Abstract
Centromere-associated protein-E (CENP-E) is a kinetochore-associated mitotic kinesin that is thought to function as the key receptor responsible for mitotic checkpoint signal transduction after interaction with spindle microtubules. We have identified GSK923295, an allosteric inhibitor of CENP-E kinesin motor ATPase activity, and mapped the inhibitor binding site to a region similar to that bound by loop-5 inhibitors of the kinesin KSP/Eg5. Unlike these KSP inhibitors, which block release of ADP and destabilize motor-microtubule interaction, GSK923295 inhibited release of inorganic phosphate and stabilized CENP-E motor domain interaction with microtubules. Inhibition of CENP-E motor activity in cultured cells and tumor xenografts caused failure of metaphase chromosome alignment and induced mitotic arrest, indicating that tight binding of CENP-E to microtubules is insufficient to satisfy the mitotic checkpoint. Consistent with genetic studies in mice suggesting that decreased CENP-E function can have a tumor-suppressive effect, inhibition of CENP-E induced tumor cell apoptosis and tumor regression.
Keywords: cancer, checkpoint, kinesin, inhibitor, mitosis
Centromere-associated protein-E (CENP-E; kinesin-7) is a kinetochore-associated kinesin motor protein with an essential and exclusive role in metaphase chromosome alignment and satisfaction of the mitotic checkpoint (1). CENP-E is a likely candidate to integrate the mechanics of kinetochore–microtubule interaction with the mitotic checkpoint signaling machinery responsible for restraining cell-cycle progression into anaphase. CENP-E is a large dimeric protein consisting of an N-terminal kinesin motor domain tethered to a globular C-terminal domain through an extended coiled-coil rod domain (2, 3). The C-terminal, noncatalytic region of CENP-E is not only sufficient to specify localization to kinetochores, but it also mediates interaction of CENP-E with the serine/threonine kinase BubR1, a key effector of mitotic checkpoint signaling that forms complexes with the checkpoint proteins Cdc20, Bub3, and Mad2 to inhibit the ubiquitin ligase activity of the anaphase promoting complex APC/CCDC20 (4–7). The combined interaction of CENP-E with microtubules and a key regulator of APC/CCDC20 has led to the hypothesis that CENP-E functions as the key kinetochore microtubule receptor responsible for silencing mitotic checkpoint signal transduction after capture of spindle microtubules. This hypothesis was further strengthened by the finding that CENP-E could stimulate the kinase activity of BubR1 in a microtubule-sensitive manner (8, 9). In vitro, the addition of CENP-E to BubR1 resulted in a stimulation of BubR1 kinase activity. The addition of microtubules suppressed this stimulatory activity, an effect thought to be mediated by the CENP-E kinesin motor domain. Although the importance of CENP-E interaction with BubR1 and the role of BubR1-mediated phosphorylation in mitotic checkpoint function remain unclear, CENP-E remains a prominent candidate to play a key role in mitotic checkpoint signal transduction.
Depletion of CENP-E from cultured human cells using antisense oligonucleotides or RNAi causes prolonged cell-cycle delay in mitosis that is characterized by an intact bipolar mitotic spindle with several chromosomes clustered close to either spindle pole (10, 11). Studies combining detailed light and electron microscopy, inhibitors of KSP/Eg5 and Aurora kinase, and siRNA-mediated depletion of CENP-E showed that loss of CENP-E correlated with a failure of monooriented chromosomes to congress to the spindle midzone (12). Microinjection of antibody directed against CENP-E (13, 14) and expression of N-terminal deletion mutants of CENP-E lacking 800 or more N-terminal residues, including the motor domain (9, 13), produced a similar failure of the minority of chromosomes to achieve metaphase alignment. Collectively, these studies indicate that CENP-E plays an important role in a cooperative process of chromosome congression to the metaphase plate and in mitotic checkpoint satisfaction in human cells. However, these studies do not specifically address the role of CENP-E kinesin motor function in chromosome alignment or mitotic checkpoint signaling.
Our interest in identifying an inhibitor of CENP-E motor function was stimulated not only by its essential and exclusive mitotic roles but also by the observation that partial loss of CENP-E function was associated with decreased tumor incidence in mice (15). Mice heterozygous for CENP-E exhibited decreased incidence of spontaneous liver tumors, decreased incidence of tumors induced by the carcinogen 9,10-dimethyl-1,2-benzanthracene (DMBA), and decreased incidence of tumors arising after homozygous deletion of the tumor suppressor p19ARF. The antitumor effect associated with reduced CENP-E function suggests that inhibition of CENP-E kinesin function might be an effective approach for the treatment of cancers.
We report here the identification and characterization of GSK923295, a first-in-class, specific, allosteric inhibitor of CENP-E kinesin motor function, and describe the effects of this unique mode of CENP-E inhibition on chromosome alignment, mitotic checkpoint satisfaction, and human tumor-cell viability in vitro and in vivo.
Results
Biochemical Mechanism of Action of GSK923295.
We screened a library of structurally diverse, small organic compounds for inhibition of CENP-E microtubule-stimulated ATPase activity. Medicinal chemistry optimization of initial inhibitors gave rise to a series of high affinity, stereoselective, specific and drug-like inhibitors of CENP-E with potent on-target activity against human cancer cells, culminating in the identification of GSK923295 and related compounds (Fig. 1 and Table S1) (16).
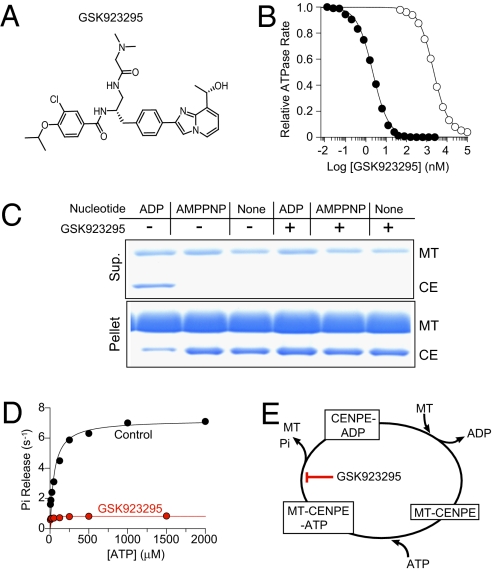
Fig. 1.
GSK923295 inhibits microtubule (MT)-stimulated ATPase of CENP-E and promotes formation of a MT-bound complex. (A) Chemical structure of GSK923295. (B) Concentration-dependent inhibition of CENP-E motor domain (1 nM) ATPase by GSK923295 in the presence (•) and absence (○) of 5 μM MT indicates a preference of GSK923295 for CENP-E-MT complex. GSK923295 inhibited CENP-E MT-stimulated ATPase with a Ki of 3.2 ± 0.2 nM in a manner uncompetitive with either MT or ATP (Table 1, Table S1, and Fig. S1). (C) Equilibrium binding and cosedimentation of CENP-E motor domain (CE; 3 μM) with MT (6 μM) in the presence or absence of the indicated nucleotides (1 mM) or in the presence or absence of 20 μM GSK923925 reveals inhibitor-induced loss of CENP-E motor domain from the supernatant and increase in the MT pellet, irrespective of nucleotide state. (Table S2). (D) Presteady-state rates of release of inorganic phosphate (Pi) from CENP-E motor domain in the presence (red) or absence (black) of GSK923295 as a function of ATP indicates that GSK923295 inhibits the production or release of Pi. (E) Summary model of the biochemical mechanism of action of GSK923295.
Using steady-state and presteady-state kinetic methodologies and equilibrium binding, we defined the biochemical potency and mechanism of GSK923295 inhibition of CENP-E motor domain (Fig. 1, Table 1, Fig. S1, and Tables S1–S3). Analysis of the rates of CENP-E ATP hydrolysis observed in the presence of varying concentrations of inhibitor and substrates [ATP and microtubules (MT)] indicated that GSK923295 is uncompetitive with both ATP and MT, inhibiting CENP-E MT-stimulated ATPase activity with a Ki of 3.2 ± 0.2 nM (Table 1, Tables S1 and S2, and Fig. S1). Among a diverse group of seven kinesins tested, GSK923295 was found to be highly selective for CENP-E (Fig. S2).
Table 1.
Sensitivity of human, murine, and canine CENP-E motor domains to GSK923295 and reduction in sensitivity of human CENP-E after mutation of I182 and T183 to corresponding murine residues
| Species | Mutation | GSK923295 Ki (nM) |
| Human | wt | 3.2 ± 0.2 |
| Canine | wt | 1.6 ± 0.1 |
| Murine | wt | 67 ± 5 |
| Human | I182L T183A | 61 ± 8 |
| Human | I182L | 14 ± 0.2 |
| Human | T183A | 13 ± 0.2 |
The greater potency of GSK923295 against microtubule-stimulated ATPase compared with basal ATPase suggested that GSK923295 exhibits a preference for CENP-E motor domain bound to MT. Typical of kinesin motor domains, the nucleotide state has a profound effect on the kinesin affinity for MT (17); nucleotide-free and ATP-bound kinesins bind MT tightly, whereas the ADP-bound forms bind MT much more weakly. We examined the interaction of CENP-E motor domain with MT in the presence and absence of GSK923295 under various nucleotide conditions by copelleting of CENP-E motor domain with MT (Fig. 1C). In the absence of GSK923295 and presence of adenosine 5′-β,γ-imido triphosphate (AMPPNP), a poorly hydrolyzable ATP analog, or in the nucleotide-free state, CENP-E motor domain bound tightly to MT. When bound to ADP, CENP-E motor domain bound loosely to MT. In contrast, the addition of GSK923295 resulted in quantitative recovery of CENP-E motor domain with the MT pellet under all nucleotide states examined. Using changes in turbidity of a solution of MT and CENP-E motor domain to monitor binding (18), we determined that GSK923295 reduced the rate of ATP-promoted dissociation of CENP-E from MT (koff, MT) by more than 50-fold (Table S2). The rate of ATP binding to CENP-E was not substantially changed. These data indicate that GSK923295 stabilizes CENP-E kinesin motor domain in a conformational state with dramatically enhanced affinity for MT.
Inhibition of the rate of MT-stimulated ATPase by GSK923295 could be caused by a slowing of any of several discrete steps in the CENP-E catalytic cycle. We measured the effects of GSK923295 on the presteady-state kinetics of release of Pi and ADP in the presence of MT using fluorescent reporters as described (19). In the absence of GSK923295, the rate of Pi release depended on concentration of ATP, and a maximum rate of release was comparable with the maximum steady-state turnover rate, suggesting that the rate-limiting step in the CENP-E catalytic cycle is release of Pi (Fig. 1D and Table S2). In the presence of GSK923295, we observed a dramatic slowing of the rate of Pi release from CENP-E (Fig. 1D and Table S2). GSK923295 exerted no effect on the rate of ATP binding to CENP-E but dramatically slowed MT-stimulated release of ADP (Table S2). Collectively, our observations indicate that GSK923295 stabilizes the ADP.Pi-bound or ATP-bound form of CENP-E, locking the motor domain in a state strongly bound to MT (Fig. 1E). Among kinesin inhibitors described to date, this mechanism is unique.
Mapping of Inhibitor Binding Site.
Efforts to cocrystallize CENP-E motor domain with GSK923295 and related inhibitors had proven unsuccessful, perhaps because of the preference of the inhibitor for MT-bound motor. To identify the region of CENP-E interacting with GSK923295, we employed photo-affinity labeling using a structurally similar inhibitor containing a photoreactive benzophenone moiety, GSK-1 (Table S1). Unlike GSK923295, GSK-1 exhibited an ATP competitive-like behavior (Table S1 and Fig. S1). Such ATP-competitive behavior had been observed during the optimization of this chemical series of CENP-E inhibitors (16). Remarkably, chemical modifications as small as a one carbon extension of a sidechain were sufficient to alter the steady-state mechanism of inhibition from ATP-uncompetitive to ATP-competitive behavior (16). Such minor changes seemed unlikely to have induced binding to a substantially different site on CENP-E motor domain; this provides reassurance that despite these differences in the steady-state mechanism of inhibition, GSK-1 probably interacted with a site on CENP-E substantially overlapping with the binding site of GSK923295.
Irradiation of CENP-E motor domain in the presence of GSK-1 and MT resulted in efficient labeling of CENP-E with GSK-1 (Fig. S3). After identification of the labeled peptide using comparative peptide mapping, we used targeted full-time electrospray ionization-liquid chromotography-tandem mass spectrometry (ESI-LC-MS/MS) to unambiguously localize the site of labeling to Met96 or Met97 (location shown in Fig. 2 and Fig. S4).
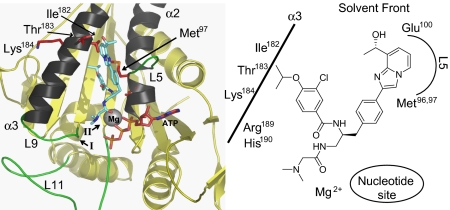
Fig. 2.
Proposed binding mode of GSK923295 to ATP-bound CENP-E motor domain GSK923295 binding site lies between helices α2 and α3, adjacent to L5 of CENP-E motor domain. GSK923295 is positioned in the center of the left figure, shown predominantly in blue. Switch I and switch II regions are indicated by roman numerals I and II. Loops L5, L9, and L11 are shown in green. Residues on helix α3 subjected to mutational analysis (I182, T183, K184) and the sites of photo-labeling with inhibitor GSK-1 (Met96 and Met97) are shown in red (Fig. S4). A simplified scheme for this proposed binding mode is shown in Right.
Comparison of the sensitivity to GSK923295 of human, murine, and canine CENP-E motor domains revealed that canine CENP-E was ∼2-fold more sensitive than human, whereas murine CENP-E was ∼20-fold less sensitive (Table 1). Thirty residues in murine CENP-E motor domain differ from both human and canine CENP-E and thus, were potential contributors to decreased sensitivity of murine CENP-E to GSK923295 (Fig. S4, black- and red-shaded residues). Three of these residues (Ile182, Thr183, and Lys184) are estimated to be within 4 Å from the site of GSK-1 photo-labeling. We mutated each of these three residues individually and in all possible combinations to the corresponding murine residue, and we measured the sensitivity of each resulting enzyme to GSK923295. Among these mutations, the combined change of Ile182 and Thr183 to the corresponding murine residues (Leu182 and Ala183) was sufficient to render the sensitivity of human CENP-E comparable with that of murine CENP-E (Table 1). All other single or double mutations shifted sensitivity ∼3–6-fold, and the triple mutant was comparably sensitive to GSK923295 as the double mutant I182L/T183A. These findings, together with GSK-1 photo-labeling results, identify the probable binding site of GSK923295 on CENP-E as sandwiched between helices α2 and α3 and adjacent to loop L5.
A proposed binding mode for GSK923295 to ATP-bound CENP-E motor domain is shown in Fig. 2. Using a CENP-E structure modeled using the available structures of ADP-bound CENP-E (20), AMPPNP-bound KSP complexed with an inhibitor closely related to the loop 5 inhibitor ispinesib, and ADP-bound KAR3R598A (21) was searched for potential favorable binding modes of GSK923295. In the model presented in Fig. 2, the benzamide moiety of GSK923295 is buried in a pocket between the central beta sheets of CENP-E, helix α3, and the base of loop L5. The N, N-dimethyl glycine amide moiety projects toward the nucleotide site, and the phenylimidazopyridinyl moiety binds between the shallow loop L5 pocket and helix α3, projecting toward the solvent front. Met96 and Met97, the residues photolabeled with GSK-1, are situated between helices α2 and α3 at the base of loop L5 and ∼10 Å away from the nucleotide binding pocket of CENP-E (Fig. 2). In this model, Ile182 forms a wall of a hydrophobic pocket where the isopropoxy group binds, and Thr183 may interact with polar functionality on the phenylimidazopyridinyl moiety. Glu100 of loop L5 may also interact with polar groups of the phenylimidazopyridinyl moiety, and the benzamide carbonyl may form a hydrogen bond with Arg189 of loop L9, a structural component involved in the γ–phosphate-sensing system of kinesin motors (22). Hydrolysis and release of γ-phosphate is associated with movement of loop L9 (23, 24), suggesting that interaction of GSK923295 with Arg189 may play an important role in inhibition of Pi release.
Cellular Response to Inhibition of CENP-E.
GSK923295 and the closely related inhibitor GSK-2 (Table S1) provided us with uniquely precise tools to investigate the role of CENP-E kinesin motor function in chromosome alignment, mitotic checkpoint satisfaction, and longer term consequences of CENP-E inhibition. Exposure of asynchronous cultured cells to GSK-2 or GSK923295 resulted in a penetrant cell-cycle delay in mitosis with a morphological phenotype very similar to that observed after antisense- or RNAi-mediated knockdown of CENP-E mRNA; this was characterized by a bipolar spindle with the majority of chromosomes positioned at the spindle midzone and several chromosomes clustered close to the spindle poles (Fig. 3 and Movie S1) (10, 11). Flow cytometry and Western blot analysis of HCC1954 breast carcinoma cells harvested at fixed times after exposure to GSK-2 revealed accumulation of cells with 4n DNA content and increases in two markers of mitosis, cyclin B, and phosphorylated histone H3 (Ser10) (Fig. 3 A and B). At longer times of exposure to GSK-2, markers of mitosis decreased as the proportion of cells with less than 2n DNA content and cleavage of poly-(ADP-Ribose) polymerase (PARP) increased (Fig. 3 A and B), indicative of apoptotic cell death.
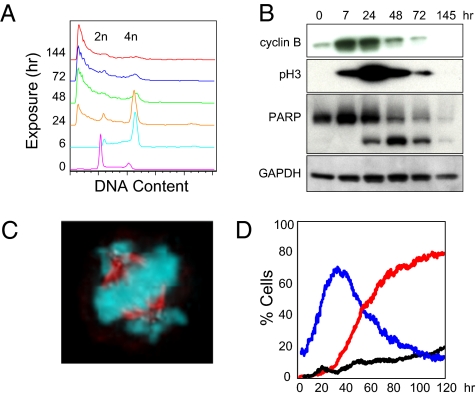
Fig. 3.
Inhibition of CENP-E motor domain ATPase causes failure to achieve metaphase chromosome alignment, mitotic cell-cycle delay, and apoptosis. (A) Flow cytometric analysis of DNA content in HCC1954 cell line (GI50 = 27 nM) at indicated times of exposure to 325 nM GSK-2 (Table S1). (B) Western blots of HCC1954 cells treated as in A are stained for mitotic markers cyclin B and phospho-histone H3 (Ser10) and for cleavage of full-length PARP (top band) to yield a PARP fragment (bottom band), a marker of apoptosis. GAPDH is included as control. (C) SKOV3 cell (GI50 = 26 nM) treated with 50 nM GSK-2 for 24 h is stained for chromatin (blue) and MT (red). Similar mitotic figures were apparent in all cell lines treated with GSK-2 or GSK923295. (D) Quantitative analysis of timelapse images of HCC1954 breast carcinoma cells stably expressing GFP-Histone H2B treated with 325 nM GSK-2. The percentage of mitotic nuclei per image in the presence of GSK-2 (blue) and the percentage of apoptotic nuclei per image in the presence (red) and absence (black) of GSK-2 (Movie S1) is shown.
Quantitative timelapse fluorescence microscopy of living HCC1954 breast carcinoma cells provided a detailed view on the kinetics of cell cycle and apoptotic response to inhibition of CENP-E motor function (Fig. 3D and Movie S1). Images of cells expressing GFP-histone-2B cultured in the presence or absence of GSK-2 were acquired every 15 min for 120 h (5 days) and subjected to image-analysis algorithms to determine the percentage of condensed, mitotic nuclei. Inclusion of propidium iodide (PI) in the culture medium allowed for simultaneous monitoring of plasma-membrane integrity; dying cells unable to exclude PI were visualized and analyzed in a manner similar to GFP–histone-labeled nuclei. Analysis of timelapse images revealed a maximal proportion of mitotic cells with spindles similar to those show in Fig. 3C after ∼32 h of exposure to CENP-E inhibitor (Fig. 3D and Movie S1). The proportion of PI-positive cells began to progressively increase from the time of maximal mitotic index until the end of the experiment (Fig. 3D). Although we have not quantitated the prevalence of individual cell fates, preliminary inspection suggests a diversity of fates, most leading to eventual apoptosis, which is consistent with previous reports detailing responses to various MT-targeted drugs and inhibitors of the mitotic kinesin KSP/Eg5 (25, 26).
These findings show a requirement for CENP-E kinesin motor ATPase function to achieve metaphase and further suggest that binding of CENP-E motor domain to MT is insufficient to satisfy the mitotic checkpoint.
Anticancer Activity of GSK923295.
To assess potential diversity in response of different tumor types to CENP-E inhibition, we assessed the growth inhibitory activity of GSK923295 across 237 tumor cell lines after 72 h of continuous exposure. The growth inhibitory activity (GI50 value) spanned more than three orders of magnitude, from 12 nM to greater than 10,000 nM, with an average GI50 of 253 nM and a median GI50 of 32 nM (Fig. 4A and Dataset S1). Two hundred twelve of 237 cell lines tested exhibited GI50 values less than 100 nM. Two cell lines, HT-3 and SNU-1, exhibited GI50 values of greater than 10,000 nM; we were unable to discern any common characteristics among these most resistant lines. Across all 237 lines, we observed no correlation between proliferation rate and GI50 value, suggesting that other, unidentified factors dominate in determining growth inhibitory effect of GSK923295.
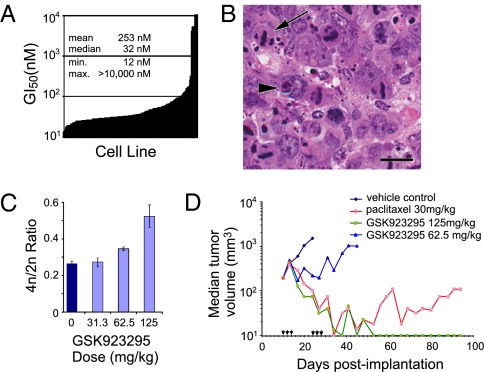
Fig. 4.
Antitumor activity of GSK923295. (A) GSK923295 is a potent inhibitor of tumor cell growth in vitro (Dataset S1). Variation in GI50 values across 237 cell lines suggests intrinsic determinants of sensitivity. (B) Representative photomicrograph of an H&E-stained section of Colo205 tumor xenografts removed 24 h after a single injection of GSK923295 (125 mg/kg). (Scale bar: 20 μm.) GSK923295 induced appearance of mitotic figures consistent with CENP-E inhibition (arrow) and scattered apoptotic bodies (arrowhead). See Fig. S5 for comparison of vehicle control and GSK923295. (C) Dose-dependent increases in the ratio of 4n to 2n nuclei present in dispersed tumor tissue from Colo205 tumor xenografts 24 h after administration of the indicated doses GSK923925. (D) Dose-dependent antitumor activity of GSK923295 administered as three daily doses on 2 consecutive weeks (arrowheads) to mice bearing xenografts of the colon carcinoma cell line Colo-205. Among five animals treated with 125 mg/kg GSK923295, four experienced partial tumor regressions (PR), and one had a complete regression (CR); 62.5 mg/kg produced tumor-growth delay and no regressions. GSK923295 failed to produce detectable host toxicity at doses as high as 500 mg/kg; the maximal tolerated dose (MTD) in mice is unknown. Paclitaxel dosed at its MTD of 30 mg/kg produced regressions in four of five mice treated (2CR and 2PR). Similar results were obtained in several other tumor xenograft models (Table S4).
To assess antitumor activity of GSK923295 in vivo, we administered inhibitor to mice bearing xenografts of the Colo205 colon tumor-cell line. GSK923295 produced clear increases in the abundance of mitotic figures and scattered apoptotic bodies in tumors that were identical in morphology to those observed in cultured cells exposed to CENP-E inhibitor (Figs. 4B and Fig. S5). We used flow cytometry of dispersed nuclei from Colo205 tumor xenografts to quantify changes in cell-cycle distribution of tumor cells in vivo, revealing a dose-dependent increase in the ratio of 4n to 2n nuclei compared with tumors from vehicle-treated animals; this is indicative of a GSK923295-induced increase in mitotic index (Fig. 4C). Longer term studies measuring tumor volume as an endpoint revealed robust, dose-dependent antitumor activity of GSK923295 against Colo205 xenografts, including partial and complete regressions at the 125 mg/kg dose (Fig. 4D). Similar results were obtained in a variety of tumor xenograft models, and tumor regressions apparent in 8 of 11 xenografts were tested (Table S4).
Discussion
We describe here the identification of potent and specific inhibitors of the MT-stimulated ATPase of CENP-E, a mitotic kinesin with a critical role in prometaphase chromosome movement and satisfaction of the mitotic checkpoint. Using a combination of photo cross-linking and mutagenesis, we have defined the inhibitor binding site on CENP-E. Our findings are consistent with binding of GSK923295 and related inhibitors to CENP-E kinesin motor domain between helices α2 and α3 and adjacent to loop L5, a region on CENP-E analogous to an inhibitor binding site on the kinesin KSP/Eg5 (27, 28). Inhibitors of KSP binding adjacent to loop L5 (e.g., monastrol and ispinesib) act by dramatically slowing release of ADP, locking KSP in a state with reduced affinity for MT (19, 27, 29). Despite binding in a location on the kinesin motor domain similar to that bound by loop 5 KSP inhibitors, GSK923295 exhibited a distinct mode of action, inhibiting release of inorganic phosphate from CENP-E motor domain and locking CENP-E in a state tightly bound to MT. GSK923295 induced tight binding of CENP-E to microtubules irrespective of nucleotide state, suggesting that inhibitor binding can cause the CENP-E motor domain to assume a conformation similar to that of an ATP-bound or nucleotide-free motor.
The specific mechanism by which GSK923295 prevents release of Pi from CENP-E could include conformational stabilization of the switch I and switch II regions and MT binding loops, thereby obstructing release of Pi. The structural components involved in the γ–phosphate-sensing system (L9, L11, switch I, and switch II) are evolutionarily conserved in the kinesin family of motors (22). Crystal structures of the kinesin family member KIF1A complexed with various transition-state nucleotide analogs revealed that rapid hydrolysis and release of γ-phosphate is associated with movement of loop L9 in switch I (23, 24). These conformational changes in loop L9 of KIF1A were accompanied by conformational changes in MT binding loops L11 and L12 in the switch II cluster, presumably causing the motor to switch from high microtubule affinity to a low-affinity binding state. The presence of these conserved structural elements in CENP-E suggests that similar changes in switch I and switch II regions are coupled with the upward movement of loops L11 and L12 during Pi release. The binding of GSK923295 between helices α2 and α3 and adjacent to loop L5 may stabilize switch I loop L9 and switch II loop L11, thereby preventing the release of Pi and forcing CENP-E to maintain a high-affinity MT binding state.
Molecular modeling of CENP-E motor domain with GSK923295 docked between helices α2 and α3 and adjacent to loop L5 suggests that the N, N-dimethyl glycine amide moiety projects toward the region of CENP-E motor domain occupied by nucleotide and Mg2+. The proximity of this region of GSK923295 to the nucleotide binding pocket provides a possible explanation for the ATP-competitive behavior of GSK-1 and other inhibitors of this chemical series that share this distinct steady-state mechanism of action (16).
Although we have not excluded the possibility that GSK923295 and related compounds might affect the function of other undetermined mitotic proteins, the general correlation of biochemical and cellular activity across this structurally related chemical series, including stereoselective inhibition of both CENP-E ATPase and cell growth, supports CENP-E as the biologically relevant target (16). These observations, coupled with the selectivity of GSK923295 for CENP-E among seven diverse kinesins tested, the similarity of phenotype to that produced by knockdown of CENP-E protein, and interaction with a structurally unique allosteric binding site, support our conclusion that CENP-E is the target responsible for GSK923295 antimitotic and antitumor activity.
The mode of inhibition of GSK923295 and GSK-2 enabled us to show not only that CENP-E kinesin ATPase is required for metaphase chromosome alignment but also that constitutive tight binding of CENP-E motor domain to MT is insufficient to satisfy the mitotic checkpoint. Our findings clearly show a requirement for CENP-E kinesin motor function in complete metaphase chromosome alignment and in satisfaction of the mitotic checkpoint in mammalian cells. Interestingly, despite continuous inhibition of CENP-E, most chromosomes seem to at least transiently achieve metaphase positioning. Although we have not identified the characteristics distinguishing those chromosomes that fail to achieve metaphase alignment from those that reach the spindle midzone, our findings are consistent with the proposed role for CENP-E in congression of monooriented chromosomes from locations near the spindle poles toward the spindle midzone along mature kinetochore fibers (12).
Similar defects in chromosome alignment have been observed on spindles formed in vitro in Xenopus egg extracts supplemented with full-length catalytically inactive CENP-E harboring a point mutation in the kinesin motor domain that results in constitutive tight binding to MT (3). In these Xenopus extracts, this mutant CENP-E was found to be localized to regions near the spindle poles. After exposure of mammalian cells to GSK-2, we observed a similar accumulation of CENP-E at broad regions near the two spindle poles. Our results indicate that although the process of chromosome alignment on spindles assembled in Xenopus egg extracts may differ from typical prometaphase congression in cultured mammalian cells, CENP-E motor function is required in both contexts.
CENP-E interaction with the BubR1 kinase has been hypothesized to be the key linkage between kinetochore–microtubule interaction and mitotic checkpoint signaling (8–10, 30, 31). Although we have not investigated the effects of CENP-E inhibitor on BubR1 kinase activity, our finding that GSK-2 and GSK923295 induce cell-cycle arrest in mitosis indicates that binding of CENP-E motor domain to MT is insufficient to satisfy the mitotic checkpoint.
We observed considerable variability in the antiproliferative and/or proapoptotic effectiveness of GSK923295 across the 237 cell lines tested in vitro and among the 11 tumor xenografts tested, which suggests the existence of intrinsic determinants of sensitivity that may prove useful in predicting tumor response to CENP-E inhibitors. Characterization of the sensitivity of a diverse group of malignant and nonmalignant breast-cancer cell lines to GSK923295 revealed that basal subtype breast-cancer cells were most sensitive, whereas nonmalignant cells are very resistant to GSK923295 (32). The details of cell cycle and apoptotic response that underlie these differences in sensitivity to GSK923295 remain unclear. Our preliminary findings are consistent with reported intra- and interline heterogeneity in response to other mitotic inhibitors (25, 26).
GSK923295 is a unique tool used to further understand the workings of the CENP-E kinesin motor domain at the atomic level, in metaphase chromosome movement, in regulation of mitotic checkpoint signal transduction, and in the viability of normal and malignant cells, and it may prove useful in the treatment of patients with cancer.
Materials and Methods
CENP-E Inhibitors.
CENP-E inhibitors were prepared as described (16).
Enzymology.
Unless otherwise specified, all methods were as described (19). Kinesin motor domains were expressed in Escherichia coli BL21(DE3) and purified as described (19, 33). CENP-E proteins included residues 2–340 with a carboxyl-terminal 6-his tag. Unless otherwise noted, all studies using MT were conducted in PEM25 buffer [25 mM PipesK+ (pH 6.8), 2 mM MgCl2, 1 mM EGTA] supplemented with 10 μM paclitaxel. The IC50 for steady-state inhibition was determined at 500 μM ATP, 5 μM MT, and 1 nM CENP-E in PEM25 buffer. Ki,app (apparent inhibitor dissociation constant) estimates of GSK923295 were extracted from the concentration-response curves with explicit correction for enzyme concentration as described (19).
CENP-E binding to MT was also assayed by an MT pelleting assay. CENP-E was mixed with MT under defined nucleotide conditions, and MT were pelleted by ultracentrifugation. The amount of CENP-E in supernatant and pellet fractions was subsequently analyzed by SDS/PAGE.
Photoaffinity Labeling.
CENP-E motor domain (5 μM) and paclitaxel-stabilized MT (10 μM) were incubated in PEM25 in the presence or absence of GSK-1 (Table S1). Photolysis was carried out for 30 min under UV light (λ = 305 nm); CENP-E motor domain was purified by SDS/PAGE and subjected to digestion with trypsin, LysC/AspN, and LysC/V8. Labeled peptide was identified by differential mapping relative to unlabeled CENP-E using LC-MS and MALDI-TOF, and the specific site of attachment was determined using LC-MS/MS sequencing.
Molecular Modeling.
Using the homology model builder in Molecular Operating Environment (MOE 2004.03), the CENP-E.ATP structure was modeled based on the available crystal structures of CENP-E.ADP (20) and the crystal structure of KSP bound to AMPPNP in complex with an inhibitor closely related to the loop 5 inhibitor ispinesib. Docking was carried out in this site with GOLD v2.1 using the standard default settings and GoldScore fitness function to search for binding modes of GSK923295 and related inhibitors (34).
Cell Culture and Growth Inhibition.
Cell-growth inhibition assays were performed by MDS in 384-well plates, and DNA content of fixed cells stained with DAPI using an Incell 1000 (GE) was analyzed. DNA content was determined 24 h after seeding (T0) and after exposure to varying concentrations of drug for an additional 72 h (T72). All T72 measurements were normalized to T0. Curves were analyzed using the XLfit curve-fitting tool to determine the concentration of GSK923295 yielding 50% growth inhibition relative to T0 and Ymax values (GI50).
Flow Cytometry.
Cultured cells treated were fixed in 85% ice-cold ethanol, stained with 10 μg/mL propidium iodide, and treated with RNase A. Tumors excised from euthanized mice 24 h after treatment were dissociated in ice-cold PBS using a 50 μM Medicon and Medimachine (BD Biosciences). DNA content of stained nuclei was analyzed using either a FACScan or FACSCalibur flow cytometer (BD Biosciences). Cell-cycle analysis of flow cytometry data from cultured cells was performed with FLOWJO (Treestar).
Fluorescence Imaging.
Cells were fixed and stained with α-tubulin antibody and with Hoechst stain and rhodamine-labeled donkey anti-rat antibody as described (33). Cells were imaged on a Deltavision model D-OL Olympus microscope (Applied Precision) using a U-plan Apo 100× Oil, 1.35 NA objective in 0.2 μM steps, which was deconvolved for 10 iterations and assembled into 2D projections.
The HCC1954 cell lines engineered to stably express a GFP fusion of Histone 2B were cultured in a 96-well plate in medium supplemented with PI at 0.2 μg/mL, and fluorescence images were acquired of GFP and PI channels every 15 min for 5 days. Image analysis was conducted with custom written software. Nuclei were identified using adaptive edge-detection algorithms. Nuclei displaying a rapid and substantial increase in GFP intensity accompanied by a decrease in size were scored as entering mitosis; the reverse was scored as exiting mitosis. Nuclei were classified as being PI positive or negative using a preset threshold applied after background subtraction.
Western Blot Analyses.
Blots of cell lysates prepared in 50 mM Tris (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 1% complete protease inhibitor mixture were resolved stained with antibodies recognizing cyclin-B and phospho-Histone H3(Ser10) (Upstate-Millipore), PARP (BD Pharmingen), and GAPDH (Santa Cruz Biotechnology). Secondary antibodies were infra-red 680/800CW Licor; signal detection and analysis were performed on a Licor-Odyssey imaging system.
Tumor Xenograft Studies and Histochemistry.
Unless otherwise noted, methods were as described (35). GSK923295 in 4% N,N-dimethylacetamide (DMA)/Cremaphore (50/50) at pH 5.6 was administered intraperitoneally in two cycles of three daily injections separated by 1 week. Paclitaxel administered iv in three doses of 30 mg/kg separated by 4 days (q4dx3) was used as a positive control. Results are reported as median tumor volume; 5-μm sections of formalin-fixed tumors were prepared and stained with H&E using standard methods. All in vivo procedures were carried out in accordance with protocols approved by the GlaxoSmithKline Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
The authors thank all members of the much larger CENP-E Program Team at Cytokinetics and GlaxoSmithKline for their contributions and also the staff at Piedmont Research Center for assistance with xenograft studies. This work was funded by GlaxoSmithKline and Cytokinetics.
Footnotes
Conflict of interest statement: All authors are current or former employees of Cytokinetics or GlaxoSmithKline and may hold stock in these companies.
See Commentary article on page 5699.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915068107/DCSupplemental.
References
- 1.Wood KW, Chua P, Sutton D, Jackson JR. Centromere-associated protein E: A motor that puts the brakes on the mitotic checkpoint. Clin Cancer Res. 2008;14:7588–7592. doi: 10.1158/1078-0432.CCR-07-4443. [DOI] [PubMed] [Google Scholar]
- 2.Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malureanu LA, et al. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 8.Weaver BA, et al. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–880. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 11.Tanudji M, et al. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol Biol Cell. 2004;15:3771–3781. doi: 10.1091/mbc.E03-07-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor TM, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaar BT, Chan GK, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwen BF, et al. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Qian X, et al. Discovery of GSK923295, the first potent and selective inhibitor of centromere-associated protein E (CENP-E) ACS Med Chem Lett. 2010 doi: 10.1021/ml900018m. ACS ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyer ML, Gilbert SP, Johnson KA. Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry. 1998;37:800–813. doi: 10.1021/bi9711184. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert SP, Webb MR, Brune M, Johnson KA. Pathway of processive ATP hydrolysis by kinesin. Nature. 1995;373:671–676. doi: 10.1038/373671a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lad L, et al. Mechanism of inhibition of human KSP by ispinesib. Biochemistry. 2008;47:3576–3585. doi: 10.1021/bi702061g. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Saez I, Yen T, Wade RH, Kozielski F. Crystal structure of the motor domain of the human kinetochore protein CENP-E. J Mol Biol. 2004;340:1107–1116. doi: 10.1016/j.jmb.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 21.Yun M, Zhang X, Park CG, Park HW, Endow SA. A structural pathway for activation of the kinesin motor ATPase. EMBO J. 2001;20:2611–2618. doi: 10.1093/emboj/20.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vale RD, Milligan RA. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 23.Kikkawa M, Okada Y, Hirokawa N. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 24.Okada Y, Hirokawa N. A processive single-headed motor: Kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 25.Orth JD, et al. Quantitative live imaging of cancer and normal cells treated with Kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol Cancer Ther. 2008;7:3480–3489. doi: 10.1158/1535-7163.MCT-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Cox CD, et al. Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP. Bioorg Med Chem Lett. 2005;15:2041–2045. doi: 10.1016/j.bmcl.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 28.Bergnes G, et al. In: Burger's Medicinal Chemistry, Drug Discovery and Development. 7th Ed. Rotella D, editor. New York: Wiley; [Google Scholar]
- 29.DeBonis S, et al. Interaction of the mitotic inhibitor monastrol with human kinesin Eg5. Biochemistry. 2003;42:338–349. doi: 10.1021/bi026716j. [DOI] [PubMed] [Google Scholar]
- 30.Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- 32.Hu Z, et al. Small molecular inhibitor of the centromere-associated protein E (CENP-E), GSK923295A inhibits cell growth in breast cancer cells. Proceedings of the 100th Annual Meeting of the American Association for Cancer Research. 2009 ; 2009 Apr 18–22; Denver, CO. Philadelphia (PA): AACR; Abstract nr. 5572. [Google Scholar]
- 33.Sakowicz R, et al. Antitumor activity of a kinesin inhibitor. Cancer Res. 2004;64:3276–3280. doi: 10.1158/0008-5472.can-03-3839. [DOI] [PubMed] [Google Scholar]
- 34.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 35.Gilmartin AG, et al. Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 2009;69:6969–6977. doi: 10.1158/0008-5472.CAN-09-0945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.