Abstract
The circadian rhythms exhibited in the cyanobacterium Synechococcus elongatus are generated by an oscillator comprised of the proteins KaiA, KaiB, and KaiC. An external signal that commonly affects the circadian clock is light. Previously, we reported that the bacteriophytochrome-like protein CikA passes environmental signals to the oscillator by directly binding a quinone and using cellular redox state as a measure of light in this photosynthetic organism. Here, we report that KaiA also binds the quinone analog 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), and the oxidized form of DBMIB, but not its reduced form, decreases the stability of KaiA in vivo, causes multimerization in vitro, and blocks KaiA stimulation of KaiC phosphorylation, which is central to circadian oscillation. Our data suggest that KaiA directly senses environmental signals as changes in redox state and modulates the circadian clock.
Keywords: biological rhythms, DBMIB, environmental signals, pseudoreceiver, quinone
Circadian rhythms of physiological processes occur in diverse prokaryotes and eukaryotes from cyanobacteria to humans (1, 2). The biochemical nature of the endogenous clock that drives these rhythms has been described in rich detail for the cyanobacterium Synechococcus elongatus, which uses a circadian oscillator that is evolutionarily and mechanistically distinct from those of eukaryotic organisms. Three proteins that comprise an oscillator, KaiA, KaiB, and KaiC, are sufficient to establish a circadian rhythm of KaiC phosphorylation in vitro when incubated in appropriate ratios in the presence of ATP (3). KaiC has both autophosphorylation and autodephosphorylation activity (4, 5), as well as a temperature-compensated ATPase activity (6). KaiA stimulates KaiC phosphorylation (7, 8), whereas KaiB attenuates the effects of KaiA (8, 9). These interactions result in a cycle of specific phosphorylation events, and presumably different conformational states, that comprise a basic circadian oscillation (10, 11). In vivo, additional components of the clock, such as the KaiC-interacting kinase SasA (12, 13), serve as output pathways to deliver temporal information to downstream cellular processes including gene expression (14) and chromosome compaction (15); input pathways transmit environmental cues to the oscillator and synchronize it with local time.
Known members of the input pathways include the redox-active members LdpA, an FeS protein (16, 17), and CikA, a histidine protein kinase (18, 19). LdpA fine-tunes the period of circadian rhythms in the cyanobacterium; in an ldpA null mutant, the circadian period is locked into the shorter end of a range of periods between 24 and 26 h that normally varies with incident light intensity, such that the period is shorter under higher light and longer under lower light (16). The bacteriophytochrome-like protein CikA is required to reset the timing of circadian peaks—the relative phasing of the rhythm—in response to a dark pulse (18). CikA mutants also have a shortened circadian period, a reduction in amplitude of gene expression rhythms, and an altered circadian pattern of KaiC phosphorylation. CikA abundance varies inversely with light intensity, with the highest levels in the dark; in an ldpA mutant, CikA abundance is locked at its lowest level independent of light intensity, and KaiA, whose levels are not light dependent, is elevated (17, 20).
We previously used the photosynthetic electron transport inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), a water-soluble halogenated analog of native plastoquinone (PQ) (21), in experiments designed to manipulate the redox state of the PQ pool in cyanobacterial cells (21). An unexpected finding was that DBMIB decreases the stability of LdpA, CikA, and, to a lesser degree, KaiA (17). The pseudoreceiver (PsR) domain of CikA was shown to bind to DBMIB and other quinone analogs directly (20, 22). The data suggest that the cyanobacterial clock obtains information regarding the light environment indirectly, through redox changes in a CikA-bound quinone (20).
Like CikA, KaiA has a PsR domain and is destabilized in vivo in the presence of DBMIB. Here, we show that oxidized, but not reduced, DBMIB binds directly to the PsR domain of KaiA and affects the oligomeric state of KaiA and its ability to stimulate the phosphorylation of KaiC. The data suggest that KaiA itself is a redox sensor and a key component of the circadian input pathway in S. elongatus.
Results
The Oxidized Form of the PQ Analog DBMIB Affects the Stability of KaiA and CikA in Vivo.
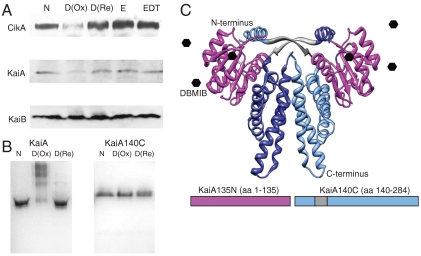
We showed previously that DBMIB decreases the abundance of LdpA, CikA, and, to a lesser degree, KaiA in vivo, but has no impact on levels of D1 (a key photosystem II protein) or PsaC (a photosystem I iron-sulfur-containing protein) (17). To investigate which redox state of DBMIB [Em = 90 mV (23)] affects the stability of KaiA and CikA, immunoblot analysis was performed using DBMIB (oxidized) and DBMIB reduced with sodium dithionite (24). Treatment of cells with oxidized DBMIB decreased the levels of KaiA and CikA, but neither protein was affected by addition of reduced DBMIB (Fig. 1A) or by the ethanol and dithionite added with DBMIB (Fig. 1A). KaiB, an internal control, was not affected by either the oxidized or reduced form of DBMIB. Although these protein levels were not affected by dithionite, this reducing agent may have other effects in the cell that were not detected by this assay.
Fig. 1.
Interaction of oxidized DBMIB with the N-terminal domain of KaiA. (A) Effects of oxidized and reduced DBMIB on the stability of CikA, KaiA, and KaiB in vivo. Cyanobacterial cells were treated for 20 min with oxidized [D(Ox)] or reduced [D(Re)] DBMIB, or with ethanol and dithionite (EDT) or ethanol (E) as controls. Soluble protein extracts were separated by SDS-PAGE and subjected to immunoblot analysis with antisera to detect CikA, KaiA, and KaiB as indicated. (B) The PsR domain of KaiA is responsible for DBMIB sensitivity. Oxidized [D(Ox)], but not reduced [D(Re)] DBMIB reduced the mobility of KaiA, but not KaiA140C, on native gels. N, no addition. (C) Structure of KaiA showing the relative positions and relevant endpoints of the N-terminal (KaiA135N) and C-terminal (KaiA140C) domains used in this study. DBMIB is depicted as a black hexagon, interacting with the N-terminal domain.
DBMIB Causes Aggregation of KaiA in Vitro That Is Dependent on the PsR Domain and DBMIB Oxidation State.
To examine the effect of DBMIB on KaiA in vitro, we performed reactions using purified KaiA incubated for 20 min with either oxidized or reduced DBMIB and subjected the samples to native polyacrylamide gel electrophoresis (PAGE). Most of the KaiA band was shifted after incubation with oxidized DBMIB, but very little was shifted when the DBMIB was reduced with dithionite (Fig. 1B). Additional experiments showed that the KaiA aggregation is noncovalent (Fig. S1) and irreversible (Fig. S2). Because quinone binding by CikA occurs in the PsR domain, we tested KaiA140C, a dimeric protein variant that is missing that domain (Fig. 1C). The migration of KaiA140C was not shifted after incubation with either oxidized or reduced DBMIB (Fig. 1B). The N-terminal PsR can be purified as a monomeric folded domain comprising the first 135 aa residues of KaiA (KaiA135N) (8). This domain also showed noncovalent aggregation upon DBMIB treatment (Fig. S1 C and D). An oxidized analog of ubiquinone (not native to Synechococcus), Q0, also caused aggregation of KaiA in vitro (Fig. S3) with some reversibility (Fig. S2). Thus, an oxidized quinone interacts with the PsR domain of KaiA and affects its migration in vitro, consistent with previous findings that the effect of DBMIB on KaiA levels in vivo is posttranslational (20).
DBMIB Binds Directly to the PsR Domain of KaiA.
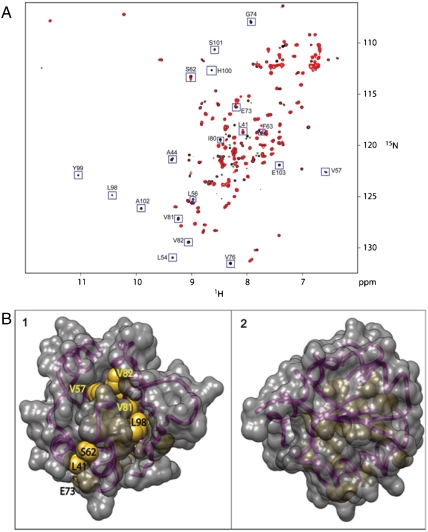
We collected two-dimensional 1H, 15N, heteronuclear single quantum correlation (HSQC) NMR spectra in the presence and absence of DBMIB to detect binding of the quinone analog to KaiA135N, controlling for small perturbations that might be induced by the ethanol solvent (0.6% vol/vol). At a 2∶1 molar ratio of KaiA135N to DBMIB significant perturbations were seen for the peaks of L41, A44, L54, L56, V57, S62, F63 E73, G74, V76, I80, V81, V82, L98-E103 and others near the crowded center of the spectrum (Fig. 2A). Mapping the perturbed residues that were assignable from the HSQC spectrum onto the NMR structure of KaiA135N revealed that the spectral perturbations are clustered together (Fig. 2B), involving sections of α2, α3, β3, β4, and preceding β5. DBMIB-induced chemical shift perturbations do not map identically onto the structures of KaiA135N and CikAPsR, suggesting that there are differences in the way DBMIB binds to these PsRs (20, 22). In contrast, a 10-fold higher concentration of DBMIB caused no changes in the peaks of CheY, Fig. S4, a canonical receiver domain (25), which was tested as a control to determine whether quinone interaction was an accidental consequence of the receiver fold. Reduced DBMIB did not affect the spectrum of KaiA135N (Fig. S5). The NMR titration results are consistent with the specific interaction of DBMIB with the PsR domain of KaiA.
Fig. 2.
Interaction of DBMIB with KaiA135N in solution. (A) 15N, 1H HSQC spectra of KaiA135N (black contours) and KaiA135N + DBMIB (red contours). The concentration of KaiA135N in both samples was 100 μM. DBMIB was added to one sample to a final concentration of 50 μM and the untreated sample was normalized with respect to ethanol concentration. Assignable peaks showing significant perturbation upon the addition of DBMIB are boxed and labeled. Asterisks denote small contaminating peaks in the spectrum. (B) Structure of KaiA135N showing the side chains of assignable residues most affected by DBMIB. (1) The surface of KaiA135N showing affected residues (labeled) not blocked from view. (2) The side of KaiA135N directly opposite to that shown in (1).
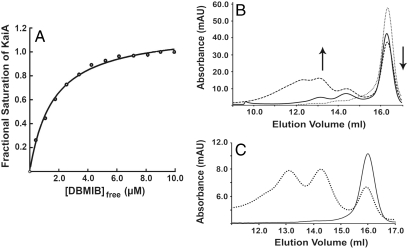
The dissociation constant for DBMIB binding to KaiA was calculated by quenching of KaiA tryptophan fluorescence at 338 nm (Fig. 3A). Nonlinear fitting of the quenching data to Eq. 2 (see Materials and Methods) indicated a Kd of oxidized DBMIB binding to KaiA of 1.8 ± 0.3 μM at a stoichiometry of approximately 1 quinone per monomer. For reduced quinone binding measurements, there was no change in the corrected fluorescence as calculated by Eq. 1 (Fig. S6). The absence of quenching suggests that reduced quinone does not bind to KaiA.
Fig. 3.
Quinone binding and KaiA aggregation state. (A) One of three equivalent trials of KaiA fluorescence quenching by titration of DBMIB. x axis, concentration of free DBMIB in solution as calculated by Eq. 4; y axis, fractional saturation of KaiA as calculated by Eq. 3. (B) Chromatogram from gel filtration analysis of KaiA135N with 0, 3, and 5 molar equivalents of oxidized DBMIB. Arrows indicate the increase in oligomeric species and the concomitant decrease in monomeric KaiA135N with increasing quinone concentration. (C) Chromatogram from gel filtration analysis of KaiA135N with 5 M equivalents of oxidized DBMIB (dotted line) and 5 M equivalents of DBMIB and sodium dithionite (solid line).
To examine the specificity of aggregation, oxidized DBMIB was added in 3 and 5 M equivalents to KaiA135N and the sample was analyzed by gel filtration. The result was an increase in the aggregation state of KaiA135N with increasing quinone concentration and a concomitant decrease in the amount of KaiA135N monomer (Fig. 3B). The aggregated material is a complex mixture with molecular weights that correspond to a trimer, a hexamer, and a nonamer for this normally monomeric domain. Reduced DBMIB added to KaiA135N at 5 M equivalents did not change the oligomeric state of KaiA135N (Fig. 3C). Thus, the PsR domain of KaiA aggregates specifically in the presence of oxidized, but not reduced quinone, suggesting a direct regulatory role for redox sensing by the circadian oscillator.
KaiC Phosphorylation Is Influenced by Oxidized Quinone.
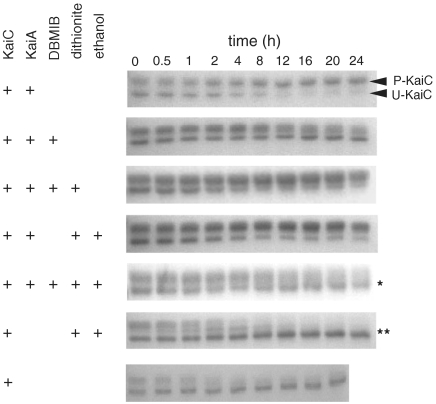
KaiC isolated from Escherichia coli is approximately 50% phosphorylated (26); over the course of 6–10 h it dephosphorylates extensively when incubated alone and phosphorylates extensively in the presence of KaiA (26). Unphosphorylated and phosphorylated forms are separable in SDS-PAGE gels, where phospho-KaiC migrates more slowly (Fig. 4). We examined the effect of oxidized and reduced DBMIB on the ability of KaiA to stimulate KaiC phosphorylation over a 24 h time course (Fig. 4). KaiC in the presence of KaiA and DBMIB increased in phosphorylation state during the first 4 h, and then KaiC started to dephosphorylate (Fig. 4). At approximately the same time, KaiC incubated with untreated KaiA, or with KaiA in the presence of both DBMIB and sodium dithionite, became increasingly phosphorylated. In the absence of DBMIB, no effect was seen on KaiC phosphorylation by addition of sodium dithionite and ethanol, which was the solvent added in trace amounts for the quinone (Fig. 4). Thus, the oxidized form of DBMIB affects KaiA stimulation of KaiC, but the reduced form of DBMIB does not. These results support the regulation of KaiC phosphorylation by redox signals through KaiA. Addition of dithionite after 12 h did not restore KaiC phosphorylation, consistent with irreversibility of aggregation (Fig. S2).
Fig. 4.
KaiC phosphorylation with KaiA in the presence of oxidized or reduced DBMIB as a function of time. KaiC was incubated with the additions indicated by a plus sign and samples were taken at the times shown across the top. SDS-PAGE was used to separate the slower migrating phosphorylated forms of KaiC (P-KaiC) from the unphosphorylated form (U-KaiC). Ethanol, used as the solvent for DBMIB, was included as a control in some reactions that lack DBMIB. *Dithionite was added after 12 h. **There was no KaiA in the reaction.
Discussion
The biological clock is typically viewed as the combined activities of a central oscillator, behavior-controlling output pathways, and synchronizing input pathways. The N-terminal PsR domain of KaiA belongs to a class of signal transduction proteins, suggesting that information impinges on the oscillator through KaiA (8). However, it was assumed that the direct sensing of light, or redox, or any other cue that acts through a cofactor, would be transmitted from an upstream partner such as LdpA and CikA. Here, we show that KaiA can sense redox signals directly. However, these data do not discount the potential roles of partner interactions in the entrainment of the clock in vivo.
Quinones are used widely as redox-active cofactors. As shown for KaiA and CikA, the ArcB (27) and RegB (28) sensor kinases from E. coli and Rhodobacter capsulatus, respectively, are inhibited by the oxidized form of exogenously added soluble quinone analogs, but not by their reduced forms. Both ArcB and RegB are integral membrane proteins. CikA and KaiA are not integral membrane proteins, yet they bind quinones, which are generally lipid-soluble electron carriers. The prospect of protein interactions at the periphery of a cellular membrane could explain this dilemma. CikA localizes to a pole of the cell, dependent on the PsR domain, consistent with peripheral membrane association (19). Soluble truncated variants of the kinases BvgS and EvgS of Bordetella pertussis and E. coli, respectively, which are missing their transmembrane spans, are inhibited specifically by oxidized Q0, a soluble analog mimic of the native ubiquinone-8 (29). Yan et al. recently showed that the crystal structure of the cytochrome b6f complex from the cyanobacterium Mastigocladus laminosus carries DBMIB (crystallized in place of PQ) in a peripheral binding site, accessible to both aqueous and membrane lipid phases, from which it likely shuttles into the Qp binding site through an intraprotein channel (30). Quinones in this interface may be accessible to hydrophobic pockets in peripheral membrane proteins.
What is the quinone that serves as a cofactor for KaiA and CikA in vivo? Photosynthetic electron transport inhibitors that modify the redox state of the PQ pool linking photosystem II and cytochrome b6f affect CikA and KaiA stability; thus, it was initially assumed that this bulk PQ pool serves as a signal (20). However, the data presented here suggest that the cofactor does not exchange with the PQ pool. Oxidized DBMIB affects KaiA stability and oligomerization, but it causes overall reduction of the PQ pool (21). Moreover, oxidation of the PQ pool by addition of the photosystem II inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (21) does not cause changes in KaiA or CikA (17, 20). Thus, it is unlikely that PQ from the bulk pool is the cofactor of these clock proteins. A tightly bound PQ is a candidate, and S. elongatus also contains a polar naphthoquinone (31). We propose that the KaiA quinone undergoes redox changes with a specific partner. CikA and LdpA, which copurify with KaiA, are good candidates for specific electron transfer partners.
KaiA is responsible for switching KaiC from an autophosphatase to an autokinase. Fig. 4 shows that oxidized DBMIB, which causes KaiA aggregation, also attenuates KaiA-mediated stimulation of KaiC kinase activity. These data support a role for a redox-active KaiA as an input mechanism (8) to synchronize the circadian oscillator with daily external cycles that affect cellular physiology.
Material and Methods
Bacterial Strains and Culture Conditions.
S. elongatus PCC 7942 strain AMC 1239 (17) was grown in 100 ml BG-11M medium (32) with 2 μg/ml of spectinomycin. The strain was cultured as described previously (33) at 30 °C under 150 μE/m2s. E. coli strain BL21(DE3) was used for KaiA and KaiB expression, and it was grown in LB medium with 200 μg/ml ampicillin at 37 °C. E. coli strain DH5α was used for KaiC expression, and it was grown in LB medium with 200 μg/ml ampicillin at 30 °C as described previously (26, 34).
Construction of the KaiA Truncation Variant and DNA Manipulation.
The gene encoding KaiA140C was amplified by PCR with Pfu polymerase and primers to create the flanking restriction sites for NcoI (forward primer) in front of the codon for KaiA residue L140 and a BamHI restriction site and the stop codon (reverse primer). After double digestion of the PCR product, the fragment was cloned into the pET32a+ vector (Novagen) and introduced into E. coli BL21(DE3) competent cells. Basic DNA manipulations were performed using standard procedures (34). Plasmid DNA was isolated using a Mini Kit (Qiagen, Inc., Chatsworth, CA) and digested by restriction enzymes from New England Biolabs, Inc. (Beverly, MA). DNA sequencing was performed by the Gene Technologies Laboratory (Texas A&M University).
Protein Expression and Purification.
Purification of polyhistidine tagged KaiA, KaiA135N, and KaiA140C from E. coli BL21(DE3)/pET32a + (kaiA), BL21(DE3)/pET32a + (kaiA135N), and BL21(DE3)/pET32a + (kaiA140C), respectively, and of glutathione-S-transferase tagged KaiC from E. coli DH5α/pGEX-6P-2(kaiC), was performed as described previously (26).
NMR Experiments.
1H, 15N HSQC spectra of KaiA135N ± DBMIB were acquired with eight scans in 39.5 min at 600 MHz. The acquisition times in both dimensions were 66(t1) and 67(t2) ms, consisting of 128 and 640 complex points, respectively, and processed such that the final digital resolutions were 4.2(F1) and 6.2(F2) Hz/pt.
Gel Filtration Analysis of Quinone Binding to KaiA135N.
Gel filtration analysis was performed using an Akta FPLC and a Superdex 200 gel filtration column from GE Healthcare. Reduced quinone gel filtration studies were performed in an M-Braun glovebox with an atmosphere that contains less than 1 ppm oxygen as monitored continuously by a Teledyne oxygen analyzer. The Superdex 200 was run with 20 mM Tris, 150 mM NaCl, 0.5 mM EDTA, and 5 mM MgCl2 buffer at pH = 8. Samples were fit to a standard curve generated from the gel filtration standard (Bio-Rad) chromatogram for molecular weight determination. Aerobic samples were prepared to contain 60 μM KaiA135N alone and with 3 and 5 M equivalents of DBMIB. For gel filtration analysis of the effects of reduced quinone binding to KaiA135N, samples contained KaiA135N with 5 M equivalents of DBMIB, with and without sodium dithionite.
Fluorescence Determination of KaiA-DBMIB Dissociation Constant (Kd).
Tryptophan quenching in KaiA by titration with DBMIB was used to determine the Kd for the complex of DBMIB-KaiA at 25 °C (35). DBMIB was mixed with KaiA for 2 min and immediately measured for fluorescence intensity at 338 nm with excitation at 280 nm. Fluorescence [FluoroMax-4 (HORIBA Jobin Yvon)] was corrected for inner filter effect using Eq. 1 and the absorbance (Agilent UV-Visible ChemStation) of the DBMIB-KaiA complex at 280(Aex) and 338(Aem) nm (35),
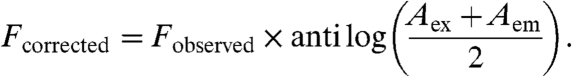 |
[1] |
The Kd for DBMIB binding was determined by nonlinear fitting (Kaleidagraph) to Eq. 2 (35),
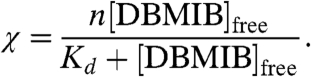 |
[2] |
Fractional saturation χ of KaiA was calculated using Eq. 3, which included corrected fluorescence with (F), without (F[DBMIB]0), and saturating (F[DBMIB] max) DBMIB concentrations (35),
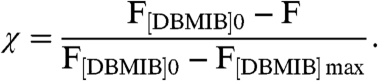 |
[3] |
Unbound DBMIB ([DBMIB]free) was determined by Eq. 4, where [DBMIB]total and [KaiA]total are the total concentrations of quinone and KaiA, corrected for dilution (35),
| [4] |
KaiC Phosphorylation Reaction.
Purified KaiA (1.5 μM) was added to a 1.5 ml centrifuge tube containing 50 μM DBMIB (Sigma-Aldrich, St. Louis, MO), autophosphorylation assay buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 5 mM ATP, pH 8.0), and/or 1 mM sodium dithionite. An equivalent volume of ethanol, the solvent for the DBMIB stock, was used instead of DBMIB as a control. After the samples were incubated at room temperature for 15 min, 3.5 μM KaiC was added into the sample. The total volume of each sample was 500 μl, and the sample was sterilized by filtration using 0.2 μm Acrodisc syringe filters (Pall, Ann Arbor, MI). The samples were incubated at 30 °C, and 40 μl aliquots were collected at each 4-h time point (0–24 h) and added to centrifuge tubes containing 6 μl of 5X loading dye. The “zero” time point sample was collected immediately after KaiC was added. The samples were analyzed using sodium dodecyl sulfate- SDS-PAGE (26).
DBMIB Treatment and Immunoblot Analysis.
To induce the expression of His-tagged LdpA in AMC 1239 strain prior to the treatment with DBMIB, 100 μM of IPTG was added to cultures for 24 h (17). DBMIB was added to cyanobacterial cultures to a final concentration of 10 μM. For the reduced DBMIB sample, excess sodium dithionite (100 μM) was added into the reaction. Since DBMIB was dissolved in ethanol, the same amount of ethanol was added to control samples. The 50 ml cyanobacterial cultures (OD750 = 0.7) were treated for 20 min, and the cells were pelleted at 1,500 × g for 10 min at 4 °C. The cell pellets were resuspended in 50 μl chilled IA lysis buffer (17). The resuspended cells were lysed by vigorous vortex mixing in the presence of glass beads in cycles of 30 s mixing and 30 s cooling on ice for 5 min. The 50 μl chilled IA buffer was added into the lysate samples. Beads were removed by a brief spin at 1,000 × g and the total sample was analyzed using SDS-PAGE.
Immunoblot analysis was performed as previously described (17, 36) except that proteins were incubated at room temperature for 1 h. Antisera were diluted with Tris buffered saline buffer with addition of 0.1% Tween 20 and 2% milk. After washing, the blots were incubated with peroxidase-conjugated goat antirabbit IgG (Calbiochem, San Diego, CA) at 1∶5,000 dilution. SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) was used for signal visualization, and the blots were exposed to X-ray film.
Native Polyacrylamide Gel Electrophoresis.
To prepare samples for native polyacrylamide gel electrophoresis, 10 μM of KaiA or KaiA140C was incubated with 100 μM DBMIB in buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, pH 8.0) for 20 min at room temperature, and then 5X loading dye was added to the samples. For the reduced DBMIB sample, 1 mM sodium dithionite was added into the reaction to keep DBMIB in the reduced form. The samples were analyzed using native PAGE (37).
Supplementary Material
ACKNOWLEDGMENTS.
We thank M. Benedik for use of an FPLC and M. Okamura and M. Paddock for helpful advice. This work was funded by the National Institutes of Health (Grants R01 GM62419 and P01 NS39546 to S.S.G.; Grant GM064576 to A.L.) and the Welch Foundation (Grant A-1647 to D.P.B.). J.B.-R. is a fellow of the NIH Chemistry Biology Interface training program 2T32GM008523. DNA sequencing was performed by the Gene Technologies Laboratory (Texas A&M University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910141107/DCSupplemental.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer Associates, Inc.; 2004. [Google Scholar]
- 2.McClung CR. The cyanobacterial circadian clock is based on the intrinsic ATPase activity of KaiC. Proc Natl Acad Sci USA. 2007;104:16727–16728. doi: 10.1073/pnas.0708757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vakonakis I, Klewer DA, Williams SB, Golden SS, LiWang AC. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J Mol Biol. 2004;342:9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 14.Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends Microbiol. 2007;15:381–388. doi: 10.1016/j.tim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama M, Kondo T, Xiong J, Golden SS. LdpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2003;185:1415–1422. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60:658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
- 20.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trebst A. Inhibitors in electron flow: Tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- 22.Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16:465–475. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prince RC, Linkletter SJG, Dutton PL. The thermodynamic properties of some commonly used oxidation reduction mediators inhibitors and dyes as determined by polarography. Biochimica et Biophysica Acta. 1981;635:132–148. doi: 10.1016/0005-2728(81)90014-1. [DOI] [PubMed] [Google Scholar]
- 24.Stidham MA, Siedow JN. Photochemical reactions of dibromothymoquinone structure and inhibitory properties of the photoproduct. Photochem Photobiol. 1983;38:537–540. [Google Scholar]
- 25.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y-I, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105:12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgellis D, Kwon O, Lin EC. Quinones as the redox signal for the Arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 28.Swem LR, Gong X, Yu CA, Bauer CE. Identification of a ubiquinone-binding site that affects autophosphorylation of the sensor kinase RegB. J Biol Chem. 2006;281:6768–6775. doi: 10.1074/jbc.M509687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bock A, Gross R. The unorthodox histidine kinases BvgS and EvgS are responsive to the oxidation status of a quinone electron carrier. Eur J Biochem. 2002;269:3479–3484. doi: 10.1046/j.1432-1033.2002.03029.x. [DOI] [PubMed] [Google Scholar]
- 30.Yan J, Kurisu G, Cramer WA. Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex. Proc Natl Acad Sci USA. 2006;103:69–74. doi: 10.1073/pnas.0504909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law A, Thomas G, Threlfall DR. 5′-Monohydroxyphylloquinone from Anacystis and Euglena. Phytochemistry. 1973;12:1999–2004. [Google Scholar]
- 32.Bustos SA, Golden SS. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278:19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Simkovic M, Frerman FE. Alternative quinone substrates and inhibitors of human electron-transfer flavoprotein-ubiquinone oxidoreductase. Biochem J. 2004;378(Pt 2):633–640. doi: 10.1042/BJ20031272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivleva NB, Golden SS. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2007. [Google Scholar]
- 37.Walker JM. Methods in Molecular Biology. Totowa, NJ: Humana Press; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.