Abstract
The genome of the fruitfly Drosophila melanogaster contains a single p53-like protein, phylogenetically related to the ancestor of the mammalian p53 family of tumor suppressors. We reasoned that a comprehensive map of the protein interaction profile of Drosophila p53 (Dmp53) might help identify conserved interactions of the entire p53 family in man. Using a genome-scale in vitro expression cloning approach, we identified 91 previously unreported Dmp53 interactors, considerably expanding the current Drosophila p53 interactome. Looking for evolutionary conservation of these interactions, we tested 41 mammalian orthologs and found that 37 bound to one or more p53-family members when overexpressed in human cells. An RNAi-based functional assay for modulation of the p53 pathway returned five positive hits, validating the biological relevance of these interactions. One p53 interactor is GTPBP4, a nucleolar protein involved in 60S ribosome biogenesis. We demonstrate that GTPBP4 knockdown induces p53 accumulation and activation in the absence of nucleolar disruption. In breast tumors with wild-type p53, increased expression of GTPBP4 correlates with reduced patient survival, emphasizing a potential relevance of this regulatory axis in cancer.
Keywords: in vitro expression cloning, p73, p63, NOG1
The p53 tumor suppressor is a transcription factor capable of sensing a wide range of stress signals and coordinating a complex response that leads to cell-cycle arrest, DNA repair, apoptosis, or senescence (1). In line with this crucial activity, development of most tumors is associated with p53 mutation (2). Because p53 activation and function involve a complex repertoire of posttranslational modifications and protein interactions (3–5), identification of p53 regulators is essential to understand cellular transformation and to identify potential drug targets for cancer therapy. When considering the p53 pathway in mammals, it is necessary to take into account two additional p53-related genes, p63 and p73, that have essential functions in embryonic development and differentiation (6, 7). Despite functional differences, all p53-family proteins play a role in genome protection and share the tumor-suppressive activity of p53 to some extent (8–10). Thus, the entire p53 family forms a complex network, whose intricacy is amplified by the fact that p53-related proteins are expressed in multiple functionally nonredundant isoforms (7, 11). Interestingly, there is a single p53-like gene in Drosophila as well as in other arthropods, nematodes, and mollusks (6, 12). By sequence alignment, invertebrate p53 is more similar to p63 than to p53 or p73. However, the single p53 in Drosophila melanogaster (Dmp53) is dispensable for normal development but fundamental for DNA damage-induced apoptosis (13, 14), in this respect being more similar to p53 than to p63 or p73. Although phylogenesis of the p53 family remains controversial, recent evidence confirmed that Dmp53 incorporates functions of multiple p53 family members (15). Thus, studying the single p53 in an invertebrate bears the potential to illuminate core properties of the network, helping us to better understand the functions of all members of the p53 family in mammals.
Results
Small Pool in Vitro Expression Cloning (IVEC) Screen for Drosophila p53 Interactors.
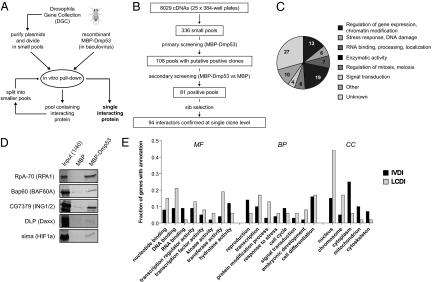
The Drosophila Gene Collection (DGC) comprises full-length annotated cDNAs of the majority of known genes in D. melanogaster (16). Clones from DGC1.0 and DGC2.0 were purified and pooled in groups of 24. Recombinant maltose binding protein (MBP)-Dmp53 fusion protein was prepared from Baculovirus-infected insect cells and used as bait for in vitro pull-down experiments with DGC pools (Fig. 1). We screened a total of 8,029 nonredundant cDNAs and identified 94 proteins that bound to MBP-Dmp53 in vitro (Fig. S1 and Table S1). At the end of the procedure, each positive hit had been repeatedly scored as Dmp53 interactant in a minimum of four independent pull-downs. One of the identified proteins was Dmp53 itself. Another clone was a putative transposon-encoded reverse transcriptase, not considered here. The remaining 92 interactors are referred to as in vitro Dmp53 interactors (IVDI). Their distribution in broad functional categories is summarized in Fig. 1C. The IVDI dataset contains five Drosophila orthologs of mammalian p53 interactors (interologs), providing proof of principle for functionality of this approach (Fig. 1D). We estimate that at least 64 additional p53 interologs were present in the screened population; these are either false negatives or proteins that do not interact with p53 in Drosophila.
Fig. 1.
Small-pool IVEC screen for Drosophila p53 interactors. (A) Schematic diagram of the small-pool Drosophila IVEC approach used to identify Dmp53 binding proteins. (B) Flow chart summarizing data relative to each step of the screening. (C) Pie chart describing 92 identified in vitro Dmp53 interactors, arbitrarily divided into functional categories. (D) Identification of expected Dmp53 interactors. Shown are in vitro pull-down experiments relative to five proteins whose human orthologs (in parentheses) are known interactors of p53. (E) Comparison of Dmp53 interactors identified in this screening (IVDI, in vitro Dmp53 interactors) with Dmp53 interactors reported in previous studies (LCDI, literature-curated Dmp53 interactors). The fractions of proteins with the indicated GO slim annotations in the IVDI (solid bars) and LCDI (shaded bars) datasets are shown. The histogram reports only the most abundant (>5%) GO slim annotations in the categories of molecular function (MF), biological process (BP), and cell compartment (CC).
The IVDI dataset was compared to known Dmp53 interactors described in the literature and protein–protein interaction databases. This list was named literature-curated Dmp53 interactors (LCDI) and comprises 48 proteins (Table S2). Notably, only one protein is common to IVDI and LCDI datasets. General features of in vitro Dmp53 interactors were determined by comparing the IVDI and LCDI datasets using Gene Ontology (GO) annotations. Whereas previous screens identified principally DNA binding and nuclear proteins, the IVEC screen enriched for RNA binding and cytoplasmic proteins (Fig. 1E). Interestingly, nine in vitro Dmp53 interactors are mitochondrial proteins. Enrichment analysis for GO terms and phenotypes revealed significant overrepresentation of genes associated with gametogenesis, in particular of the female germ line (Table S3).
Evolutionary Conservation of Dmp53 Interactions.
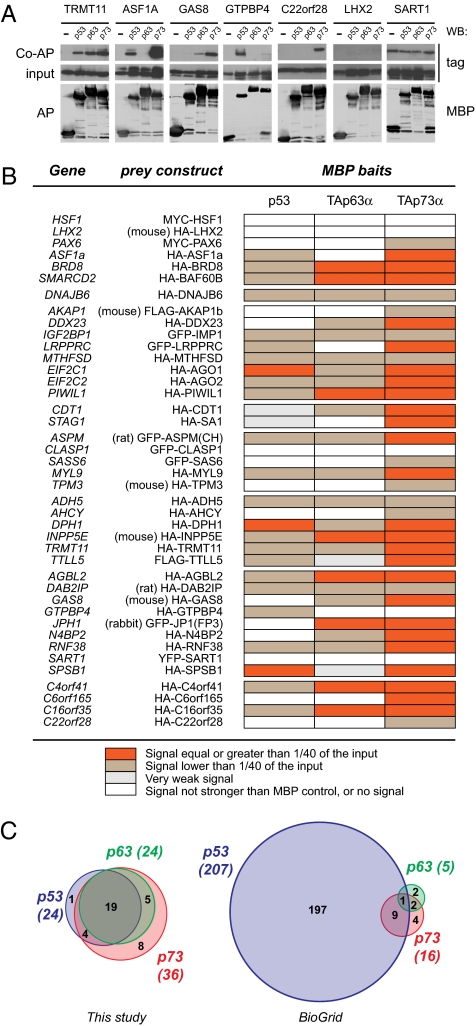
Of 92 identified Dmp53 interactors, 60 have an ortholog in Homo sapiens (Table S4). We collected epitope-tagged expression constructs for 41 such orthologs. Plasmids were transfected in human cells, and expression of encoded proteins was confirmed by immuofluorescence, revealing a variety of intracellular localizations (Fig. S2). To test interaction with human p53, TAp63α, and TAp73α, we performed coaffinity purification (co-AP) assays in 293T cells (Fig. 2 and Fig. S3). Using this assay, 37 of 41 proteins bound to one or more p53-family members. Nineteen mammalian orthologs bound to all baits, whereas the remaining 18 interactors displayed varying degrees of specificity. All but one interacted with p73, whereas the nucleolar protein GTPBP4 bound selectively to p53.
Fig. 2.
Binding of human orthologs of Dmp53 interactors to p53 and p53-related proteins. (A) Seven representative co-AP experiments. Shown are examples of preys that interacted with all three p53-family proteins (TRMT11), preys that bound to selected members of the p53 family (ASF1A, GAS8, GTPBP4, and C22orf28), and preys that were scored as not interacting (LHX2 and SART1). (Top) Tagged prey proteins after coaffinity purification. (Middle) Expression of prey in total lysates (1/40th of the input). (Bottom) MBP-tagged baits after affinity purification. The Top and Middle are cropped from the same autoradiography (i.e., have the same exposure). (B) Table summarizing the results of co-AP assays for 41 mammalian orthologs of Dmp53 interacting proteins. Expression plasmids encoding the indicated tagged proteins were cotransfected with pcDNA3-MBP, pcDNA3-MBP-p53, pcDNA3-MBP-TAp63α, or pcDNA3-MBP-TAp73α in 293T cells. MBP fusion proteins (baits) were purified on amylose beads, and copurified proteins were detected by immunoblotting. Strength of interaction was scored according to the fraction of the input prey protein that copurified with MBP baits. (C) Venn diagrams summarizing protein interactions associated with p53 family members in mammals. The results of this study are compared to current data available in protein–protein interaction databases (BioGrid release 2.0.45).
Functional Validation.
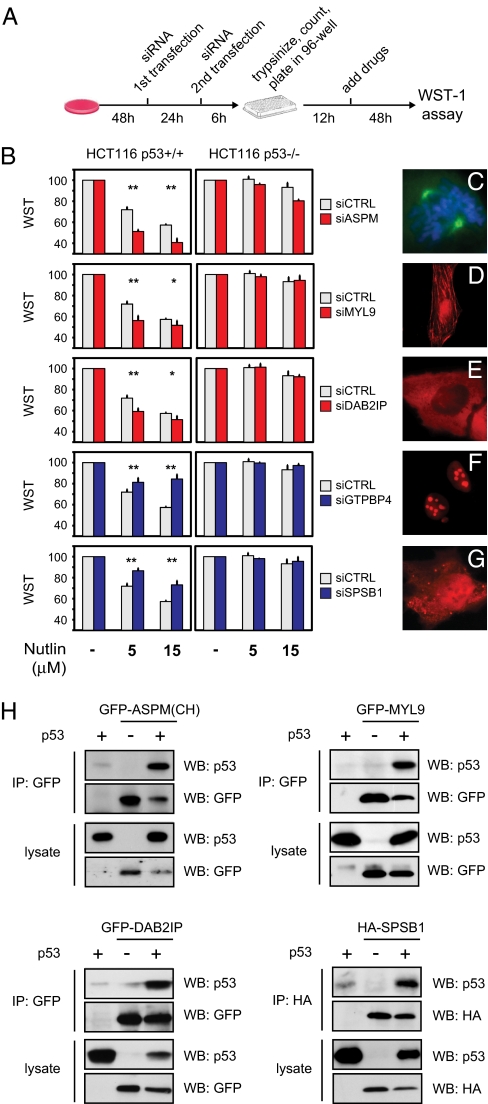
To evaluate the functional relevance of new potential interactions, we focused on p53. Human HCT116 colon cancer cells bearing wild-type p53 (HCT116 WT) were treated with Nutlin-3, a drug that prevents p53 degradation by Mdm-2, and cell viability was measured using WST-1 colorimetric assay. Under these conditions, Nutlin-3 induces a dose-dependent decrease in WST signal, indicative of growth arrest (Fig. 3). The same setup was used to analyze p53-dependent responses to DNA-damaging drugs (Fig. S4). siRNA pools targeting each of the 24 new potential p53-binding proteins were transfected and assayed in this cellular model. The p53 dependency of any effects was verified in HCT116 p53−/− cells.
Fig. 3.
Functional validation of p53 interactors by RNAi. (A) Schematic diagram of the experimental procedure. p53-dependent growth inhibition was assayed after knockdown of newly identified potential p53 interactants. After siRNA transfection, cells were trypsinized and counted, and identical numbers of cells were seeded in 96-well plates. Drugs were added 12 h later, and WST-1 activity was measured after an additional 48 h. (B) Transient knockdown of five putative p53 interactors affects the response of HCT116 WT cells to Nutlin-3. To control p53 dependency, the same experiment was done in HCT116 p53−/− cells (*, P < 0.05; **, P < 0.01; n = 3). (C–G) Cellular localization of functionally validated p53 interactors. C shows localization of endogenous ASPM in a mitotic U2OS cell, with Hoechst counterstaining for metaphase chromosomes. D–G show immunofluorescence of the indicated HA-tagged proteins transiently transfected in U2OS cells. (H) Coimmunoprecipitation of p53 with four functionally validated interactors. Expression plasmids encoding the indicated proteins were cotransfected in p53-null H1299 cells. After cell lysis, tagged proteins were immunoprecipitated using antibodies cross-linked to protein G beads and analyzed by immunoblotting. Expression of transfected proteins in the lysate (1/40th of the input) is shown in the Lower part.
As shown in Fig. 3, silencing of MYL9, DAB2IP, or ASPM resulted in more efficient growth arrest, suggesting they may be negative p53 modulators; silencing of GTPBP4 or SPSB1 resulted in less efficient growth arrest, suggesting they may be positive p53 modulators or downstream effectors. However, additional experiments revealed that GTPBP4 behaves in fact as a negative p53 modulator (see below). Expression levels of these genes, and efficiency of siRNA-mediated knockdown, were verified by RT-qPCR (Fig. S4). Physical interaction of these proteins with p53 was confirmed by coimmunoprecipitation (Figs. 3H and 4 A and B).
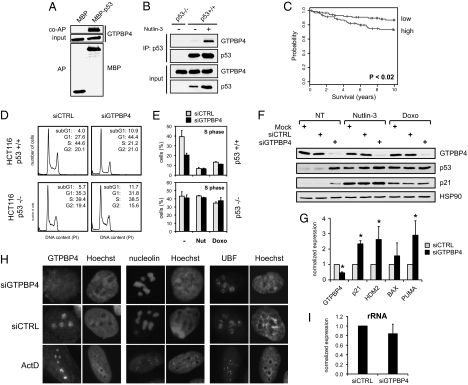
Fig. 4.
Evidence of a functional link between the nucleolar protein GTPBP4 and p53. (A) Exogenous p53 binds endogenous GTPBP4. Coaffinity purification of GTPBP4 with overexpressed MBP-p53 in U2OS cells is shown. (Top) Endogenous GTPBP4 after coaffinity purification. (Middle) Expression of GTPBP4 in total lysates (1/40th of the input). (Bottom) MBP and MBP-p53 proteins after affinity purification. (B) Interaction between endogenous GTPBP4 and p53. Coimmunoprecipitation is shown of GTPBP4 with p53 in HCT116 WT cells untreated or treated for 12 h with 5 μM Nutlin-3. (Upper) Proteins immunoprecipitated using the anti-p53 monoclonal antibody (DO-1). (Lower) Endogenous p53 and GTPBP4 proteins in the lysates (1/40th of the input). As negative control, immunoprecipitation was performed in HCT116 p53−/− cells. (C) Cumulative survival (Kaplan–Meier) curves showing that higher expression of GTPBP4 correlates with reduced survival in breast cancer bearing wild-type p53 (30). Probesets corresponding to GTPBP4 were averaged and divided into low expressers (88 samples) and high expressers (93 samples), using the median GTPBP4 expression as cutoff. The two groups display a significant difference in survival rates (P = 0.0193). (D) Transient knockdown of GTPBP4 induces p53-dependent inhibition of cell proliferation. FACS analysis is shown of HCT116 WT and HCT116 p53−/− cells transfected with control or GTPBP4 siRNA. (E) Knockdown of GTPBP4 reduces S-phase in cells bearing p53. Average S-phase fraction is shown of HCT116 WT and HCT116 p53−/− cells transfected with control or GTPBP4 siRNA and treated for 24 h with 5 μM Nutlin-3 or 0.05 μM Doxorubicin. Values are means ± SD (n = 3). (F) Knockdown of GTPBP4 induces accumulation of p53 and p21 proteins. Immunoblotting is shown of HCT116 WT cells transfected with control or GTPBP4 siRNA and left untreated (NT) or treated for 24 h with Nutlin-3 (5 μM) or Doxorubicin (0.05 μM). A specific antibody to GTPBP4 confirmed efficient knockdown of the endogenous protein. Hsp90 was detected in the same lysates as a loading control. (G) Knockdown of GTPBP4 promotes transcription of p53-target genes. RT-qPCR shows up-regulation of p53-target genes in HCT116 WT cells transfected for 48 h with GTPBP4 siRNA. mRNA expression is normalized to GAPDH. Analysis of GTPBP4 mRNA in the same samples confirms the efficiency of knockdown (*, P < 0.05; n = 3). (H) GTPBP4 knockdown does not affect localization of key nucleolar proteins. Immunofluorescence analysis is shown of GTPBP4, Nucleolin, and UBF in U2OS cells transfected for 48 h with either control or GTPBP4 siRNA. The same proteins were analyzed in U2OS cells treated with 5 nM Actinomycin D for 12 h. Nuclei are counterstained with Hoechst. (I) GTPBP4 knockdown does not affect rRNA transcription. HCT116 WT cells were transfected with control or GTPBP4 siRNA, and prerRNA levels were quantified by RT-qPCR after 48 h, using primers specific for the 5′ external transcribed spacer of pre-rRNA and normalizing for GAPDH. Values are means ± SD.
The functionally validated p53 interactors are unrelated proteins. Abnormal spindle-like primary microcephaly (ASPM) is the human ortholog of the Drosophila gene abnormal spindle. ASPM is essential for mitotic spindle function in embryonic neuroblasts, being required for proliferative division of neuroepithelial cells during brain development (17). In humans, ASPM mutation is associated with primary autosomal microcephaly (18). MYL9 is a myosin light chain that localizes to the contractile ring in mitosis, and to stress fibers in interphase, thus controlling cytokinesis and motility (19). Disabled 2 interactive protein (DAB2IP), also known as ASK-interacting protein 1 (AIP1), mediates TNF-induced activation of ASK1-JNK signaling pathways (20, 21). Interestingly, DAB2IP is methylated in a variety of tumors (22). SPSB1 belongs to a family of proteins with a central SPRY (repeats in splA and RyR) domain and a C-terminal suppressor of cytokine signaling (SOCS) box, whose molecular function is elusive (23). Drosophila SPSB1, gustavus, is required for posterior localization of polarity determinants in developing oocytes (24). GTPBP4 is the human ortholog of yeast Nog1, a monomeric GTPase that plays a crucial role in 60S ribosome biogenesis (25–27). GTPBP4 is the only p53 interactor that does not bind p63 or p73, so we further explored its relationship with p53.
Evidence of a Functional Link Between GTPBP4 and p53.
Interaction between GTPBP4 and p53 was confirmed in various conditions. Endogenous GTPBP4 was copurified with MBP-p53 in transfected U2OS cells (Fig. 4A), and endogenous GTPBP4 was coimmunoprecipitated with endogenous p53 from HCT116 WT cells treated with Nutlin-3 (Fig. 4B). Using purified proteins, we found that GTPBP4 can bind to p53 in vitro (Fig. S5). To evaluate a possible role in cancer, we analyzed public datasets to determine if GTPBP4 might have predictive value in terms of survival. Using Cox univariate analysis, we found that increased GTPBP4 expression correlates with reduced patient survival in three large breast cancer datasets: NKI (P = 0.00025), Pawitan (P = 0.00024), and Miller (P = 0.0015) (28–30). In the Miller study, where the status of p53 is known for each sample, we found that GTPBP4 levels correlate negatively with survival in tumors with wild-type p53 (Fig. 4C).
To define the functional link between GTPBP4 and p53, we analyzed the cell cycle profile of cells transfected with GTPBP4 siRNA, before and after drug treatment. As shown in Fig. 4, depletion of GTPBP4 inhibited cell proliferation, causing a marked p53-dependent reduction in the S-phase fraction. This result apparently contradicts WST experiments indicating a less efficient arrest of GTPBP4 knockdown cells (see Fig. 3B), but can be explained considering that cells depleted of GTPBP4 tend to arrest in the absence of drug treatment. Under these conditions, p53 activation does not induce a significant further drop in cell proliferation. This phenomenon is specific for GTPBP4, because FACS analysis was consistent with WST results for all other functional hits (Fig. S4). Immunoblotting revealed that silencing of GTPBP4 induces a marked increase in p53 levels and accumulation of the p53 target p21Waf1 (Fig. 4F). GTPBP4 depletion caused p53 accumulation in other cell lines, so this effect is not cell-context specific (Fig. S5). To test if p53 is also transcriptionally active, we analyzed selected p53-target genes by RT-qPCR, revealing increased mRNA levels of p21, Hdm2, and Puma (Fig. 4G).
Because nucleolar stress induces p53 stabilization and activation (31, 32), we asked if GTPBP4 knockdown might cause nucleolar dysfunction. We used RT-qPCR to analyze transcription of rRNA and found no significant alterations in pre-rRNA levels (Fig. 4I). We used immunofluorescence to study nucleolar markers; localization of Nucleolin and UBF (Fig. 4H), as well as B23/Nucleophosmin and Fibrillarin (Fig. S5), was not affected. On the contrary, treatment with actinomycin D altered localization of all markers analyzed, including GTPBP4 (Fig. 4H). These data indicate that GTPBP4 knockdown does not disrupt nucleolar structure. However, ribosomal stress can cause p53 stabilization in the absence of evident nucleolar alteration (33). We therefore measured p53 half-life after GTPBP4 knockdown in HCT116 WT cells; notably, no stabilization was detected (Fig. S5). Moreover, GTPBP4 depletion did not impair p53 accumulation after DNA damage (Fig. S5), confirming that p53 can still be stabilized in response to specific signals. Together, these data indicate that tran-sient GTPBP4 knockdown does not disrupt nucleolar structure or function, suggesting a more direct effect in regulating p53 levels.
Discussion
Taking advantage of the small size of the fly proteome, we studied the protein interaction profile of Drosophila p53 by IVEC. This approach identified many potential interactions that had escaped detection in previous studies, including high-throughput whole-genome yeast-two-hybrid screens (34, 35). This result suggests that current protein interaction databases are probably far from saturation and that exhaustive protein interaction maps can be generated only by combining multiple experimental approaches. GO term analysis of the in vitro Dmp53 interactome revealed enrichment of genes involved in gametogenesis. Because p53 mutant flies accumulate an aberrant number of primordial germ cells ectopic to the gonads (36), and p53 and p63 are involved in genoprotective apoptotic responses during mammalian gametogenesis (9, 37, 38), novel interactors may help dissect this functional role of the p53 family. Another notable feature of the in vitro Dmp53 interactome is the high representation of cytoplasmic and mitochondrial proteins. Mammalian p53 has an important transcription-independent function at mitochondria (39, 40); our results suggest that this function may be evolutionarily ancient, and we shall look for mitochondrial activities of all p53 family paralogs.
We tested binding of mammalian orthologs of Dmp53 interactors to p53, p63, and p73. Notably, 90% bound to one or more p53-family members in coaffinity purification experiments, suggesting that these interactions are evolutionarily conserved. Surprisingly, most orthologs of in vitro Dmp53 interactors bound efficiently to TAp73α. If confirmed, these data may have provocative implications for p53 phylogenesis (6, 12, 15).
We functionally validated potential p53 interactors using RNAi. Of five genes whose knockdown affected the p53 response, we focused on the nucleolar protein GTPBP4. We found that GTPBP4 knockdown induces accumulation of p53, resulting in reduced cell proliferation. Because GTPBP4/Nog1 is essential for biogenesis of the 60S ribosomal subunit (25–27), we cannot exclude that p53 accumulation after GTPBP4 siRNA is the indirect consequence of a defect in ribosome biogenesis triggering a stress pathway (31, 41, 42). However, we provide evidence that nucleolar structure and function are not altered by GTPBP4 depletion, at least in the time frame of our experiments. Importantly, we find that p53 half-life is not increased in these cells, thus indirectly excluding ribosomal stress pathways that act via inhibition of Mdm2 (31, 41, 42). A mechanistic understanding of how p53 is activated by GTPBP4 depletion will require significant additional work, but may have relevant implications; in fact, in line with the observation that high GTPBP4 expression correlates with reduced survival in breast tumors, increased levels of GTPBP4 might contribute to dampen p53 activities in cancer cells.
In conclusion, starting from an in vitro interaction screen with Drosophila proteins, we uncovered an evolutionarily conserved p53 interactor that might be relevant in cancer. This result supports the potential impact of our approach. Despite the large amount of available knowledge, we still lack an organic model of how the p53 network responds to specific signals in diverse cellular contexts, how p53-family proteins are fine tuned to achieve a wide spectrum of cellular responses, and how these are functionally integrated with other signaling pathways. Given the emerging importance of all p53 family members in tumor suppression, our list of evolutionarily conserved potential interactors may constitute a valuable platform to gain additional insights into the mammalian p53 network.
Methods
Plasmids.
Many expression vectors for mammalian orthologs of Dmp53 interactors were kindly provided by other investigators. Alternatively, full-length ESTs were purchased from I.M.A.G.E. clone distributors; the coding regions were amplified by PCR and cloned in frame with the HA tag in a modified version of the pCS2 expression vector. More detailed information on plasmids and primers is available upon request.
Antibodies, Immunoblotting, and Immunofluorescence.
For immunoblotting experiments, HCT116 cells were lysed in RIPA buffer [150 mM NaCl, 50 mM Tris·HCl (pH8), 1 mM EDTA, 1% Nonidet P-40, 0.5% Na-deoxycholate] with proteases inhibitors. Protein concentration was measured by Bradford assay (Bio-Rad). Lysates were resolved by SDS/PAGE and transferred to nitrocellulose (Millipore). Antibodies were detected by chemiluminescence (ECL; GE Healthcare). For immunofluorescence, cells were fixed 24 h posttransfection in 4% paraformaldehyde at room temperature, and permeabilized in PBS plus 0.1% Triton X-100. Proteins were visualized directly for GFP-tagged clones or using specific antibodies. Images were captured using a Leica DM4000B epifluorescence microscope. Antibodies used are mouse anti-Flag (M2; Sigma), mouse anti-Myc (9E10), mouse anti-HA (12CA5), mouse anti-HSP90 (Santa Cruz), mouse anti-p53 (DO-1; Santa Cruz), mouse anti-p21 (Santa Cruz), mouse anti-UBF1 (F-9; Santa Cruz), mouse anti-Nucleolin (Zymed), goat anti-GST (GE Healthcare), rabbit anti-GFP (self-produced), rabbit anti-MBP (self-produced), and rabbit anti-GTPBP4 (Proteintech).
Coaffinity Purifications.
Co-AP experiments were performed as described (43), with minor changes. Plasmids were transfected in human 293T cells. Forty-eight hours after transfection cells were lysed in coaffinity purification buffer [50 mM Tris·HCl (pH 8), 150 mM NaCl, 1 mM EDTA (pH 8), 1 mM DTT, 5% glycerol, protease inhibitors (Sigma), 1 mM PMSF, 5 mM NaF, 1 mM beta-glycerolphosphate] for 30 min on ice and cleared by centrifugation for 30 min at 13,000 × g at 4 °C, and protein complexes were collected on amylose beads. After extensive washes, purified complexes were separated by SDS/PAGE, detected by immunoblotting, and visualized by ECL. Purified MBP baits were visualized by DAB staining (Sigma).
Cell Viability and Proliferation Assays.
A custom library containing three siRNAs (Ambion Silencer) for each p53 interactor was purchased from Applied Biosystems. For viability assays, HCT116 colon cancer cells were plated at low density in six-well plates. Thirty-six hours postseeding, cells were transfected with siRNA pools (60 nM) using RNAiMAX reagent (Invitrogen). A second round of siRNA transfection was performed 24 h later using Lipofectamine 2000 (Invitrogen). After an additional 6 h, cells were detached, counted, and dispensed into 96-well plates. Drugs were added 12 h later, and after 48 h, cell viability was assayed using the WST-1 reagent (Roche). For FACS analysis, both adherent and floating cells were collected and fixed in ethanol. After rehydration, cells were treated with 200 μg/mL RNAseA and 40 μg/mL propidium iodide and analyzed on a flow cytometer (FacsCalibur; BD). FACS data were processed using FlowJo software, and cell cycle profiles were determined using the Watson pragmatic model (Tree Star).
Additional methods are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank L. Carlsson, C. Y. Choi, R. Crosbie, G. Denis, A. Dutta, T. Fujita, N. Galjart, P. Gonczy, T. Hollenberger, W. G. Kaelin, Jr., A. I. Lamond, S. Lin-Chao, L. Liu, J. Loturco, C. Mitchell, R. I. Morimoto, F. Cilius Nielsen, S. Ove Boe, J.-M. Peters, H. Takeshima, S. Taylor, S. Rosenbaum, R. Samant, S. Stoney-Simons, Jr., N. Watanabe, and D. Wieczorek for providing expression plasmids for many of the mammalian proteins used in this work. We thank Stefano Gustincich and members of his lab for sharing reagents and equipment. We acknowledge the important contribution of Fabio Fais and Stefania Luppi, who helped during the IVEC screening. This work was supported by grants from the Italian Association for Cancer Research, the Italian Ministry for University and Research, and Regione Friuli Venezia Giulia (LR26/2005) (to L.C.) and by grants from the Italian Association for Cancer Research, the Italian Ministry for University and Research, European Community FP6 contracts 503576 and 502983 (to G.D.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002447107/DCSupplemental.
References
- 1.Vousden KH, Lu X. Live or let die: The cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Hainaut P, et al. Database of p53 gene somatic mutations in human tumors and cell lines: Updated compilation and future prospects. Nucleic Acids Res. 1997;25:151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantovani F, et al. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol. 2007;14:912–920. doi: 10.1038/nsmb1306. [DOI] [PubMed] [Google Scholar]
- 4.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 7.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 8.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Suh EK, et al. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- 10.Rosenbluth JM, Pietenpol JA. The jury is in: p73 is a tumor suppressor after all. Genes Dev. 2008;22:2591–2595. doi: 10.1101/gad.1727408. [DOI] [PubMed] [Google Scholar]
- 11.Bourdon JC, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu W-J, Amatruda JF, Abrams JM. p53 ancestry: Gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 13.Sogame N, Kim M, Abrams JM. Drosophila p53 preserves genomic stability by regulating cell death. Proc Natl Acad Sci USA. 2003;100:4696–4701. doi: 10.1073/pnas.0736384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, et al. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 2003;550:5–10. doi: 10.1016/s0014-5793(03)00771-3. [DOI] [PubMed] [Google Scholar]
- 15.Fan Y, et al. Dual roles of Drosophila p53 in cell death and cell differentiation. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.182. 10.1038/cdd.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stapleton M, et al. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bond J, et al. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki T, Murata-Hori M, Ishitobi S, Hosoya H. Diphosphorylated MRLC is required for organization of stress fibers in interphase cells and the contractile ring in dividing cells. Cell Struct Funct. 2001;26:677–683. doi: 10.1247/csf.26.677. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, et al. AIP1/DAB2IP, a novel member of the Ras-GAP family, transduces TRAF2-induced ASK1-JNK activation. J Biol Chem. 2004;279:44955–44965. doi: 10.1074/jbc.M407617200. [DOI] [PubMed] [Google Scholar]
- 21.Min W, et al. AIP1 recruits phosphatase PP2A to ASK1 in tumor necrosis factor-induced ASK1-JNK activation. Circ Res. 2008;102:840–848. doi: 10.1161/CIRCRESAHA.107.168153. [DOI] [PubMed] [Google Scholar]
- 22.Yano M, et al. Aberrant promoter methylation of human DAB2 interactive protein (hDAB2IP) gene in lung cancers. Int J Cancer. 2005;113:59–66. doi: 10.1002/ijc.20531. [DOI] [PubMed] [Google Scholar]
- 23.Kleiber ML, Singh SM. Divergence of the vertebrate sp1A/ryanodine receptor domain and SOCS box-containing (Spsb) gene family and its expression and regulation within the mouse brain. Genomics. 2008;93:358–366. doi: 10.1016/j.ygeno.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–876. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- 25.Lapik YR, Misra JM, Lau LF, Pestov DG. Restricting conformational flexibility of the switch II region creates a dominant-inhibitory phenotype in Obg GTPase Nog1. Mol Cell Biol. 2007;27:7735–7744. doi: 10.1128/MCB.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen BC, Wang Q, Kifer CT, Parsons M. The NOG1 GTP-binding protein is required for biogenesis of the 60S ribosomal subunit. J Biol Chem. 2003;278:32204–32211. doi: 10.1074/jbc.M304198200. [DOI] [PubMed] [Google Scholar]
- 27.Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol Genet Genomics. 2007;278:105–123. doi: 10.1007/s00438-007-0233-1. [DOI] [PubMed] [Google Scholar]
- 28.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 29.Pawitan Y, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opferman JT, Zambetti GP. Translational research? Ribosome integrity and a new p53 tumor suppressor checkpoint. Cell Death Differ. 2006;13:898–901. doi: 10.1038/sj.cdd.4401923. [DOI] [PubMed] [Google Scholar]
- 33.Fumagalli S, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 35.Stanyon CA, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada Y, Davis KD, Coffman CR. Programmed cell death of primordial germ cells in Drosophila is regulated by p53 and the outsiders monocarboxylate transporter. Development. 2008;135:207–216. doi: 10.1242/dev.010389. [DOI] [PubMed] [Google Scholar]
- 37.Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency develop-mental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 38.Yin Y, Stahl BC, DeWolf WC, Morgentaler A. p53-mediated germ cell quality control in spermatogenesis. Dev Biol. 1998;204:165–171. doi: 10.1006/dbio.1998.9074. [DOI] [PubMed] [Google Scholar]
- 39.Murphy ME, Leu JI, George DL. p53 moves to mitochondria: A turn on the path to apoptosis. Cell Cycle. 2004;3:836–839. [PubMed] [Google Scholar]
- 40.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun XX, Dai MS, Lu H. 5-Fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 43.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.