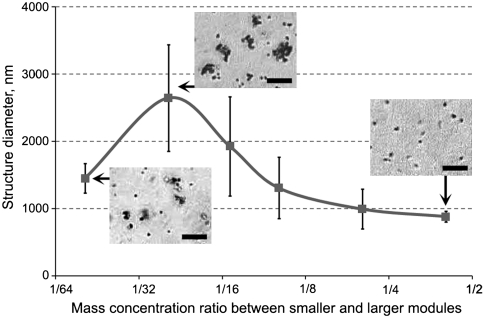
A remarkable property of the protein-assisted nanoassembler is the dependence of stable structure size on the ratio of modules in the mixture. In some experiments assembling two modules of different sizes, along with single structures consisting of one larger module, we observed assembly of much larger structures. Their size proved to depend on the concentration ratio of the modules in the initial mixture. The assembly process can be explained as follows. In the case of 1∶∞ ratio of two different modules used for assembly, every “unique” particle is quickly covered by a shielding layer of the complementary modules and such structure becomes inert, i.e., does not interact with other particles in the system. However, if two modules of the same particles covered by different BBS proteins are mixed in 1∶1 ratio, the modules stick to each other because of equal probability for binding with either of the modules due to the absence of the “shielding layer.” In this process the structures incorporate many modules and are much larger than in the previous case. This theoretical consideration was proven by assembly of 500 nm magnetic particles (MagSense) and 53 nm fluorescent polystyrene nanoparticles (Spherotech) over a wide range of concentration ratios between the modules. To eliminate the effect of size change during measurements due to further assembly or agglomeration, the size of all superstructures was measured within minutes after long overnight incubation. The experimentally observed dependence of the average sizes of the superstructures on the concentration ratio of modules is shown in Fig. 5. The data demonstrate that with a fixed concentration of larger modules, the structures become larger for a lower concentration of smaller modules, which should be opposite for nonspecifically agglomerating particles. The left point falls out of this dependence because of an insufficient relative concentration of smaller modules to form the structures. Statistical size dispersion of the structures also depends on the ratio between smaller and larger modules. For the left and right points of Fig. 5 the major part of roughly 10% size dispersion can be attributed to the size variation of modules, i.e., the particles of which they consist, whereas almost 40% dispersion for the points in the midrange of Fig. 5 is due to the assembly process. More descriptive explanation of the size dispersion can be found in SI Text. This experiment again proves that the protein-assisted self-assembly of the superstructures is tight, specific, and can be employed to assemble the structures with not only programmable functionality but also of various sizes from the same modules.
Fig. 5.
Dependence of average size of the superstructures after overnight incubation on mass ratio of modules based on 500 nm magnetic particles and 53 nm fluorescent polystyrene nanoparticles. Error bars show standard deviation calculated based on information about 20 structures for every point. (Scale bar: 10 μm.)
In conclusion, the protein-assisted nanoassembler method could be regarded not as a substitution of other most general approaches, (strept)avidin-biotin system (13), and complementary DNA assembled scaffolds (26, 27), but as an alternative that broadens the range of available nanoagents. It can be employed either on its own or in combination with other techniques allowing self-assembly of a wide variety of superstructures with in-advance programmable properties such as specific functionality, size, and composition. A remarkable feature of the strategy is the opportunity to attach any protein in its functional form to the key components, barnase and barstar, by gene engineering methods to construct recognition, visualization, or cytotoxic modules. Their assembly with nanoparticles of different biochemical and physical nature using the same generic approach could be an easy and convenient way to create desirable multifunctional superstructures.
Materials and Methods
Construction and Expression of 4D5scFv-barnase-4D5scFv.
Genetic engineering manipulations, cell culture, and cell lysis followed the standard protocols. The DNA fragment encoding the 4D5scFv-barnase protein was amplified from a pSD-4D5scFv-barnase plasmid (18) using primers 5′-ATCGTTATGACCCAGTCTC-3′ and 5′-ATCGGCGCCACTAGTATGGGTGGTATCACCCAGTGGGGTACCTCTGATTTTTGTAAAGG-3′ (encoding flexible human IgG3 hinge-like peptide linker). The amplification product was cloned into the Eco RV site of pSD-4D5scFv (with precloned barstar) (18). The orientation of 4D5scFv-barnase gene was checked by restriction using Eco RI endonuclease.
To produce the recombinant protein, Escherichia coli SB536 strain was transformed with pSD-4D5scFv-barnase-4D5scFv and grown in LB at 25 °C. At OD550 = 1 the culture was induced with 1 mM IPTG and then incubated at 25 °C for 12 h. Purification of 4D5scFv-barnase-4D5scFv fusion protein was carried out according to the procedure described earlier (18). The protein homogeneity was confirmed by SDS-PAGE analysis in 10% polyacrylamide gel according to the standard protocol.
Other Proteins.
Barstar and barnase were produced in E. coli cells and purified as described in our previous work (18). BSA was purchased from Sigma.
Particles.
500 nm and 1 μm SiMAG (Chemicell) and 500 nm Carboxyl magnetic particles (MagSense), Qdot® 605 ITK™ carboxyl quantum dots (Invitrogen), and Carboxyl Fluorescent Pink 53 nm particles (Spherotech) with -COOH groups available for conjugation were used as functional parts of modules in the assembly process.
Conjugation.
Binding of the proteins with particles was achieved using EDC (28) (Fluka) enhanced by Sulfo-NHS (Sigma–Aldrich) according to the particle manufacturer recommendations. For 1 mg of magnetic and polystyrene particles 1–40 mg of EDC and 0.6–26 mg of Sulfo-NHS were used for activation. In the case of barnase, 1 mg of protein per 1 mg of particles was added for conjugation, and 0.1–1 mg of protein per 1 mg of particles was added for barstar. For QD, molar ratios were QD∶EDC∶Protein = 1∶1000∶40. The conjugation efficiency was studied by RNA-hydrolyzing activity of barnase and inhibitary properties of barstar (29).
Visualization and Measurements of the Structure Size.
Carl Zeiss Axiovert 200 fluorescent microscope was used to take images of the particles and cells. 100x and 400x magnifications were used. To get Fig. 3B and C, multiple overlapping photos at 100x magnification were taken and then digitally combined into a single photograph to visualize individual cells over a large field of view.
Supplementary Material
Acknowledgments.
The work was supported by Russian Academy of Sciences Programs Molecular and Cellular Biology and Nanotechnologies and Nanomaterials, Russian Foundation of Basic Research Grants #09-04-12284, 09-04-01201, 09-04-01665, and 09-02-12195, and the Russian Federal Agency for Science and Innovation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001142107/DCSupplemental.
References
- 1.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan TT, et al. Encapsulation of organic molecules in calcium phosphate nanocomposite particles for intracellular imaging and drug delivery. Nano Lett. 2008;8:4108–4115. doi: 10.1021/nl8019888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michalet X, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–44. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang YR, et al. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat Nanotechnol. 2008;3:284–288. doi: 10.1038/nnano.2008.99. [DOI] [PubMed] [Google Scholar]
- 5.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;1:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 6.Frey NA, Peng S, Cheng K, Sun S. Magnetic nanoparticles: Synthesis, functionalization, and applications in bioimaging and magnetic energy storage. Chem Soc Rev. 2009;38:2532–2542. doi: 10.1039/b815548h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikitin MP, Vetoshko PM, Brusentsov NA, Nikitin PI. Highly sensitive room-temperature method of non-invasive in vivo detection of magnetic nanoparticles. J Magn Magn Mat. 2009;321:1658–1661. [Google Scholar]
- 8.Nikitin MP, Torno M, Chen H, Rosengart A, Nikitin PI. Quantitative real-time in vivo detection of magnetic nanoparticles by their nonlinear magnetization. J Appl Phys. 2008;103:07A304. [Google Scholar]
- 9.Dames P, et al. Targeted delivery of magnetic aerosol droplets to the lung. Nat Nanotechnol. 2007;2:495–499. doi: 10.1038/nnano.2007.217. [DOI] [PubMed] [Google Scholar]
- 10.Barnett BP, et al. Magnetic resonance-guided, real-time targeted delivery and imaging of magnetocapsules immunoprotecting pancreatic islet cells. Nat Med. 2007;13:986–991. doi: 10.1038/nm1581. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Piao Y, Hyeon T. Multifunctional nanostructured materials for multimodal imaging, and simultaneous imaging and therapy. Chem Soc Rev. 2009;38:372–390. doi: 10.1039/b709883a. [DOI] [PubMed] [Google Scholar]
- 12.Liong M, et al. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randall ME, Hui SS, Bappaditya S, Vincent MR, Benjamin BY. Magnetic assembly of colloidal superstructures with multipole symmetry. Nature. 2009;457:999–1002. doi: 10.1038/nature07766. [DOI] [PubMed] [Google Scholar]
- 14.Quarta A, Di Corato R, Manna L, Ragusa A, Pellegrino T. Fluorescent-magnetic hybrid nanostructures: preparation, properties, and applications in biology. IEEE T Nanobiosci. 2007;6:298–308. doi: 10.1109/tnb.2007.908989. [DOI] [PubMed] [Google Scholar]
- 15.Shi W, et al. A general approach to binary and ternary hybrid nanocrystals. Nano Lett. 2006;6:875–881. doi: 10.1021/nl0600833. [DOI] [PubMed] [Google Scholar]
- 16.Liong M, et al. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS Nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley RW. Barnase-barstar interaction. Methods Enzymol. 2001;341:599–611. doi: 10.1016/s0076-6879(01)41179-7. [DOI] [PubMed] [Google Scholar]
- 18.Deyev SM, et al. Design of multivalent complexes using the barnase-barstar module. Nat Biotechnol. 2003;21:1486–1492. doi: 10.1038/nbt916. [DOI] [PubMed] [Google Scholar]
- 19.Deyev SM, Lebedenko EN. Multivalency: the hallmark of antibodies used for optimization of tumor targeting by design. BioEssays. 2008;30:904–918. doi: 10.1002/bies.20805. [DOI] [PubMed] [Google Scholar]
- 20.Buckle AM, Schreiber G, Fersht AR. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0 Å resolution. Biochemistry. 1994;33:8878–89. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- 21.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 22.Ilinskaya ON, et al. Binase induces apoptosis of transformed myeloid cells and does not induce T-cell immune response. Biochem Biophys Res Commun. 2007;361:1000–1005. doi: 10.1016/j.bbrc.2007.07.143. [DOI] [PubMed] [Google Scholar]
- 23.Predonzani A, Arnoldi F, López-Requena A, Burrone OR. In vivo site-specific biotinylation of proteins within the secretory pathway using a single vector system. BMC Biotechnol. 2008;8:41. doi: 10.1186/1472-6750-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenyuk EG, et al. Expression of single-chain antibody-barstar fusion in plants. Biochimie. 2007;89:31–38. doi: 10.1016/j.biochi.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Mastroianni AJ, Claridge SA, Alivisatos AP. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds. J Am Chem Soc. 2009;131:8455–8459. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S-K, Maye MM, Zhang Y-B, Gang O, van der Lelie D. Controllable g5p-Protein-Directed Aggregation of ssDNA-Gold Nanoparticles. Langmuir. 2009;25:657–660. doi: 10.1021/la803596q. [DOI] [PubMed] [Google Scholar]
- 28.Hermanson GT. Bioconjugate Techniques. 2nd Ed. London: Academic; 2008. [Google Scholar]
- 29.Edelweiss E, et al. Barnase as a new therapeutic agent triggering apoptosis in human cancer cells. PLoS One. 2008;3:e2434. doi: 10.1371/journal.pone.0002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serebrovskaya EO, et al. Targeting cancer cells by using an antireceptor antibody-photosensitizer fusion protein. Proc Natl Acad Sci USA. 2009;106:9221–9225. doi: 10.1073/pnas.0904140106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willuda J, et al. Tumor targeting of mono-, di-, and tetravalent anti-p185(HER-2) miniantibodies multimerized by self-associating peptides. J Biol Chem. 2001;276:14385–14392. doi: 10.1074/jbc.M011669200. [DOI] [PubMed] [Google Scholar]
- 32.Slamon D, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 33.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.