Abstract
We present a method termed mining developmentally regulated genes (MiDReG) to predict genes whose expression is either activated or repressed as precursor cells differentiate. MiDReG does not require gene expression data from intermediate stages of development. MiDReG is based on the gene expression patterns between the initial and terminal stages of the differentiation pathway, coupled with “if-then” rules (Boolean implications) mined from large-scale microarray databases. MiDReG uses two gene expression-based seed conditions that mark the initial and the terminal stages of a given differentiation pathway and combines the statistically inferred Boolean implications from these seed conditions to identify the relevant genes. The method was validated by applying it to B-cell development. The algorithm predicted 62 genes that are expressed after the KIT+ progenitor cell stage and remain expressed through CD19+ and AICDA+ germinal center B cells. qRT-PCR of 14 of these genes on sorted B-cell progenitors confirmed that the expression of 10 genes is indeed stably established during B-cell differentiation. Review of the published literature of knockout mice revealed that of the predicted genes, 63.4% have defects in B-cell differentiation and function and 22% have a role in the B cell according to other experiments, and the remaining 14.6% are not characterized. Therefore, our method identified novel gene candidates for future examination of their role in B-cell development. These data demonstrate the power of MiDReG in predicting functionally important intermediate genes in a given developmental pathway that is defined by a mutually exclusive gene expression pattern.
Keywords: B-cell differentiation, microarray, gene expression, human, mouse
Precursor cells differentiate to their terminal progeny through a series of developmental intermediates and a network of gene expression changes that gradually establish lineage commitment and the identity of the mature cell type. The identification of genes that are involved in this process has largely been dependent upon the physical isolation and characterization of gene expression patterns within these developmental intermediates. Current methods such as genetic and biochemical experiments to identify developmentally regulated genes are time-consuming, costly, and technically challenging. Array-based approaches examining differential expression between populations are expensive, require highly pure starting populations, and are narrow in scope, as only gene expression levels among the arrayed populations are compared (1–4). Thus, when intermediate steps are unknown for a particular cellular differentiation pathway, the identification of genes that are developmentally regulated in that pathway can be difficult.
In this paper, we present a bioinformatics method called mining developmentally regulated genes (MiDReG), which mines the massive repertoire of publicly available microarray data to identify a specific subset of developmentally regulated genes whose expression patterns change from either low to high or high to low significantly during the course of development. In the case of B-cell development, many important genes including KIT, CD19, and PAX5 fall in this category. MiDReG does not require that arrays of the intermediate populations exist, only the knowledge of two or more genes within a developmental pathway, of which at least one is expressed in the stem or progenitor and at least one is expressed in the mature lineage. For this study, we chose B-cell development, an already well-studied system, to exemplify and validate MiDReG, but the method is widely applicable to other developmental pathways including those that are poorly characterized.
Results
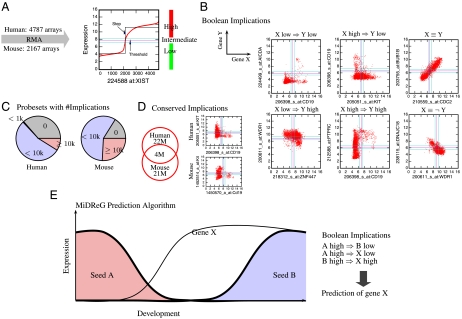
Previously, we described a method to process and analyze all publicly available microarray gene expression datasets on the Gene Expression Omnibus database, as outlined in Fig. 1A (5). Within these datasets we identified expression relationships between pairs of genes (represented by probesets on the arrays) that follow simple “if-then” rules such as “if gene A is high, then gene B is low,” or more succinctly, “A high⇒B low” (“A high implies B low”). We call these relationships “Boolean implications.” Fig. 1B outlines the six different types of Boolean implications discovered among the probesets of the human and mouse datasets. More than 60% of the probesets from either mouse or human arrays have more than one thousand Boolean implications (Fig. 1C). We checked for conservation among these Boolean implications by comparing homologous genes between the human and mouse datasets and identified 15,199 human probesets and 10,695 mouse probesets that have corresponding homologs. These human and mouse probesets contain 22 million and 21 million Boolean implications, respectively. Of those, four million Boolean implications (approximately 18%) are preserved between homologous genes of the human and mouse datasets and are thus considered “conserved” (Fig. 1D). Additionally, Boolean implications can also be extended to logical combinations of genes as described in Methods. For example, the Boolean implication “X⇒Y” can be discovered where X and Y are either single gene conditions (e.g., A high) or logical combinations of multiple genes (e.g., A high AND B high).
Fig. 1.
Computational prediction of developmental genes using Boolean implications. (A) BooleanNet algorithm on 4,787 Affymetrix U133 Plus 2.0 human microarrays and 2,167 Affymetrix 430 2.0 mouse arrays that were downloaded from NCBI’s Gene Expression Omnibus. (B) The scatter plots show six different types of Boolean implications between X and Y in human datasets. (C) The pie charts show the percentage of probesets with the indicated number of Boolean implications (0, < 1,000, < 10,000, and ≥10,000) in human and mouse datasets. More than 60% of the probesets have greater than 1,000 Boolean implications. (D) The Venn diagram shows the number of Boolean implications that are conserved across humans and mice. The mouse homologs were identified by using the euGene database: 15,199 human probesets and 10,695 mouse probesets have corresponding homologs. There are 4 M conserved Boolean implications out of 22 M in the human dataset. A conserved Boolean implication, KIT high⇒CD19 low is shown on the right. (E) MiDReG algorithm. It uses two seed genes: A, which is expressed early in development, and B, which is expressed later in the development, and identifies gene X by using Boolean implications, which is hypothesized to be expressed earlier than gene B and its expression is maintained throughout further development.
Computational Prediction of Developmental Genes Using Boolean Implications.
MiDReG uses Boolean implications to identify developmentally regulated genes. This method is based on the hypothesis that if a Boolean implication holds in sufficiently large amounts of existing data derived from a sufficient variety of different cell types, then it likely holds for cell types not represented in that data. The MiDReG algorithm requires only two seed conditions involving known developmentally regulated genes: one that holds early in development and another that holds late in development. The seed conditions can be single genes or logical combinations of genes.
For example, suppose that there are two seed genes, “A” and “B,” and that during development, gene A becomes down-regulated as gene B becomes up-regulated (Fig. 1E). Genes A and B would necessarily have the relationship “A high⇒B low” (high expression is mutually exclusive) in cells from the developmental path. The expression of these genes does not have to be restricted to the developmental pathway of interest, provided their Boolean implication holds in all other biological samples in the gene expression datasets. MiDReG searches for genes X that are expressed during development and satisfy the implications “A high⇒X low” and “B high⇒X high” (Fig. 1E), which represents the pattern of expression we expect for genes that are not expressed early in development when A is highly expressed (“A high⇒X low”) and then up-regulated later in development when B is also up-regulated (“B high⇒X high”). Because both genes A and B are developmentally regulated, the genes X that satisfy the above conditions are likely candidates for factors that become stably expressed at a developmental stage occurring after the repression of gene A but before the expression of gene B. Importantly, MiDReG does not require microarray datasets representing the developmental intermediates that exist during this transition to identify these genes. Further, to reduce false positive prediction, MiDReG focuses only on genes with conserved Boolean implications, i.e., genes that have the same Boolean implications with the seed genes in both human and mouse datasets. These conserved relationships increase the applicability of mouse results in humans.
Validation of B-Cell Precursor Genes Based on KIT and CD19.
To validate the method with an experimentally tractable developmental system, we applied it to the B-cell development pathway downstream of hematopoietic stem cells (HSCs). B-cell development is a well-characterized pathway in hematopoiesis, and methods to identify and isolate many of the intermediates are known. For the first seed gene, we chose KIT, the gene for the receptor tyrosine kinase c-kit that is expressed in HSCs in both humans and mice and whose expression is maintained in many progenitor cells within the bone marrow (6–9). For the second seed gene, we chose CD19, a membrane protein whose expression is confined exclusively to cells of the B lineage and is expressed after KIT expression is extinguished in the course of development (10, 11). Fig. 1D displays the “KIT high⇒CD19 low” implication—in other words, KIT and CD19 are very rarely coexpressed in the same sample used for microarray analysis. Whereas KIT is expressed in many different cell types unrelated to hematopoiesis and/or B-cell development, including mast cells, bone marrow stromal cells, melanocytes, interstitial cells of Cajal, and thymocyte progenitors in the thymus, or malignant tissues (12), the mutually exclusive relationship between KIT and CD19 is maintained in all the samples. This implication is also conserved between human and mouse datasets.
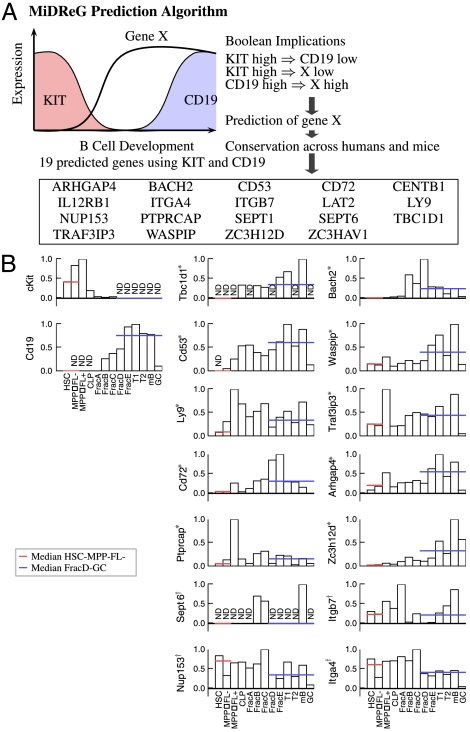
Having established a clear Boolean implication between KIT and CD19, we used MiDReG to identify such genes as shown in Fig. 2. To improve the quality of the results, the gene list was filtered by considering only those genes that are identified from these Boolean implications in both humans and mice (i.e., are conserved). The algorithm identified 19 such genes using KIT and CD19 as shown in Fig. 2A. Fig. 2A shows a schematic diagram of the known expression patterns of KIT and CD19 at sequential stages of B-cell differentiation (13). KIT is highly expressed in HSCs and multipotent progenitor (MPPFL- and MPPFL+) stages, whereas CD19 transcripts are not detected in these stages (Fig. 2B). CD19 is expressed from the fraction B (Fr.B) stage through the germinal center (GC) B-cell stage, whereas KIT transcripts are not detected from the Fr.D to the GC stages (Fig. 2B). To determine if the identified genes follow the expected expression patterns, median expression levels from HSCs to MPPFL- and Fr.D to GC stages were computed for 14 genes (see Fig. S1 for purification strategies). The expected expression levels follow a pattern in which the median level from HSC to MPPFL- is less than the median level from the Fr.D to GC stages. Strikingly, 10 out of the 14 identified genes passed this test (false discovery rate = 14.7%) (Fig. 2B). The bottom four genes are not consistent with our prediction because they are expressed in most stages of differentiation including HSC. Thus, our method has a success rate of 71% (10/14) for the prediction of genes that are developmentally regulated during B-cell differentiation.
Fig. 2.
Validation of B-cell precursor genes based on KIT and CD19. (A) B-cell precursor genes were predicted by using KIT and CD19 as seed genes. KIT is expressed early in the development, and CD19 is expressed in the mature B cell. The Boolean implication KIT high⇒CD19 low indeed reflects this situation. The identified genes turning on between KIT and CD19 are genes X such that KIT high⇒X low and CD19 high⇒X high. The list of genes is filtered by intersecting results from both human and mouse datasets. (B) The MiDReG algorithm identified 19 B-cell precursor genes by using KIT and CD19. Quantitative RT-PCR (qRT-PCR) was performed on 13 purified hematopoietic populations at different stages of B-cell differentiation: HSC, MPPFL- (multipotent progenitors Flk2-), MPPFL+ (multipotent progenitors Flk2+), CLP (common lymphoid progenitors), Frac A (Pre-Pro-B), Frac B (Pro-B), Frac C (large pre-B), Frac D (small pre-B), Frac E (immature B), T1 (Transitional 1), T2 (Transitional 2), mature B, and GC (germinal center B cells). The bar plot shows relative gene expressions from the qPCR result of 16 genes including the seed genes: KIT and CD19. The gene expressions are displayed as a percentage to the maximum gene expression level. The expression level of KIT is high, and none of the CD19 transcripts are detected from HSC to MPPFL+ stages. The expression level of CD19 is high, and none of the KIT transcripts are detected from FracD to GC stages. Therefore, for each of the 14 experimental genes the median expression level from HSC to MPPFL- stages is compared against the median expression level from FracD to GC stages. The results show that 10 out of 14 genes (indicated with *) have higher median expression levels from FracD to GC stages compared to the HSC and MPPFL- stages (FDR = 14.7%). These genes have low expression or turn off at HSC to MPPFL-; then they turn on between MPPFL+ to Frac C and are highly expressed in FracD to GC stages. The bottom four genes (indicated with †) did not pass the above test.
Validation of B-Cell Precursor Genes Based on KIT, AICDA, and CD19.
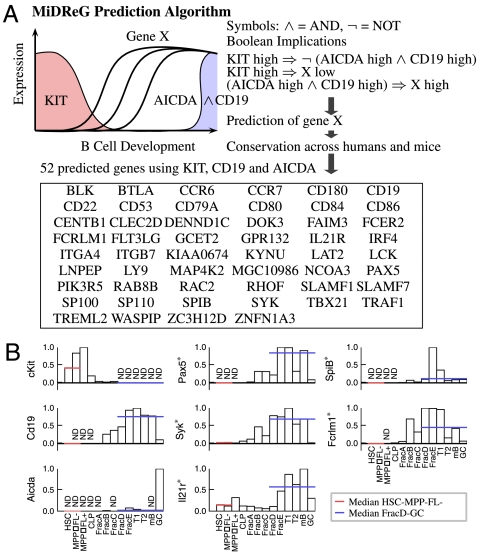
Because CD19 expression begins rapidly after KIT expression turns off, relatively few developmentally regulated genes can be identified in the intervening stages. In order to develop a more comprehensive list of B-cell precursor genes, we used the logical combination of both CD19 and AICDA expression as a seed because simultaneous high expression levels of both these genes are specific to GC B cells (14), which are developmentally downstream of mature B cells. In this case Boolean implications are derived from the logical combination of genes “(CD19 high AND AICDA high)” as described in Methods. We computed genes X such that “KIT high⇒X low” and “(CD19 high AND AICDA high)⇒X high.” The list of genes was filtered for being conserved across humans and mice, as before. There are 52 such genes, 8 of which are in common with the 19 genes identified when only CD19 was used as the (mature expressed gene) seed (Fig. 3A). Whereas it may seem counterintuitive that the addition of AICDA to MiDReG would expand the list of identified genes, this combination (CD19 AND AICDA) specifies a later stage of development than CD19 alone, and thus the number of genes that are up-regulated between KIT-expressing progenitors and AICDA-expressing GC B cells is increased. Several known genes encoding B cell-specific transcription factors were found in this list of 52 genes including Pax5 and SpiB (15, 16). SYK, which encodes a tyrosine kinase that is a critical component of B-cell receptor signaling, is also identified (17). Fig. 3B shows the qRT-PCR results of 8 genes including the 3 seed genes as controls (Kit, Cd19, and Aicda) and 5 other selected genes. The qRT-PCR results clearly show that the expression of these genes is established early during B-cell differentiation after the HSC/MPP stages and is maintained stably through the GC B cell. The only exception is that the expression level of SYK is low at the GC stage. A detailed list of the predicted B-cell genes with their Affymetrix ID in both human and mouse platform can be found in Table S1.
Fig. 3.
Validation of B-cell precursor genes based on KIT, AICDA, and CD19. (A) B-cell precursor genes were predicted by using KIT as the first seed gene and a combination of CD19 and AICDA as the second seed gene. The list of genes is filtered by using conservation across both human and mouse datasets. The combination of CD19 and AICDA expression levels are specific to a narrow region in the later stages of B-cell development, so the MiDReG algorithm is expected to return more genes than the earlier results using CD19 only. The MiDReG algorithm predicted 52 B-cell precursor genes by using KIT, CD19, and AICDA. These genes are hypothesized to be expressed after the c-kit+ progenitor cell stage and remain expressed through CD19+AICDA+ GC B cells. (B) qRT-PCR results for Pax5, Syk, Il21r, Spi-B, and Fcrlm1 are shown. The results show that all five genes indicated with * have higher median expression levels from FracD to GC stages compared to the HSC and MPPFL- stages, which suggests that the expression patterns for these genes are indeed stably maintained through GC B cells.
Classification of the Predicted B-Cell Genes.
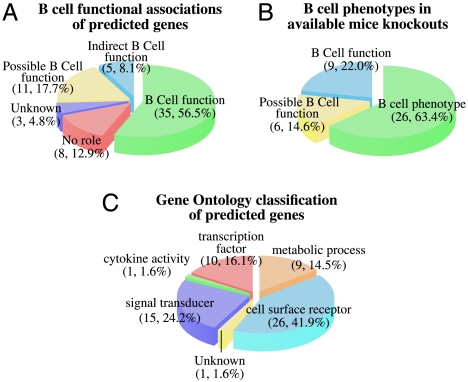
To estimate the ability of MiDReG to identify functionally significant genes, we examined the published literature for knockout phenotypes or other evidence of B-cell function among the resulting genes (Fig. 4 and Table S2). The classifications of the identified genes are described in detail in Table S3. Of the 62 genes identified by MiDReG in Figs. 2 and 3 to be up-regulated in B-cell development, 34 had published B-cell functions (56.5%), and another 5 genes had indirect connections to B-cell function (8.1%), for example. Combined, these data indicate that MiDReG had a 64.6% success rate for predicting genes with known B-cell functions (Fig. 4A). Furthermore, in 11 of the remaining genes (17.7%), we either could confirm expression in B cells (8 genes) or found knockout phenotypes or functions in other tissues and/or pathways similar to B cells [e.g., T cell trafficking, Toll-like receptor (TLR) signaling, cytoskeletal rearrangements]. We would therefore predict that if interrogated, B-cell functions would be identified for many of those genes. Last, only 3 genes were completely unstudied (4.8%), and 8 genes had no role in any pathway related to B-cell function (12.9%). Knockout mice have been published for 41 of the 62 genes, of which 26 genes (63.4%) had an identified B-cell phenotype (Fig. 4B). Also, according to the gene ontology annotations (18), these 62 genes include 26 surface receptors, 15 signal transducers, 10 transcription factors, 9 metabolic genes, 1 cytokine, and 1 unknown (Fig. 4C). Many of the genes identified by MiDReG were related to B-cell receptor signaling, as either cell surface receptors or signal transducers. There were 6 genes related to NF-κB signaling (ARHGAP4, BTLA, CENTB1, DOK3, TRAF1, TRAF3IP3, and ZC3HAV1), of which most were attenuators. There were also four members of the slam family of surface receptors (CD84, LY9, SLAMF1, and SLAMF7). Two genes (WASPIP and GCET2) are known prognostic markers of B-cell lymphomas (19–21). Of the nine genes with no obvious connection to B-cell function, three belonged to the GLUT4 glucose uptake pathway (LNPEP, RAB8B, and TBC1D1), a pathway by which muscle cells can rapidly uptake glucose in response to insulin signals. In summary, these 62 genes are highly enriched for important B-cell related activities or promising candidates for future functional studies.
Fig. 4.
Classification of the predicted B-cell genes. (A) Predicted B-cell genes are grouped according to reported B-cell functions in the literature. Out of 62 genes, 35 (56.5%) genes are associated with known B-cell function, 5 (8.1%) genes are indirectly related to the B cell through interacting proteins, 3 (4.8%) genes are unknown, 8 (12.9%) genes have other roles, and 11 (17.7%) genes could have a B-cell function based on their expression in the B cell and reported other hematopoietic functions. (B) Predicted B-cell genes with available mice knockouts are grouped according to reported B-cell phenotypes in the literature. Out of 62 genes, 41 genes have been knocked out in mice. Out of these 41 mice knockouts, 26 (63.4%) genes show defects in B-cell function and differentiation, 9 (22.0%) genes are associated with known B-cell function according to other experiments, and 6 (14.6%) genes could have a B-cell function based on their expression in the B cell and reported other hematopoietic functions. (C) Predicted B-cell genes grouped according to gene ontology classification. Out of 62 genes, 26 (41.9%) genes are cell surface receptors, 15 (24.2%) genes are associated with signal transduction, 10 (16.1%) genes are transcription factors, 9 (14.5%) genes are associated with other metabolic process, 1 (1.6%) unknown gene, and 1 (1.6%) cytokine.
Discussion
Comparison with Existing Approaches.
The identification of genes that are involved in establishing a cellular lineage can be a technically difficult process. Investigators will often take empirical approaches such as functional genetic screens or biochemical characterizations of their cell type of interest to identify the regulators of fate decisions and lineage commitment. For instance, many of the transcriptional regulators involved in B lineage commitment were first identified as factors that were bound to immunoglobulin enhancer elements in cell lines that were readily available for biochemical studies (15, 22). For myriad technical reasons, however, these types of approaches are often not feasible for the study of a variety of other important developmental processes. In particular, when few of the developmental intermediate steps are known for a particular lineage, the identification of genes involved in lineage commitment and differentiation can be extremely challenging. With the advent of microarray technology, comparisons between two or more populations could reveal developmentally regulated genes (1–4, 23). However, because the purity of the isolated populations is proportional to the quality of the data, this comparison approach faces the proverbial “chicken-and-egg” problem, because microarrays are needed to identify markers to better purify populations. Whereas this chicken-and-egg problem can be partially solved by repeated rounds of purification and microarray usage, the high cost of microarrays makes this approach somewhat prohibitive. But perhaps the greatest weakness of this approach is the narrow scope, where only closely related populations are compared. In comparisons of more distantly related populations, the number of differentially regulated candidates becomes enormous and unwieldy. However, a complete characterization of differentially regulated genes can be performed once microarrays of the intermediate populations of a developmental pathway are available. Similarly, a thorough characterization of B-cell GC genes has been performed (23). Additionally, there are other approaches to identify important genes by building regulatory networks such as relevance networks (24), the algorithm for the reconstruction of accurate cellular networks (25), Bayesian networks (26), and Inferelator (27). It is not obvious how these methods can be applied to identify genes similar to MiDReG. Our technique takes advantage of Boolean implication relationships mined from the large publicly available repositories of microarray data to identify developmentally regulated genes, even when few of the intermediate stages are known.
Advantages and Limitations of MiDReG.
Our method assumes only minimal knowledge of candidate seed genes, by using c-kit (KIT) as a gene known to be expressed in HSCs and later extinguished and CD19 as a B-cell-specific gene expressed after the c-kit+ progenitor stages. MiDReG can identify important genes in B-cell development that are conserved in humans and mice. Therefore, it opens a possibility of translating the complex mouse genetics results to humans. Many of the MiDReG-identified genes have not been analyzed for B-cell function in the literature. Some of these genes have been shown to be expressed in B cells, and they have other hematopoietic function (Table S3). A possible unexpected link between the GLUT4 pathway and B-cell function is described in detail in Table S4. Therefore, these genes are perfect candidates for future B-cell functional experiments. As a test for the power of this method in hematopoietic lineage analysis, in a companion paper MiDReG was used to identify a gene that encodes a cell surface molecule present in cells called common lymphocyte progenitors (CLPs) (28). This gene identified a subpopulation that is committed to the B lineage and is the earliest precursor yet found in that lineage, whereas the other subpopulation is capable of differentiating to T, B, natural killer (NK), and dendritic cells (DCs) (28).
The ability of MiDReG to identify markers of developmental stages in hematopoiesis, which is in many ways a paradigmatic developmental system, opens the possibility of better understanding less well-characterized developmental systems. An important advantage of MiDReG is that it uses all publicly available microarray data and it does not require additional microarrays to be performed on pure populations at the beginning, end, or intermediate stages of the developmental pathways under investigation. Moreover, the genes identified by MiDReG in B-cell development were based on a minimal number of known seed genes; adding additional seed genes can enhance the resolution by broadening or narrowing the scope, as we show elsewhere (28).
One of the important limitations of our method is that it does not identify genes that are expressed only transiently during development, such as RAG1 and RAG2, which are required for antigen receptor recombination but are shut off after productive rearrangement (29, 30). However, genes that are critically important for maintaining B-cell identity, such as Pax5, are known not to be transient (31, 32). There are also limitations in identifying conserved Boolean implications using orthologous human and mouse genes, because these are entirely based on the current annotations. The inaccuracy in annotations will most likely result in important genes missing (false negatives), because our random permutation experiment on BooleanNet shows no conserved Boolean implications. It is important to note that the reliability of the results depends entirely on the choice of the seed genes and the existence and quality of their corresponding probesets on existing microarrays. MiDReG requires at least two probesets that represent developmentally significant seed genes, and those two probesets must have a Boolean relationship. Given the quantity of Boolean implications identified for any given gene (Fig. 1C), we are confident that many developmental pathways will contain multiple seed choices. Indeed, our previous studies demonstrated that logical Boolean implications are made in other developmental systems, such as the mutually exclusive expression relationship between HoxA13 and HoxD3 (5). However, we do not know whether such conditions exist for all developmental pathways. Thus, the applicability of MiDReG to any developmental pathway should be approached on a case-by-case basis.
As a tool for gene discovery, MiDReG can complement existing array-based methods by independently identifying candidate genes. As we show for B-cell development, over half of the genes predicted by MiDReG are known to be functionally relevant to B-cell biology. Provided that two seed conditions exist, MiDReG may be able to predict pathway-related genes not only on the differentiation pathways from stem cells to mature cells as we describe here, but also on developmental pathways from pluripotent stem cells to specific tissue or on disease pathways according to malignancy stages or chronic to acute phases. We feel that MiDReG can serve as a useful addition to the toolbox of developmental biologists searching for developmentally regulated genes.
Methods
Data Collection and Preprocessing.
Raw data files (“.cel” files) for 4,787 Affymetrix U133 Plus 2.0 human microarrays and 2,167 Affymetrix 430 2.0 mouse arrays were downloaded from Gene Expression Omnibus (33). These array types were chosen because they are widely used and because results from different arrays can be compared more reasonably than results from two-channel arrays. The datasets were normalized and probeset level expressions were generated by using the standard robust multichip average algorithm (Fig. 1A) (34). The human U133 Plus 2.0 platform has 54,677 probesets, and the mouse 430 2.0 platform has 45,101 probesets. Boolean implications between pairs of probesets were extracted from these data (5). A database of all Boolean implications was created for each platform.
MiDReG Algorithm Using Two Seed Conditions.
A seed condition is described by using either a single gene logical condition or logical combinations of multiple genes. For single gene seed conditions A high and B high, we first check if there is a Boolean implication A high⇒B low between genes A and B. Then, the algorithm identifies all genes C such that A high⇒C low and B high⇒C high by intersecting the list of genes that have high-low implication with A (e.g., A high⇒C low) and high-high implication with B (e.g., B high⇒C high). Optionally, the algorithm filters the candidate C genes by insisting that the implications are conserved across humans and mice. For the conservation analysis, the probesets in U133 Plus 2.0 and mouse 430 2.0 were matched by using the ortholog functional annotation file from the Affymetrix web site.
When logical combinations of multiple genes are used in the seed conditions, the algorithm computes Boolean implications from those seed conditions to all the probesets in the datasets. In order to check if a Boolean implication “seed A⇒X low” is significant, BooleanNet computes the number of arrays satisfying the following four conditions: (seed A, X low), (seed A, X high), (negation of seed A, X low), and (negation of seed A, X high). Then it checks if (seed A, X high) is sparse by using the statistic described in the BooleanNet paper (5). For the identification of up-regulated genes, MiDReG searches for all genes C such that seed A⇒C low and seed B⇒C high. Similarly, for the identification of down-regulated genes, MiDReG computes all genes C such that seed A⇒C high and seed B⇒C low.
Validation of MiDReG.
The candidate B-cell precursor genes were identified by the MiDReG algorithm by using KIT and CD19 as seed genes. These genes are low when KIT is high and high when CD19 is high. Therefore, these genes are hypothesized to turn on after KIT turns off and before CD19 turns on. To identify more genes, a similar experiment is repeated by using KIT as one of the seed genes and by using high expression levels of both AICDA and CD19 as the other seed. In this case a virtual gene is created whose expression level is high when both AICDA and CD19 expression levels are high. Boolean implications between this virtual gene and other genes are computed in the same way as before (5). The identified genes here are supposed to be expressed after the c-Kit+ progenitor cell stage and maintained through CD19+AICDA+ GC B cells.
Statistical Tests for the Validation of B-Cell Precursor Genes.
The qRT-PCR data are arranged as genes in the rows and 13 different stages of B-cell development with three replicates each in the columns. The median expression level from HSC to MPPFL- stages is compared against the median expression level from FracD to GC stages. The test is successful if the median expression level from FracD to GC stages is higher than the median expression level from HSC and MPPFL- stages. The columns are then permuted randomly 100,000 times while keeping the correlation between genes the same. The percentage of times these random tests exceeds the original number of successes is recorded as the false discovery rate.
Animals.
All animal procedures were approved by the International Animal Care and Use Committee and the Stanford Administrative Panel on Laboratory Animal Care. C57Bl/Ka-Thy1.1 mice were derived and maintained at Stanford University. Bone marrow and spleen cells were obtained from mice aged 10–12 weeks.
Antibodies.
A complete list of all antibodies used in the study is shown in Table S5.
Fluorescence-Activated Cell Sorting.
All cells were sorted and data collected on a BD FACS-Aria (Becton Dickinson). FlowJo software (TreeStar) was used for flow cytometric data analysis. HSCs, MPPs, CLPs, and Fr.A–E cells were stained and harvested from the marrow as described. T1, T2, and mature B cells were harvested from the spleen, and GC B cells were harvested from the spleens of mice immunized with 100 μg alum-precipitated 4-Hydroxy-3-nitrophenylacetyl hapten conjugated to chicken gamma globulin lysine through amide bonds (Biosearch Technologies) at 14 days postimmunization as previously described (35).
Quantitative PCR for B-Cell Precursor Validation.
Cells were sorted into TRIzol (Invitrogen Life Technologies), and RNA was isolated according to manufacturer’s instruction. cDNA was synthesized by using the Superscript III kit (Invitrogen Life Technologies) using random hexamers. Amplifications were performed by using SYBR Green (SYBR is a registered trademark of Molecular Probes, Inc.) PCR core reagents (Applied Biosystems), and transcript levels were quantified by using an ABI 7900 Sequence Detection System (Applied Biosystems). The mean ct value of triplicate reactions was normalized against the mean ct value of β-actin. Primers were used at 400 nM. A complete list of primers sequences is shown in Table S6.
Supplementary Material
Acknowledgments.
This investigation was supported by National Institutes of Health Grants 5U56CA112973 (to S.K.P.), 5R01AI047457 (to I.L.W.), and 5R01AI047458 (to I.L.W.) and a grant from Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation. D.B. was supported by a fellowship from the Cancer Research Institute (T32AI0729022) and from the National Institutes of Health (K01DK078318). J.S. was supported by a fellowship from the California Institute for Regenerative Medicine (T1-00001). M.A.I. is supported by Public Health Service Grant CA09151, awarded by the National Cancer Institute, and a fellowship from the California Institute for Regenerative Medicine (T1-00001).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913635107/DCSupplemental.
References
- 1.Lee KH, Yu DH, Lee YS. Gene expression profiling of rat cerebral cortex development using cDNA microarrays. Neurochem Res. 2008;34:1030–1038. doi: 10.1007/s11064-008-9867-6. [DOI] [PubMed] [Google Scholar]
- 2.Jochheim A, et al. Multi-stage analysis of differential gene expression in BALB/C mouse liver development by high-density microarrays. Differentiation. 2003;71:62–72. doi: 10.1046/j.1432-0436.2003.700606.x. [DOI] [PubMed] [Google Scholar]
- 3.Master SR, et al. Functional microarray analysis of mammary organogenesis reveals a developmental role in adaptive thermogenesis. Mol Endocrinol. 2002;16:1185–1203. doi: 10.1210/mend.16.6.0865. [DOI] [PubMed] [Google Scholar]
- 4.Forsberg EC, et al. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genet. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahoo D, Dill DL, Gentles AJ, Tibshirani R, Plevritis SK. Boolean implication networks derived from large scale, whole genome microarray datasets. Genome Biol. 2008;9:R157. doi: 10.1186/gb-2008-9-10-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa M, et al. Expression and function of c-kit in hemopoietic progenitor cells. J Exp Med. 1991;174:63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikuta K, Weissman IL. Evidence that hematopoietic stem cells express mouse c-kit but do not depend on steel factor for their generation. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons PJ, et al. C-kit is expressed by primitive human hematopoietic cells that give rise to colony-forming cells in stroma-dependent or cytokine-supplemented culture. Exp Hematol. 1994;22:157–165. [PubMed] [Google Scholar]
- 9.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 10.Li YS, Wasserman R, Hayakawa K, Hardy RR. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa M, ten Boekel E, Melchers F. Identification of CD19(-)B220(+)c-kit(+)Flt3/Flk-2(+)cells as early B lymphoid precursors before pre-B-I cells in juvenile mouse bone marrow. Int Immunol. 2000;12:313–324. doi: 10.1093/intimm/12.3.313. [DOI] [PubMed] [Google Scholar]
- 12.Ashman LK. The biology of stem cell factor and its receptor C-kit. Int J Biochem Cell Biol. 1999;31:1037–1051. doi: 10.1016/s1357-2725(99)00076-x. [DOI] [PubMed] [Google Scholar]
- 13.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 14.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 15.Liao F, Giannini SL, Birshtein BK. A nuclear DNA-binding protein expressed during early stages of B cell differentiation interacts with diverse segments within and 3′ of the ig H chain gene cluster. J Immunol. 1992;148:2909–2917. [PubMed] [Google Scholar]
- 16.Su GH, et al. Defective B cell receptor-mediated responses in mice lacking the ets protein, spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gold MR, Chan VW, Turck CW, DeFranco AL. Membrane ig cross-linking regulates phosphatidylinositol 3-kinase in B lymphocytes. J Immunol. 1992;148:2012–2022. [PubMed] [Google Scholar]
- 18.Ashburner M, et al. Gene ontology: Tool for the unification of biology the gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lossos IS, Alizadeh AA, Rajapaksa R, Tibshirani R, Levy R. HGAL is a novel interleukin-4-inducible gene that strongly predicts survival in diffuse large B-cell lymphoma. Blood. 2003;101:433–440. doi: 10.1182/blood-2002-06-1931. [DOI] [PubMed] [Google Scholar]
- 20.Lossos IS, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 21.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 22.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 23.Klein U, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butte AJ, Kohane IS. Mutual information relevance networks: Functional genomic clustering using pairwise entropy measurements. Pac Symp Biocomput. 2000;5:415–426. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 25.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 26.Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 27.Bonneau R, et al. The inferelator: An algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schatz DG, Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988;53:107–115. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- 30.Grawunder U, et al. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 31.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 32.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110–113. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, et al. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya D, et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.