Abstract
The evolution of floral zygomorphy is an important innovation in flowering plants and is thought to arise principally from specialization on various insect pollinators. Floral morphology of neotropical Malpighiaceae is distinctive and highly conserved, especially with regard to symmetry, and is thought to be caused by selection by its oil-bee pollinators. We sought to characterize the genetic basis of floral zygomorphy in Malpighiaceae by investigating CYCLOIDEA2-like (CYC2-like) genes, which are required for establishing symmetry in diverse core eudicots. We identified two copies of CYC2-like genes in Malpighiaceae, which resulted from a gene duplication in the common ancestor of the family. A likely role for these loci in the development of floral zygomorphy in Malpighiaceae is demonstrated by the conserved pattern of dorsal gene expression in two distantly related neotropical species, Byrsonima crassifolia and Janusia guaranitica. Further evidence for this function is observed in a Malpighiaceae species that has moved to the paleotropics and experienced coincident shifts in pollinators, floral symmetry, and CYC2-like gene expression. The dorsal expression pat-tern observed in Malpighiaceae contrasts dramatically with their actinomorphic-flowered relatives, Centroplacaceae (Bhesa paniculata) and Elatinaceae (Bergia texana). In particular, B. texana exhibits a previously undescribed pattern of uniform CYC2 expression, suggesting that CYC2 expression among the actinomorphic ancestors of zygomorphic lineages may be much more complex than previously thought. We consider three evolutionary models that may have given rise to this patterning, including the hypothesis that floral zygomorphy in Malpighiaceae arose earlier than standard morphology-based character reconstructions suggest.
Keywords: CYCLOIDEA, development, gene duplication, Malpighiaceae, phylogeny
Most flowers are either bilaterally symmetrical (i.e., zygomorphic) and have a single plane of symmetry or radially symmetrical (i.e., actinomorphic) and have several planes of symmetry (1). Floral zygomorphy has evolved independently at least 38 times (2–4) and is a hallmark feature of the most diverse angiosperm clades, including Asteraceae (23,600 spp.), Orchidaceae (21,950 spp.), Fabaceae (19,400 spp.), and Lamiales (23,275 spp.) (5). Plant evolutionary biologists therefore propose that the origin of floral zygomorphy may have been a key innovation for promoting speciation throughout the course of angiosperm evolution (6). The driving force behind the origin of floral zygomorphy has long been thought to be a consequence of selection by specialization on certain insect pollinators (1, 7), which has recently gained experimental support (8).
The tropical plant family Malpighiaceae exhibits a strong association between floral zygomorphy and insect pollinator attraction. The floral morphology of the more than 1,000 New World species of this clade is very distinctive and highly conserved, especially with regard to symmetry and pollinator reward. The single upright/dorsal banner petal is strongly differentiated from other petals in the corolla whorl, and appears to help orient and attract an extremely limited suite of pollinators, principally female bees of the tribes Centridini and Tapinotaspidini (Fig. 1) (9, 10–12). Furthermore, the very narrowed base of the petals provides the bees access to oil glands, which are borne in pairs on the abaxial surface of the sepals. The stereotypical floral morphology of New World Malpighiaceae, despite tremendous variation in vegetative and fruit morphology, led Anderson (9) to hypothesize that floral uniformity in the group results from their specialization on these oil-bee pollinators.
Fig. 1.
Comparative floral morphology of Malpighiaceae and their closest actinomorphic flowered relatives, Elatinaceae and Centroplacaceae. Banisteriopsis argyrophylla, Bergia texana, and Bhesa paniculata represent Malpighiaceae, Elatinaceae, and Centroplacaceae, respectively. Dotted lines indicate planes of symmetry. Note: the flower of B. texana was forced opened for illustration. The exact floral orientation of B. paniculata is unclear given the congested nature of their inflorescences.
Interpreting the origin and maintenance of this unique floral morphology in a comparative evolutionary framework, however, has remained elusive, in large part because of our lack of understanding of the closest phylogenetic relatives of Malpighiaceae. Fortunately, this problem has recently been resolved: the family is successively sister to two species-poor clades that possess actinomorphic flowers, Elatinaceae and Centroplacaceae [Fig. 1; Malpighiaceae (1,300 spp.), Elatinaceae (35 spp.), Centroplacaceae (6 spp.)] (13–16). These findings, together with morphology-based character state reconstructions of floral symmetry in the group, demonstrate that Malpighiaceae evolved from actinomorphic-flowered ancestors (Fig. S1). Moreover, these results suggest that the origin of Malpighiaceae and their unique flowers appear to correspond with a dramatic shift in speciation rates (14, 15). During the course of this radiation, there appears to have been seven subsequent dispersal events from the New World that gave rise to Old World Malpighiaceae (17–19). The Old World tropics lack the oil-bee pollinators that visit New World Malpighiaceae (ref. 20, p. 913), and these geographic transitions have resulted in alterations in both floral symmetry and petal morphology, as well as the loss of the oil gland morphology among Old World members of the family (17, 18, 21). Thus, the majority of Old World species possess flowers that are either truly actinomorphic or zygomorphic in a manner that is very divergent from the pattern exhibited by New World species. These zygomorphic Old World species possess two dorsal petals, two lateral petals, and one ventral petal. Malpighiaceae therefore provide a rare opportunity to elucidate the ways in which important morphologies originate and are alternatively maintained or remodeled following changes in a selective regime, such as a shift in pollination system.
One way to approach this problem is to investigate the floral developmental genetic basis for these shifts, both in terms of the origin and maintenance of zygomorphy in New World Malpighiaceae, and the secondary loss of this conserved morphology among Old World species. Fortunately, the developmental genetics of floral zygomorphy is being elucidated at a rapid pace. Genes belonging to the recently defined CYC2 clade (22) of the TCP [Teosinte Branched 1, CYCLOIDEA (CYC), and PCF] transcription factor family have been shown to play a critical role in the development of zygomorphy. In Antirrhinum, the two closely related CYC2-like genes, CYC and DICHOTOMA (DICH), demarcate the dorsal region of the flower by differentially regulating the rate of cell growth in the developing floral organs (23, 24). CYC/DICH expression is continuous throughout floral development in the dorsal region of this species. In cyc/dich double mutants, however, zygomorphy is lost—the dorsal petals become ventralized, resulting in actinomorphic corollas. The emerging paradigm from this and numerous subsequent studies is that zygomorphy has evolved independently through the repeated recruitment of CYC2-like genes in several phylogenetically diverse clades (reviewed in refs. 25, 26).
The present study builds on this strong developmental framework and suggests that CYC2-like genes are similarly associated with the origin and maintenance of zygomorphy in Malpighiaceae. We identified two forms of CYC2-like genes in Malpighiaceae, which resulted from a gene duplication in the common ancestor of the family. Both gene copies, CYC2A and CYC2B, are expressed in the dorsal region of the flowers in distantly related New World species with the stereotypical floral morphology (i.e., Byrsonima crassifolia Kunth and Janusia guaranitica A. Juss.). Further evidence of the function of CYC2 in establishing floral symmetry in Malpighiaceae derives from expression patterns in an Old World species that has lost its association with the New World oil-bee pollinators and its stereotypical morphology. In the zygomorphic-flowered species Tristellateia australasiae A. Rich., CYC2A expression mirrors a dramatic shift in the plane of floral symmetry, whereas CYC2B has been lost. The dorsal patterning of gene expression we observed in Malpighiaceae contrasts dramatically with their actinomorphic-flowered relatives, Centroplacaceae (Bhesa paniculata Arn.) and Elatinaceae (Bergia texana Seub. ex Walp.). Results from the latter species suggest unique insights on CYC2 patterning in actinomorphic-flowered species, and highlight the importance of investigating CYC2-like genes in other actinomorphic core eudicot clades beyond the model species Arabidopsis thaliana.
Results and Discussion
Gene Duplication and Loss.
Thirty-three species from 29 genera of Malpighiaceae representing all major clades of the family, two species from both genera of Elatinaceae, and one species from Centroplacaceae, were used for assembling the CYC2 genealogy (Table S1). Oxalis herrerae R. Knuth was used as an outgroup (16). One to six copies of the CYC2-like genes were isolated from each taxon. As with previous studies (e.g., ref. 22), the TCP and R domains were highly conserved, but the intervening coding region was variable across all taxa. Partial TCP and R domains (i.e., 26 of 60 and two of 18 total amino acid sequences were obtained for each region, respectively) and the entire intervening coding region were included in our analyses. The aligned CYC2 matrix included 78 sequences and was 384 bp in length; 78 of these bps were constant and 278 were parsimony-informative.
The phylogenetic relationships inferred from the CYC2 homologues mirror our understanding of accepted species tree relationships (14, 15, 17, 19) (Fig. 2 and Fig. S2). Centroplacaceae and Elatinaceae are successive sisters to Malpighiaceae with 100% maximum likelihood (ML) bootstrap support (BP) and 100% Bayesian posterior probability (PP; reported for simplicity here as percentages) and 70% BP/75% PP, respectively. A comparison of the CYC2-like gene tree with accepted species tree relationships indicates that the origin of Malpighiaceae coincided with a duplication in the CYC2 clade, which yielded two major copies, CYC2A and CYC2B. Both copies receive moderate to high support (CYC2A, 57% BP/65% PP; CYC2B, 80% BP/99% PP). We inferred additional gene duplications in CYC2A in Galphimia, Hiptage, Tristellateia, and Verrucularia, and in CYC2B in Banisteriopsis, Flabellariopsis, Hiptage, Janusia, Ptilochaeta, Ryssopterys, and Spachea. CYC2 duplications were also inferred in the outgroup taxa Bergia texana and Elatine minima (Elatinaceae). These results are consistent with previous analyses that have uncovered CYC2 duplications across diverse phylogenetic groups (22). Furthermore, they demonstrate that CYC2 gene duplications have occurred independently in Malpighiaceae and Elatinaceae, with the former duplication corresponding to the origin of Malpighiaceae and their unique floral zygomorphy (Fig. S1). Importantly, this duplication is not likely to be the result of polyploidization, as there is no evidence of genome doubling associated with the origin of Malpighiaceae (27, 28).
Fig. 2.
Phylogeny of CYC2-like genes inferred using maximum likelihood (ML) and Bayesian analyses. Majority rule consensus shown depicting clades with >50% bootstrap support and >60% Bayesian posterior probability depicted above lines, respectively. ML bootstrap support <50% indicated with a hyphen. Black circles indicate inferred gene duplication events. Inferred gene tree is reflective of accepted species tree relationships (13-16, 19) (Fig. S2). Accessions shown in boldface were additionally analyzed for gene expression using RT-PCR. Asterisks indicate Old World clades. Species identities and voucher information can be found in Table S1. (O, Oxalidaceae; C, Centroplacaceae.)
Similarly, we uncovered evidence for the loss of CYC2A or B in various Malpighiaceae clades, which has also been observed in other groups (22). Nearly all species of New World Malpighiaceae possess CYC2A and CYC2B, but some species, especially Old World Malpighiaceae, appear to retain only one copy (Table S1). We further verified the copy number of CYC2-like genes by Southern hybridization in a subset of taxa (Fig. S3 and S4). Bergia texana, Byrsonima crassifolia, J. guaranitica, and T. australasiae have six, two, four, and one copy of CYC2-like genes respectively, which is consistent with our initial assessment of gene copy number ascertained from PCR and clone screening. These results suggest that our PCR/clone screens provide a reliable estimate of copy number in taxa for which Southern hybridizations were not conducted.
Recruitment of CYC2 and the Maintenance of Floral Zygomorphy in Malpighiaceae.
To determine if the evolution of floral zygomorphy in Malpighiaceae is likely a result of changes in the regulation of CYC2 homologues, we investigated the pattern of CYC2 expression using locus-specific RT-PCR. RT-PCR was conducted on J. guaranitica and B. crassifolia (New World Malpighiaceae), T. australasiae (Old World Malpighiaceae), B. texana (Elatinaceae), and B. paniculata (Centroplacaceae; Fig. 3). Each of these taxa was carefully selected based on its phylogenetic affinities to elucidate changes in gene expression related to (i) the origin of zygomorphy in Malpighiaceae, (ii) the conservation of floral morphology of New World Malpighiaceae, and (iii) the loss of the stereotypical New World floral morphology in Old World Malpighiaceae.
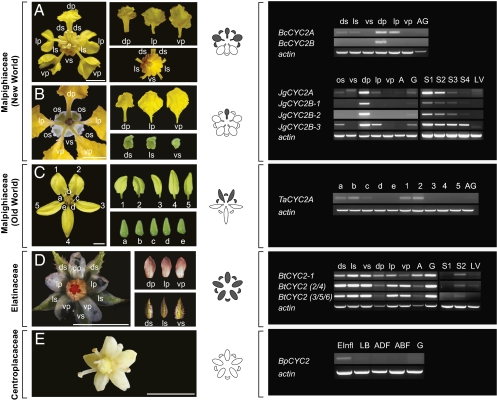
Fig. 3.
CYC2-like gene expression in Malpighiaceae, Elatinaceae, and Centroplacaceae. RT-PCR was examined in whole flower buds at early developmental stages, dissected floral organs at later stages, and leaves. The stereotypical flowers of New World Malpighiaceae are represented here by Byrsonima crassifolia (A) and J. guaranitica (B). T. australasiae (C) is an Old World species of Malpighiaceae that has lost the stereotypical New World morphology and instead has two dorsal petals and eglandular sepals. Flowers of Bergia texana (Elatinaceae, D) and Bhesa paniculata (Centroplacaceae, E) are actinomorphic and eglandular. Gray highlighting on flower diagrams indicates the spatial pattern of CYC2 expression in the corolla and calyx. Actin-specific primers were used as a positive control. ds, dorsal sepal; ls, lateral sepal; vs, ventral sepal; dp, dorsal petal; lp, lateral petal; vp, ventral petal; AG, androecium and gynoecium; os, sepal with oil glands; A, androecium; G, gynoecium; S, developmental stages of floral buds (S1, <1 mm in diameter; S2, 1–2 mm; S3, 2–3 mm; S4, 4–5 mm); LV, leaves; b, dorsal sepal; a, c, lateral sepal; d, e, ventral sepal; 1, 2, dorsal petal; 3, 5, lateral petal; 4, ventral petal; Elnfl, early inflorescence; LB, large buds (2–3 mm); ADF, adaxial flower (including sepals, petals, and stamens); ABF, abaxial flower. Note: our sequence analysis confirmed that the larger fragment amplified from the ventral sepal of J. guaranitica is not a TCP family gene. (Scale bars, 5 mm.)
Several studies from phylogenetically diverse species indicate that zygomorphy has evolved independently via the repeated recruitment of CYC2-like genes (reviewed in ref. 25). CYC2 expression in Malpighiaceae similarly suggests that these genes are responsible for the unique floral symmetry of New World Malpighiaceae (Fig. 3). During the later stages of floral development, CYC2A and CYC2B are differentially expressed along the dorsoventral plane of symmetry in Byrsonima crassifolia and J. guaranitica, which mirrors their floral morphology. B. crassifolia has two CYC2 genes: CYC2A (BcCYC2A) and CYC2B (BcCYC2B; Fig. 2). J. guaranitica has four CYC2 genes: CYC2A (JgCYC2A) and three copies of CYC2B (JgCYC2B-1, -2, and -3). BcCYC2A of B. crassifolia and JgCYC2A and JgCYC2B-3 of J. guaranitica are expressed broadly in the dorsal region of the flower, i.e., in the dorsal and lateral petals and sepals, and weakly in the gynoecium (Fig. 3). In contrast, BcCYC2B of B. crassifolia and JgCYC2B-1 and JgCYC2B-2 of J. guaranitica are expressed only in the single dorsal banner petal, and weakly in the gynoecium. Whether differential expression of these CYC2-like genes affects the symmetry of the calyx is unclear. In J. guaranitica, CYC2-like genes are differentially expressed in the dorsal and lateral sepals with oil glands, but not in the eglandular ventral sepal. This raises the possibility that CYC2 might be involved in the development of the dorsoventral asymmetry of the calyx. However, all of the sepals of B. crassifolia are identical and bear oil glands, despite the fact that CYC2 expression was detected only in the dorsal and lateral sepals.
Overlapping expression patterns of duplicated CYC2 homologues, as observed here in Malpighiaceae, are common in numerous zygomorphic-flowered groups (23–25, 29, 30). Without functional assays, however, it is difficult to discern if the CYC2A and 2B paralogues have significant functional redundancy, as in Antirrhinum (23, 24), or have more differentiated functions, as in Fabaceae, where one copy is more broadly expressed and the other is specific to the dorsal flag petal (29, 30). Regardless, duplication and functional divergence of CYC2-like genes may have contributed to the evolution of the unique zygomorphy in New World Malpighiaceae. The two species examined here, B. crassifolia and J. guaranitica, span the basal node of crown group Malpighiaceae, and diverged from one another at least 64 Mya (17), yet their pattern of gene expression is identical. This highly conserved pattern provides a likely molecular explanation for the stereotypical floral morphology in New World Malpighiaceae, which is in turn the basis of a well documented plant-pollinator mutualism.
An important test of our hypothesis that CYC2 controls floral zygomorphy in Malpighiaceae is to examine gene expression in those clades that have lost the stereotypical New World morphology. T. australasiae is an Old World species with zygomorphic flowers that has lost its association with the oil-bee pollinators, and the characteristic New World morphology (Fig. 3). Instead, T. australasiae possesses two dorsal, two lateral, and one ventral petal. Although this species superficially resembles a resupinate New World flower, its altered symmetry does not result from resupination. If this were the case, the innermost petal of T. australasiae, which is homologous with the dorsal flag/banner petal of all New World species (31), would initiate in the dorsal position and terminate in the ventral position during development. This is not observed: T. australasiae produces an innermost petal that begins and ends in the dorsal position. Hence, the shift of floral zygomorphy in T. australasiae is best explained as a 36° rotation in the plane of floral symmetry, rather than a 180° resupination (cf. ref. 32; Figs. S5 and S6). This is likely a developmental feature that is established very early in floral development. How this happens remains an open question that we will seek to address in future studies.
We have shown that T. australasiae has lost the CYC2B gene and that CYC2A (TaCYC2A) expression closely mirrors its dramatic shift in the dorsoventral plane of floral symmetry (Fig. 3). TaCYC2A is still expressed in the dorsal region of the flower, but now the domain encompasses the two dorsal petals and one dorsal and two lateral sepals. We hypothesize that changes in the regulation of TaCYC2A expression contributed to the shift in flower morphology of Tristellateia, and may reflect adaptations to a different pollination strategy. Tristellateia is visited by xylocopine bees whose reward for visiting the flowers appears to be pollen, rather than oil (14, 15, 21).
Additional expression studies of the CYC2 homologues across the six remaining Old World Malpighiaceae clades, plus two small New World clades that have lost the stereotypical New World floral morphology, will provide further insights into the genetic and developmental basis of how floral zygomorphy has been altered or lost in this group. This will enable us to begin to determine whether convergent floral morphologies have evolved via convergent genetic changes in what appears to be a broadly conserved developmental program.
CYC2 and the Actinomorphic-Flowered Relatives of Malpighiaceae, Centroplacaceae, and Elatinaceae.
Our results demonstrate that the two actinomorphic sister clades of Malpighiaceae, represented by B. paniculata (Centroplacaceae) and B. texana (Elatinaceae), have different patterns of CYC2-like gene expression, suggesting that different processes may be involved in controlling similar floral symmetries. The single CYC2-like gene of B. paniculata (BpCYC2) is expressed only during early stages of flower development (Fig. 3), which is similar to the well studied actinomorphic-flowered rosid A. thaliana (L.) Heynh. For the Arabidopsis TCP1 gene, dorsal gene expression is detected in the young floral meristem but greatly declines before the floral organ primordia are initiated (33). It remains to be determined whether early BpCYC2 expression is similarly localized in the dorsal region of the floral meristem in Bhesa.
In contrast, a unique temporal and spatial pattern of CYC2-like gene expression was discovered in the actinomorphic flowers of Bergia texana (Fig. 3). The six CYC2-like genes in B. texana exhibit a high degree of sequence similarity (i.e., 78–95%), suggesting that they are most likely recently evolved paralogues (Fig. 2). As sequence variation among some of these paralogues was especially minor (i.e., ≤5%), we distinguished three groups for RT-PCR analysis: (i) BtCYC2-1, (ii) BtCYC2-2 and BtCYC2-4, and (iii) BtCYC2-3, BtCYC2-5, and BtCYC2-6. All six BtCYC2 genes exhibit no sign of early expression, but are expressed uniformly across the floral organs during later stages of development. Given the apparent uniformity of gene expression in these paralogues, it is most likely that the expression pattern we observed in B. texana evolved before gene duplication. In terms of the temporal pattern of CYC2 expression, our result most closely resembles that of the zygomorphic-flowered species Iberis amara L. (Brassicaceae), in which early expression of IaTCP1 has been lost but late-stage expression is present. Spatial expression in I. amara is differential, however, which is consistent with the fact that its flowers are zygomorphic (34). In this respect, the uniform spatial expression of BtCYC2s throughout the corolla of B. texana is similar to the pattern in Cadia purpurea Forssk., an actinomorphic-flowered member of the otherwise zygomorphic-flowered Fabaceae (35). The uniform expression of LgCYC1B in Cadia is thought to be the result of a homeotic transformation, which accounts for the apparent morphological reversal to actinomorphy, i.e., gene expression in the lateral and ventral petals has become dorsalized. The main difference between CYC2-like gene expression in Cadia and Bergia is that LgCYC1B expression appears to be corolla-specific whereas expression of BtCYC2s in Bergia is not restricted to the corolla, and is instead expressed throughout all of the floral organs examined (Fig. 3).
Interpreting the Origin of Floral Zygomorphy in Malpighiaceae.
Differential expression of the CYC2-like genes during the late stage of floral development is required for establishing zygomorphic flowers in several phylogenetically diverse rosid [Iberis, Lotus, Lupinus, and Pisum (29, 30, 34, 35)] and asterid [Antirrhinum, Gerbera, Linaria, and Senecio (23, 24, 36–38)] clades, suggesting that the recruitment of CYC2-like function at later developmental stages is necessary for evolving zygomorphic flowers. To better understand the evolution of gene expression that likely contributed to the origin of zygomorphy in Malpighiaceae, we reconstructed the evolutionary pattern of late stage CYC2 expression in New World Malpighiaceae [i.e., which represents the ancestral floral morphology for the family (9, 19, 27)] and their closest actinomorphic-flowered relatives (Fig. S7).
Our results indicate that the most recent common ancestor of Centroplacaceae–Elatinaceae–Malpighiaceae likely exhibited the pattern of gene expression observed in Centroplacaceae (B. paniculata), which is also similar to the actinomorphic-flowered rosid, A. thaliana (Fig. 4 and Fig. S7). CYC2 expression in the most recent common ancestor of Malpighiaceae–Elatinaceae, however, is equivocal.
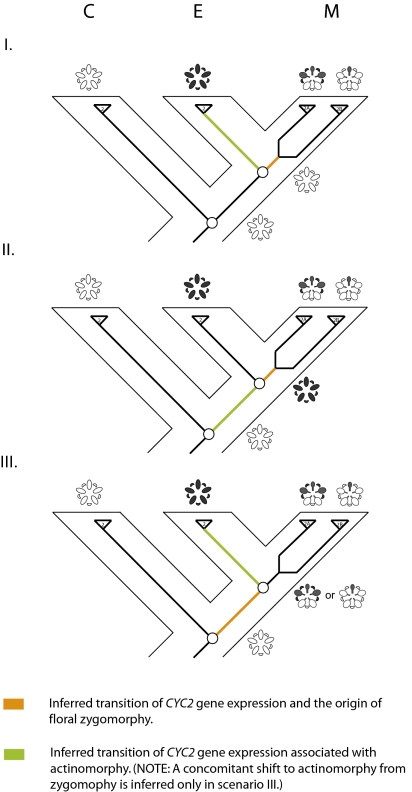
Fig. 4.
Hypothesized scenarios for the origin of floral zygomorphy in Malpighiaceae. Gray highlighting in the flower diagram indicates the spatial pattern of CYC2 expression. Orange coloring on the tree indicates the inferred transition to differential CYC2 expression and floral zygomorphy; green indicates the transition to actinomorphic CYC2 expression in actinomorphic-flowered species. In scenario (I), floral zygomorphy evolved in stem lineage Malpighiaceae from an actinomorphic-flowered ancestor with no late stage CYC2 expression. In scenario (II), floral zygomorphy evolved in stem lineage Malpighiaceae from an actinomorphic-flowered ancestor with actinomorphic late stage CYC2 expression. In scenario (III), floral zygomorphy and differential CYC2 expression evolved from an actinomorphic-flowered ancestor with no CYC2 expression along the stem lineage leading to the most recent common ancestor of Malpighiaceae and Elatinaceae. (C, Centroplacaceae; E, Elatinaceae; M, Malpighiaceae.)
In view of the uncertain pattern of CYC2-like gene expression in the most recent common ancestor of Malpighiaceae–Elatinaceae, we propose three alternative, and equally parsimonious, scenarios to explain the origin of floral zygomorphy in Malpighiaceae (Fig. 4). In the first scenario, the most recent common ancestor of Malpighiaceae–Elatinaceae had actinomorphic flowers with no late-stage CYC2 expression. Under this scenario, floral zygomorphy and differential CYC2 expression evolved in the common ancestor of Malpighiaceae and the broad expression in Elatinaceae must have evolved independently. The second scenario proposes that the most recent common ancestor of Malpighiaceae–Elatinaceae was actinomorphic and exhibited uniform CYC2 expression across the floral meristem. Under this scenario, floral zygomorphy in Malpighiaceae originated through the loss of ventral gene expression. The third scenario proposes that the most recent common ancestor of Malpighiaceae–Elatinaceae possessed zygomorphic flowers with differential CYC2 expression. Under this scenario, floral zygomorphy and differential CYC2 expression evolved in the common ancestor of Malpighiaceae–Elatinaceae. Actinomorphic flowers in Elatinaceae would therefore have been secondarily derived from a zygomorphic ancestor. Support for this model derives from the Cadia example discussed earlier, which is what we hypothesize here to explain the gene expression pattern observed in Elatinaceae. This scenario is especially intriguing because it is one that we would not have previously predicted given our character state reconstructions of floral evolution based on standard morphological characters (Fig. S1). Those analyses clearly indicate that zygomorphy arose in the most recent common ancestor of Malpighiaceae and that actinomorphy in Elatinaceae was plesiomorphic. The precedent established by Cadia, combined with these analyses, however, raises the distinct possibility that actinomorphy in Elatinaceae may instead be better explained as a result of a homeotic transformation from a zygomorphic-flowered ancestor, which ultimately gave rise to Malpighiaceae.
Overall, this complex set of alternatives demonstrates that analyzing molecular data, and gene expression data in particular, in an explicit phylogenetic framework enhances comparative studies and can add a level of richness to our understanding of morphological evolution. Determining which of these models is most likely, however, will require additional comparative expression data, particularly from other actinomorphic rosids, as well as functional studies of CYC2-like genes in Elatinaceae and Malpighiaceae. It is important to note here that most studies of CYC2 homologues have focused exclusively on zygomorphic-flowered groups (e.g., Asteraceae, Dipsacales, Fabaceae, Lamiales), meaning that we actually know very little about comparative patterns of CYC2-like gene expression in actinomorphic-flowered groups. This is especially relevant given the diverse actinomorphic-flowered ancestors that gave rise to these zygomorphic clades. Our results, as well as those from Busch and Zachgo (34), indicate that further expression studies are needed to determine if early dorsal expression followed by gene repression is the ancestral condition for many actinomorphic angiosperm clades as is commonly assumed (cf. Arabidopsis), or if the ancestral condition is, in fact, highly variable across the angiosperms.
Materials and Methods
Sequence Alignments and Phylogenetic Analyses.
CYC2-like gene sequences we obtained from Malpighiaceae–Elatinaceae–Centroplaceae–Oxalidaceae (SI Materials and Methods) were assembled with Sequencher 4.7 (Gene Codes) and aligned by eye with reference to the translated amino acid sequences using MacClade 4.06 (39). We applied the WAG + G model of amino acid evolution to the aligned CYC2 data set as determined by the Akaike Information Criterion in ProtTEST (40). One hundred ML bootstrap replicates were conducted using RAxML-VI-HPC (41). Bayesian analyses were implemented in MrBayes version 3.1.2 (42) under the same model using default priors for the rate matrix, branch lengths, and γ-shape parameter. A Dirichlet distribution was used for the base frequency parameters and an uninformative prior was used for the starting tree topology. Four chains were initiated with a random starting tree and run for two million generations sampled every 1,000 generations. Following a burn-in of 1,000 trees as determined by Tracer version 1.4.1 (http://tree.bio.ed.ac.uk/software/tracer/), trees were sampled from the posterior distribution to calculate clade posterior probabilities.
RT-PCR.
Floral organs were dissected directly in the field from flower buds ranging in size from 1 mm in diameter to open flowers, and preserved immediately in cryodewars or RNAlater (Ambion). For the samples prepared in RNAlater, the floral organs were finely chopped on sterile Petri dishes to facilitate penetration of the preservative into the tissues. These materials were processed in our laboratory using the RNAqueous kit (Ambion). RT-PCR was performed as described (43) using locus-specific primers to examine the expression of each CYC2-like gene copy (Table S2). The sequence identity of RT-PCR fragments was confirmed by sequencing.
Supplementary Material
Acknowledgments
We thank L. Ahart, W. Anderson, B. Bartholomew, M. Bartlett, D. Boufford, M. Davenport, J. Davit, L. Gómez, P. Griffith, L. Holappa, D. Howarth, D. Lee, S. Manickam, R. McFarland, C. Morse, M. Opel, C. Specht, E. Voss, K. Wong, S. Yee, and members of the Davis and Kramer laboratories. This work was funded by National Science Foundation Grants DEB-0544039 and AToL EF 04-31242 (to C.C.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU982187–GU982264).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910155107/DCSupplemental.
References
- 1.Neal PR, Dafni A, Giurfa M. Floral symmetry and its role in plant-pollinator systems: terminology, distribution, and hypotheses. Annu Rev Ecol Syst. 1998;29:345–373. [Google Scholar]
- 2.Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 1998;3:311–317. [Google Scholar]
- 3.Stebbins GL. Flowering plant: evolution above the species level. Cambridge, MA: Harvard Univ. Press; 1974. [Google Scholar]
- 4.Westerkamp C, Classen-Bockhoff R. Bilabiate flowers: the ultimate response to bees? Ann Bot (Lond) 2007;100:361–374. doi: 10.1093/aob/mcm123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevens PF. Angiosperm Phylogeny Website; version 9, June 2008. 2001. Available at http://www.mobot.org/MOBOT/research/APweb/. Accessed January 5, 2010.
- 6.Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proc R Soc Lond B Biol Sci. 2004;271:603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endress PK. Evolution of floral symmetry. Curr Opin Plant Biol. 2001;4:86–91. doi: 10.1016/s1369-5266(00)00140-0. [DOI] [PubMed] [Google Scholar]
- 8.Gomez JM, Perfectti F, Camacho JPM. Natural selection on Erysimum mediohispanicum flower shape: insights into the evolution of zygomorphy. Am Nat. 2006;168:531–545. doi: 10.1086/507048. [DOI] [PubMed] [Google Scholar]
- 9.Anderson WR. Floral conservatism in neotropical Malpighiaceae. Biotropica. 1979;11:219–223. [Google Scholar]
- 10.Vogel S. Ölblumen und ölsammelnde Bienen. Trop Subtrop Pflanzenwelt. 1974;7:283–547. [Google Scholar]
- 11.Sigrist MR, Sazima M. Pollination and reproductive biology of twelve species of neotropical Malpighiaceae: stigma morphology and its implications for the breeding system. Ann Bot (Lond) 2004;94:33–41. doi: 10.1093/aob/mch108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araújo VA, Antonini Y, Araújo APA. Diversity of bees and their floral resources at altitudinal areas in the Southern Espinhaço Range, Minas Gerais, Brazil. Neotrop Entomol. 2006;35:30–40. doi: 10.1590/s1519-566x2006000100005. [DOI] [PubMed] [Google Scholar]
- 13.Davis CC, Chase MW. Elatinaceae are sister to Malpighiaceae; Peridiscaceae belong to Saxifragales. Am J Bot. 2004;91:262–273. doi: 10.3732/ajb.91.2.262. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Kramer EM, Davis CC. Exploring the developmental genetic basis of independent reversals in floral symmetry in Malpighiaceae. Botany & Mycology 2009: Annual Meeting of the Botanical Society of America in Snowbird, Utah. 2009. Available at http://2009.botanyconference.org/engine/search/index.php?func=detail&aid=851.
- 15.Zhang W, Kramer EM, Davis CC. CYCLOIDEA2 and the origin and maintenance of floral zygomorphy in Malpighiaceae. Botany & Mycology 2009: Annual Meeting of the Botanical Society of America in Snowbird, Utah, USA. 2009. Available at http://2009.botanyconference.org/engine/search/index.php?func=detail&aid=546.
- 16.Wurdack KJ, Davis CC. Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 2009;96:1551–1570. doi: 10.3732/ajb.0800207. [DOI] [PubMed] [Google Scholar]
- 17.Davis CC, Bell CD, Mathews S, Donoghue MJ. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proc Natl Acad Sci USA. 2002;99:6833–6837. doi: 10.1073/pnas.102175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis CC, Fritsch PW, Bell CD, Mathews S. High-latitude tertiary migrations of an exclusively tropical clade: evidence from Malpighiaceae. Int J Plant Sci. 2004;165:S107–S21. [Google Scholar]
- 19.Davis CC, Anderson WR, Donoghue MJ. Phylogeny of Malpighiaceae: evidence from chloroplast ndhF and trnL-F nucleotide sequences. Am J Bot. 2001;88:1830–1846. [PubMed] [Google Scholar]
- 20.Michener CD. The bees of the world. Baltimore: Johns Hopkins Univ Press; 2000. [Google Scholar]
- 21.Davis CC. Madagasikaria (Malpighiaceae): a new genus from Madagascar with implications for floral evolution in Malpighiaceae. Am J Bot. 2002;89:699–706. doi: 10.3732/ajb.89.4.699. [DOI] [PubMed] [Google Scholar]
- 22.Howarth DG, Donoghue MJ. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA. 2006;103:9101–9106. doi: 10.1073/pnas.0602827103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo D, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/s0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- 24.Luo D, et al. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 25.Preston JC, Hileman LC. Developmental genetics of floral symmetry evolution. Trends Plant Sci. 2009;14:147–154. doi: 10.1016/j.tplants.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Kramer EM. Understanding the genetic basis of floral diversity. Bioscience. 2007;57:479–487. [Google Scholar]
- 27.Cameron KM, Chase MW, Anderson WR, Hills HG. Molecular systematics of Malpighiaceae: evidence from plastid rbcL and matK sequences. Am J Bot. 2001;88:1847–1862. [PubMed] [Google Scholar]
- 28.Anderson WR. Chromosome numbers of neotropical Malpighiaceae. Contrib Univ Mich Herb. 1993;19:341–354. [Google Scholar]
- 29.Wang Z, et al. Genetic control of floral zygomorphy in pea (Pisum sativum L.) Proc Natl Acad Sci USA. 2008;105:10414–10419. doi: 10.1073/pnas.0803291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng X, et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA. 2006;103:4970–4975. doi: 10.1073/pnas.0600681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson WR. Malpighiaceae (Malpighia family) In: Smith N, Mori SA, Henderson A, Stevenson DW, Heald SV, editors. Flowering plants of the Neotropics. Princeton, NJ: Princeton Univ Press; 2004. pp. 229–232. [Google Scholar]
- 32.Vogel S. History of the Malpighiaceae in the light of pollination ecology. Mem N Y Bot Gard. 1990;55:130–142. [Google Scholar]
- 33.Cubas P, Coen E, Zapater JMM. Ancient asymmetries in the evolution of flowers. Curr Biol. 2001;11:1050–1052. doi: 10.1016/s0960-9822(01)00295-0. [DOI] [PubMed] [Google Scholar]
- 34.Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci USA. 2007;104:16714–16719. doi: 10.1073/pnas.0705338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Citerne HL, Pennington RT, Cronk QCB. An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA. 2006;103:12017–12020. doi: 10.1073/pnas.0600986103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broholm SK, et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA. 2008;105:9117–9122. doi: 10.1073/pnas.0801359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cubas P, Vincent C. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 38.Kim M, et al. Regulatory genes control a key morphological and ecological trait transferred between species. Science. 2008;322:1116–1119. doi: 10.1126/science.1164371. [DOI] [PubMed] [Google Scholar]
- 39.Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Sunderland, MA: Sinauer; 2003. version 4.0.6. [DOI] [PubMed] [Google Scholar]
- 40.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 41.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 42.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 43.Kramer EM, Di Stilio VS, Schluter PM. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int J Plant Sci. 2003;164:1–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.