Abstract
Insects are the most diverse form of life on the planet, dominating both terrestrial and freshwater ecosystems, yet no species has a life stage able to breath, feed, and develop either continually submerged or without access to water. Such truly amphibious insects are unrecorded. In mountain streams across the Hawaiian Islands, some caterpillars in the endemic moth genus Hyposmocoma are truly amphibious. These larvae can breathe and feed indefinitely both above and below the water's surface and can mature completely submerged or dry. Remarkably, a molecular phylogeny based on 2,243 bp from both nuclear (elongation factor 1α and carbomoylphosphate synthase) and mitochondrial (cytochrome oxidase I) genes representing 216 individuals and 89 species of Hyposmocoma reveals that this amphibious lifestyle is an example of parallel evolution and has arisen from strictly terrestrial clades at least three separate times in the genus starting more than 6 million years ago, before the current high islands existed. No other terrestrial genus of animals has sponsored so many independent aquatic invasions, and no other insects are able to remain active indefinitely above and below water. Why and how Hyposmocoma, an overwhelmingly terrestrial group, repeatedly evolved unprecedented aquatic species is unclear, although there are many other evolutionary anomalies across the Hawaiian archipelago. The uniqueness of the community assemblages of Hawaii's isolated biota is likely critical in generating such evolutionary novelty because this amphibious ecology is unknown anywhere else.
Keywords: evolution, Hyposmocoma, molecular clock, phylogeography, amphibious
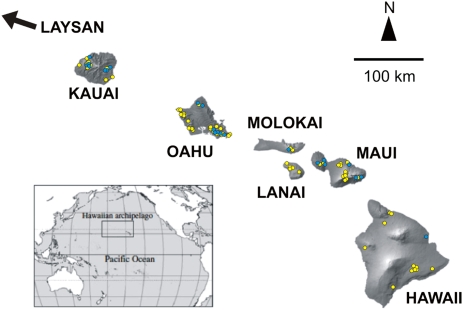
Aquatic and terrestrial environments present divergent challenges for all animals, particularly in their need to breathe, move, and feed. Although there are many aquatic insects that can tolerate extended periods of desiccation through aestivation or diapause (1–4) and terrestrial species that tolerate extended inundation through dormancy (5–7), there are no species known to function equally well in both underwater and dry environments for extended periods. The Hawaiian Islands are the most isolated archipelago on Earth and have fostered many unusual evolutionary novelties, particularly among the arthropods (8–11). Although Lepidoptera, with more than 150,000 described species, is one of the four most diverse orders of insect, only 0.5% of all caterpillars are truly aquatic (breathing directly from the water), and none of these aquatic species can develop out of water (12, 13). However, we report the discovery of caterpillars in the endemic Hawaiian moth genus Hyposmocoma that are previously uncharacterized and restricted to the mountains of the Hawaiian Islands (Fig. 1) and that are able to thrive in both environments indefinitely and represent the only insects able to do so.
Fig. 1.
Map of the Hawaiian Islands. The locations of collecting sites are indicated by yellow dots for terrestrial Hyposmocoma larvae and by blue dots for aquatic Hyposmocoma larvae.
In addition to foraging and resting above and under water, the larvae may also pupate under water, entailing weeks of constant inundation (Fig. S1). As a further adaptation to living around flowing water, they withstand extremely high and fast floodwaters, which can scour the rainforest streambeds for days during frequent storm events. Through a combination of specialized shelter-seeking behavior and use of silk tie-downs and drag lines (Movie S1), larvae avoid exposure to the strongest currents and quickly return themselves to the substrate if dislodged (14). Amphibious larvae exclusively use volcanic rocks with small holes, and the larvae almost always rest communally in the holes on the downstream side of their rocks (Fig. S1). These amphibious species occur along streams on the islands of Kauai, Oahu, Molokai, Maui, and Hawaii (Fig. 1). They may remain submerged for weeks or live on dry rocks meters from any body of water, grazing on dried algae and lichens (Movie S2). Dissection of the cases while under water reveals no air bubble to mediate gas exchange, and the larvae possess no gills or plastron (Fig. S2), common structures for underwater respiration in other insects (13). When submerged, they likely rely on direct diffusion of oxygen through the hydrophilic skin along their abdomens (Fig. S2). Perhaps as a result of their need for direct diffusion, the caterpillars occur only in fast-flowing, well-oxygenated streams, and quickly die in stagnant water.
Here we report the unique and specialized ecology of these amphibious caterpillars, which represent only a small fraction of the more than 350 otherwise strictly terrestrial species in Hyposmocoma (15), and we use both mitochondrial and nuclear gene sequence to construct a phylogeny revealing the evolution of these newly discovered amphibious species in the context of their strictly terrestrial sister taxa. We use this systematic analysis to determine the number of different amphibious species, whether they represent a single invasion of the water that then diversified or multiple independent invasions, and finally a molecular clock to estimate how long ago these aquatic shifts could have occurred.
Results
Phylogenetic Relationships and Divergence Time.
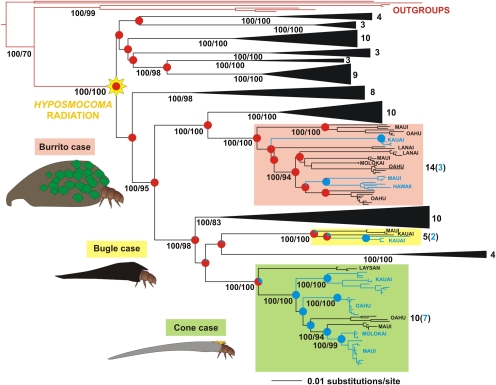
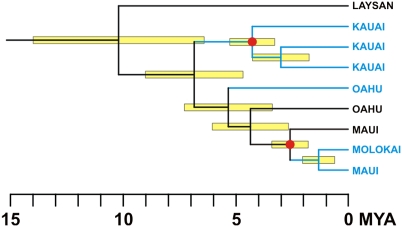
Across the Hawaiian Archipelago we found three previously unknown lineages of moths in the endemic genus Hyposmocoma (Fig. 2), each with multiple species whose caterpillars can develop both under water in streams and on dry land with no access to water: they breathe and feed with no difficulty above and below the surface of the water. This previously unrecorded phenomenon makes these species truly amphibious in a way unmatched by any other insect. We conducted phylogenetic analyses on 216 moths reared from larvae representing 89 putative species collected in the field to confirm amphibious or terrestrial ecologies. The dataset consisted of 2,243 bp from two nuclear and one mitochondrial gene, and both maximum likelihood (ML) and Bayesian inference supported the same topology (Fig. S3). The ML tree is shown in Fig. 3. All 12 lineages representing the various amphibious and terrestrial species of Hyposmocoma case types are monophyletic and were strongly supported in both analyses, as is the basal radiation that gave rise to this remarkable diversity (Bayesian posterior probabilities 1.0 and bootstrap values >80%). The aquatic species occur in three different lineages, including larvae with the “burrito,” “bugle,” and “cone” case types. Our estimate for the divergence time of the basal split in the cone lineage (Fig. 4) that occurred on the Northwestern Hawaiian Island of Laysan was 10.2 million years (95% confidence interval, 6.4–14.4), and the aquatic ecology in this lineage evolved at 6.8 million years (95% confidence interval, 4.7–9.0).
Fig. 2.
Underwater activity of larvae representing each of the three amphibious case-bearing caterpillar lineages belonging to the endemic Hawaiian moth genus Hyposmocoma. (A) “Burrito”-shaped case larvae attached to substrate with silk line. (B) “Cone”-shaped case larvae feeding on algae. (C) “Bugle”-shaped case larvae crawling on submerged rocks. See SI for videos of larvae moving in and out of water (Movies S1 and S2).
Fig. 3.
Phylogeny of Hyposmocoma moths based on molecular data with the three lineages including amphibious species highlighted. Shown is the maximum likelihood tree based on combined analysis of three genes: the mitochondrial gene cytochrome oxidase subunit I and the nuclear genes elongation factor 1α and carbomoylphosphate synthase. Bayesian posterior probabilities ≥95 and nonparametric bootstrap supports ≥70 are given under each corresponding node and clade. This topology is congruent with the Bayesian topology (Fig. S3). Numbers on the right of the different terrestrial clades represent species per clade, with blue numbers in brackets representing total of aquatic species. Proportional likelihoods of ancestral states are mapped onto each node of interest (red = terrestrial; blue = aquatic); exact values are given in Fig. S6. Branch lengths are drawn to scale, as indicated by scale bars.
Fig. 4.
Maximum clade credibility chronogram inferred using the uncorrelated relaxed clock model of rate evolution. Error bars represent 95% posterior credibility intervals and are only given for nodes that were present on more than 50% of the posterior sampled trees. Red filled circles represent the two nodes used for calibration. Ages are given in millions of years before present.
Evolution of Amphibious Species.
Although such a flexible amphibious ecology is previously unrecorded (13), we discovered 12 species of Hyposmocoma in three different case type lineages able to develop above or under water (Fig. 3). Because this life history is unrecorded, it would be parsimonious to assume that it arose once in the genus and that the lineage diversified into the 12 species and three lineages in this study. Surprisingly, the amphibious Hyposmocoma are not monophyletic, and each of the case types represents an independent aquatic invasion; the clades of amphibious Hyposmocoma are separated by groups of strictly terrestrial species, and amphibious species from different clades are sympatric with each other across most of the high islands (Fig. 3). Further, even within the burrito case type there is paraphyly with two unrelated lineages with similar burrito case shapes, each having larvae that independently invaded Hawaiian streams. This phylogenetic pattern suggests multiple independent aquatic invasions with a probable preadaptation for true amphibious life histories in the genus Hyposmocoma. Alternatively, the intervening, strictly terrestrial, sister species between the aquatic lineages could represent multiple losses of the aquatic life history and thus reversions to the ancestral terrestrial state, also held by more than 300 other species of Hyposmocoma (15) (Fig. 3). Either scenario is remarkable because they represent the repeated acquisition or loss of a truly amphibious lifestyle not recorded anywhere else.
Discussion
All caterpillars in the genus Hyposmocoma spin silk cases embedded with minute objects from their environment (e.g., pebbles, diatoms, algae, and lichens) to protect and camouflage their bodies, serving as essential shelter both in and out of the water; larvae quickly perish when removed from their cases. Because at least three different case types (Figs. 2 and 3) have independently derived both terrestrial and amphibious species, it is unlikely that specific lineages or case types are necessary to support the amphibious life history. Instead, these amphibious larvae may be an example of ecological diversification promoting speciation; similar patterns of parallel evolution in which related taxa independently derive similar ecologies has also been found in Hawaiian birds and damselflies (16, 17). Such diversification and parallel evolution may be possible for the caterpillars owing to particular characteristics of Hyposmocoma but more likely involves features of this insect lineage working in conjunction with Hawaii's extreme isolation. Hawaiian streams lack nearly all of the insect orders or families that dominate continental aquatic ecosystems (13, 15), and this ecological opportunity may have had a prominent role in the evolution of Hyposmocoma’s amphibious larvae.
According to molecular clock calibrations, the diversity of endemic amphibious Hyposmocoma on Kauai, and their derived relationship to the terrestrial Northwestern Hawaiian Island species, the aquatic ecology evolved approximately 6.8 million years ago (mya), long before the formation of Kauai. However, because there is no longer flowing water on the Northwestern Hawaiian Islands, the relictual, ancestral species that occurred there may have evolved amphibious life histories in the different lineages when those islands were still high enough to support streams. The present day Northwestern Hawaiian Island species would thus be the descendents of the first aquatic lineages, which spread to the younger, higher islands as they arose, but the species that were marooned on the original Northwestern islands must have subsequently reverted to the ancestral, strictly terrestrial state as their once-lush islands eroded into arid atolls. Thus, the advent of amphibious larvae on Laysan Atoll, which supports the oldest endemic Hyposmocoma in the same clade as the aquatic species in the Hawaiian Archipelago, could be as old as 10.2 million years and suggests a very different habitat in the younger stages of that island's history (Fig. 4).
The mountains of the Hawaiian Islands represent some of the wettest places on Earth (18). The extreme conditions of stream-living in montane Hawaii require that the larvae breathe, feed, and crawl for indefinite periods in fast-moving water, including enduring frequent, severe, flash flood events. How and why certain species of Hyposmocoma have overcome the physiological limitations of breathing directly from both water and air will be an important line of future research.
Each amphibious species of Hyposmocoma is endemic to a specific volcano, where they inhabit only relatively unmodified, fast-flowing streams and adjacent riparian zones. This habitat has become increasingly rare as water is diverted from watersheds into culverts and dikes for human use. Nearly all amphibious species are now limited to montane rainforest streams, although forest streams with intact habitat at sea level support larvae, suggesting that many species have undergone drastic reductions in range due to development and that others may already be extinct. The unique ecology and remarkable systematics of amphibious Hyposmocoma highlights the need for proactive conservation, particularly in places like Hawaii, which have high rates of extinction and habitat loss paired with high levels of regional endemism and unparalleled evolutionary phenomena.
Materials and Methods
Sampling.
All caterpillars for this study were collected in situ in streams (amphibious) or on land (terrestrial) and reared in the laboratory from case-making caterpillars. We collected across the islands of Kauai, Oahu, Molokai, Maui, and Hawaii. We also collected terrestrial caterpillars from the above islands, as well as the dry island of Lanai and the Northwestern Hawaiian Island of Laysan. We held each collection of Hyposmocoma larvae in Petri dishes, and larvae fed on carrot and commercial fish food (TetraMin). For each field collection, a unique data log entry was made including date and location, digital pictures of larvae and case, record of larval behavior, date of adult moth emergence, and digital picture of adult moth. Adult moths were then frozen at −80 °C for molecular studies.
Amphibious Ecology.
On multiple occasions between 2004 and 2008, we placed dozens of larvae found in or near streams in aerated 37.84-L aquaria with no emergent rocks. These larvae survived and fed underwater for between 2 weeks and 1 month, after which the experiment was discontinued. In the field, larvae were observed crawling or resting on and under submerged rocks in the middle of streams more than 3 to 4 m from the nearest emergent rock or shore. On the basis of observed crawl rates this would indicate several days of directed movement before such larvae could emerge. In the laboratory, hundreds of these aquatic larvae were raised to adulthood in dry Petri dishes with no water. Additionally, larvae found in terrestrial environments were placed in the same aquaria with the amphibious larvae. These larvae drowned and were unable to perambulate, feed, or breathe under water.
Molecular Work and Phylogenetic Analyses.
We extracted DNA from 216 adult moth specimens, followed by amplification and sequencing of the three concerned genes for a total of 2,243 bp, according to published protocols (14). The samples included 12 aquatic moth species and 77 terrestrial species. The final data set consists of 773 bp of elongation factor 1α (EF1α), 704 bp of carbomoylphosphate synthase (CAD), and 762 bp of cytochrome oxidase I (COI). Outgroup taxa were chosen from different genera in the Cosmopteriginae, which includes the genus Hyposmocoma. Sequences were easily assembled manually, edited, and aligned with BIOEDIT 7.0.5 (19), and alignments were unambiguous. Phylogenetic analyses were performed using the programs PhyML 2.4.6 (20) for ML analysis and MrBayes 3.2 (21) for Bayesian posterior probability analysis. We also ran Garli 0.94 (22) and RAxML 7.0.4 (23) to compare with the results of PhyML. We combined all three genes, and all parameters were estimated under a general time-reversible model with proportion of invariable sites and rate variation among sites (GTR+I+Γ8) selected by Modeltest 3.04 (24) for the combined data set. Informative sites, nucleotide bias, and gamma and invariant site estimates for the whole dataset, as well as for the ingroup, are indicated in Table S1. MrBayes was run with four simultaneous Markov chains in parallel twice for 2 million generations, and the first 200,000 initial trees were discarded as burn-in. MrBayes was also set to estimate model parameters independently and simultaneously for each gene partition. We assessed confidence in tree topologies as posterior probabilities calculated for each node as implemented in MrBayes. ML analysis on the combined dataset was performed in PhyML, by applying the model of substitution selected by Modeltest and estimating all model parameters. Nonparametric bootstrap support values (25) were calculated in PhyML with 1,000 replicates. The ML analysis in Garli was executed on the combined dataset applying a GTR+I+Γ8 model and 100 nonparametric bootstrap replicates. All other parameters in the control file were kept as default. Finally, RAxML was run on the three genes both separately and combined, each partition with a GTR+I+Γ model and all model parameters estimated, executing 1,000 rapid bootstrap inferences before a thorough ML search (Fig. S4). This permitted us to check for congruence between the three different genes. Separate analyses for each gene gave tree topologies (Fig. S5) similar to the tree reconstructed from the combined dataset. Although COI was more informative within larval case lineages, EF1α and CAD gave a better resolution for the deeper nodes in the phylogeny. COI differed in relationships between larger groups to each other, but relationships within groups, including the aquatic and terrestrial species, were consistent across all genes. Furthermore, although there was enough information in the nuclear genes to reconstruct a tree independently, combining all three genes increased the support values for most of the nodes of interest. All phylogenetic analyses were run through the Bioportal web-based service platform for phylogenomic analysis at the University of Oslo, Norway.
Ancestral State Reconstruction.
Ancestral state reconstruction was performed using an ML approach to map ecological traits on the Hyposmocoma molecular phylogeny in the Mesquite software package 2.72 (26) and executed on the ML tree with branch length information (Fig. 3 and Fig. S6), assuming that a correlation exists between the rate of genetic change and the rate of ecological change (27). Aquatic and terrestrial states were categorically coded as binary characters. The proportional likelihoods of ancestral states were calculated for each node under the double-rate AsymmMK likelihood model implemented in Mesquite that allows a bias in gain vs. losses.
Molecular Clock Calculations for Dating the Divergence of Aquatic Clades.
For the molecular dating analysis, we used the Bayesian relaxed molecular clock approach as implemented in BEAST 1.5.1 (28) on a subset of the cone lineage, because of the chronologic arrangement found between the different species inside the clade and the islands they inhabit. Two internal calibration points corresponding to the geologic ages for the emergence of different islands (29) were used in the analysis as soft constraints following a standard normal distribution (30). These dates correspond to the formation of the island of Kauai (4.7 ± 0.5 mya), and of the islands of the Maui Nui complex (2.2 ± 0.5 mya). We used as priors the uncorrelated lognormal molecular clock model with a Yule process for model of speciation, whereas GTR+I+Γ8 was used to describe the substitution model. The Markov chain Monte Carlo (MCMC) was run for 10 million generations following a burn-in of 1 million generations and sampled every 1,000 generations. Two replicate runs were performed to check the convergence of the MCMC. The measures of effective sample sizes (ESS) were used to determine the Bayesian statistical significance of each parameter (ESS >200). We constrained certain clades to be monophyletic to facilitate the placement of calibration points and increase the efficiency of the MCMC search. These were clades strongly supported as such by the aforementioned analyses. Mean parameter estimates and 95% highest posterior densities were determined through analyzing the combined BEAST tree files. BEAST analyses were run through the Computational Biology Service Unit Web Computing Interface at Cornell University.
Supplementary Material
Acknowledgments
We thank W. Haines and P. Krushelnycky for critical discussion; B. Gagné, G. Kawakami, and J. Cumming, Division of Forestry and Wildlife, Department of Land and Natural Resources, the staff of the Kokee Museum, E. Gordon, R. Loh, P. Welton, National Park Service, and S. Gon, P. Bily, The Nature Conservancy, for permits and assistance in the field; C. King, J. Eiben, W. Haines, B. Holland for collecting assistance; and C. Pong, T. Tengan, D. Nitta, J. Winhall-Rice for rearing larvae. We thank C. Simon and two anonymous reviewers for comments which greatly improved the manuscript. Movie S1 was taken in cooperation with NHK, Japan, which we gratefully acknowledge. This research was supported in part by grants from the National Geographic Society's Committee for Research and Exploration, and by the State of Hawaii's U.S. Fish and Wildlife Service State Wildlife Grant (T-3-P). Additional funding was provided by the National Science Foundation Project No. DEB-0918341. P.S. was also supported in part by a Swiss National Science Foundation postdoctoral grant (PBGEA/119-332).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU560194–GU560727).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912501107/DCSupplemental.
References
- 1.Resh VH, Rosenberg DM. The Ecology of Aquatic Insects. New York: Praeger Scientific; 1984. [Google Scholar]
- 2.Plum N. Terrestrial invertebrates in flooded grassland: A literature review. Wetlands. 2005;25:721–737. [Google Scholar]
- 3.Dietz-Brantleyab SE, Taylora BE, Batzerbc DP, DeBiaseaa AE. Invertebrates that aestivates in dry basins of Carolina Bay wetlands. Wetlands. 2002;22:767–775. [Google Scholar]
- 4.Suemoto T, Kawai K, Imabayashi H. Dried-up zone as a temporal stock of chironomid larvae: Survival periods and density in a reservoir bank. Hydrobiologia. 2005;545:145–152. [Google Scholar]
- 5.Zerm M, Walenciak O, Val AL, Adis J. Evidence for anaerobic metabolism in the larval tiger beetle, Phaeoxantha klugii (Col. Cicindelidae) from a Central Amazonian floodplain (Brazil) Physiol Entomol. 2004;29:483–488. [Google Scholar]
- 6.Topps W, Ring RA. Adaptations of Coleoptera to the marine environment II. Observations on rove beetles (Staphylinidae) from rocky shores. Can J Zool. 1988;66:2469–2474. [Google Scholar]
- 7.Webb MR, Pullin AS. Effects of submergence by winter floods on diapausing caterpillars of a wetland butterfly, Lycaena dispar batavus. Ecol Entomol. 1998;23:96–99. [Google Scholar]
- 8.Rubinoff D, Haines WP. Web-spinning caterpillar stalks snails. Science. 2005;309:575. doi: 10.1126/science.1110397. [DOI] [PubMed] [Google Scholar]
- 9.Gillespie RG. Impaled prey. Nature. 1992;355:212–213. [Google Scholar]
- 10.Montgomery SL. Biogeography of the moth genus Eupithecia in Oceania and the evolution of ambush predation in Hawaiian caterpillars (Lepidoptera: Geometridae) Entomol Gen. 1982;8:27–34. [Google Scholar]
- 11.Fleischer RC, Helen FJ, Olson SL. Convergent evolution of Hawaiian and Australo-Pacific honeyeaters from distant songbird ancestors. Curr Biol. 2008;18:1927–1931. doi: 10.1016/j.cub.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 12.Mey W, Speidel W. Global diversity of moths (Lepidoptera) in freshwater. Hydrobiologia. 2008;595:521–528. [Google Scholar]
- 13.Merritt RW, Cummins KW, Berg MB. An Introduction to the Aquatic Insects of North America. Dubuque, IA: Kendall/Hunt; 2008. [Google Scholar]
- 14.Rubinoff D. Phylogeography and ecology of an endemic radiation of Hawaiian aquatic case-bearing moths (Hyposmocoma: Cosmopterigidae) Philos Trans R Soc Lond B Biol Sci. 2008;363:3459–3465. doi: 10.1098/rstb.2008.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman EC. Insects of Hawaii. Honolulu, HI: Univ Press of Hawaii; 1978. [Google Scholar]
- 16.Reding DM, Foster JT, James HF, Pratt HD, Fleischer RC. Convergent evolution of ‘creepers’ in the Hawaiian honeycreeper radiation. Biol Lett. 2009;5:221–224. doi: 10.1098/rsbl.2008.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan S, Simon C, Polhemus D. Molecular systematics and adaptive radiation of Hawaii's endemic Damselfly genus Megalagrion. Syst Biol. 2003;52:89–109. doi: 10.1080/10635150390132803. [DOI] [PubMed] [Google Scholar]
- 18.Giambelluca TW, Nullet MA, Schroeder TA. Rainfall Atlas of Hawaii. Honolulu, HI: State of Hawaii; 1986. [Google Scholar]
- 19.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 20.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck JP, Ronquist F. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 22.Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation. at Austin, TX: University of Texas; 2006. [Google Scholar]
- 23.Stamatakis A, Hoover PJ, Rougemont A. Rapid bootstrap algorithm for the RAxML Web-Servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 24.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 26.Maddison WP, Maddison DR. 2009 Mesquite: A modular system for evolutionary analysis. Version 2.72. Available at: http://mesquiteproject.org. Accessed February 1, 2010. [Google Scholar]
- 27.Omland KE. Correlated rates of molecular and morphological evolution. Evolution. 1997;51:1381–1393. doi: 10.1111/j.1558-5646.1997.tb01461.x. [DOI] [PubMed] [Google Scholar]
- 28.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price JP, Clague DA. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc R Soc Lond B Biol Sci. 2002;269:2429–2435. doi: 10.1098/rspb.2002.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho SYW. Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biol. 2007;38:409–414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.