Abstract
Noncoding regulatory microRNAs (miRNAs) of cellular and viral origin control gene expression by repressing the translation of mRNAs into protein. Interestingly, miRNAs are secreted actively through small vesicles called “exosomes” that protect them from degradation by RNases, suggesting that these miRNAs may function outside the cell in which they were produced. Here we demonstrate that miRNAs secreted by EBV-infected cells are transferred to and act in uninfected recipient cells. Using a quantitative RT-PCR approach, we demonstrate that mature EBV-encoded miRNAs are secreted by EBV-infected B cells through exosomes. These EBV-miRNAs are functional because internalization of exosomes by MoDC results in a dose-dependent, miRNA-mediated repression of confirmed EBV target genes, including CXCL11/ITAC, an immunoregulatory gene down-regulated in primary EBV-associated lymphomas. We demonstrate that throughout coculture of EBV-infected B cells EBV-miRNAs accumulate in noninfected neighboring MoDC and show that this accumulation is mediated by transfer of exosomes. Thus, the exogenous EBV-miRNAs transferred through exosomes are delivered to subcellular sites of gene repression in recipient cells. Finally, we show in peripheral blood mononuclear cells from patients with increased EBV load that, although EBV DNA is restricted to the circulating B-cell population, EBV BART miRNAs are present in both B-cell and non-B-cell fractions, suggestive of miRNA transfer. Taken together our findings are consistent with miRNA-mediated gene silencing as a potential mechanism of intercellular communication between cells of the immune system that may be exploited by the persistent human γ-herpesvirus EBV.
Keywords: intercellular communication, exosomes, Epstein–Barr virus, small RNA, gene repression
We propose that microRNAs (miRNAs) transferred through exosomes may have an important role in intercellular communication by mediating repression of critical mRNA targets in neighboring or more distant recipient cells. Cellular and viral miRNAs control gene expression by repressing the translation of mRNAs into protein (1, 2), a process that is tightly regulated in healthy cells but is deregulated in cancerous and virus-infected cells (3, 4). Curiously, miRNAs are not strictly cellular but are secreted through the release of small vesicles called “exosomes” and therefore present extracellularly in the peripheral blood and in cell-culture media (5–7). It has been suggested that exosome-associated miRNAs have a role in intercellular communication, although concrete evidence for this has been lacking (6–9). For example, the dynamics of miRNA secretion through exosomes and the proposed transfer mechanisms remain poorly understood. In addition, it is unclear whether miRNAs are secreted in physiologically relevant amounts, and it remains to be determined whether exogenous exosome-associated miRNAs access the molecular machinery of miRNA-mediated gene repression upon transfer into recipient cells.
To investigate the possibility of functional miRNA transfer, we chose EBV infection as a model for miRNA transference. This model is advantageous, because EBV-miRNA–mediated repression of target genes in noninfected cells can be distinguished from host cellular miRNA-mediated repression but functions through similar mechanisms (10). EBV is a common, potentially oncogenic, γ-herpesvirus, the first virus known to encode miRNAs (EBV-miRNAs) (11), and exploits host cellular pathways for its own benefit (12). EBV-miRNAs are separated into three clusters of the viral genome: BHRF1 and cluster 1 and cluster 2 BARTs (13, 14), which are abundantly expressed in EBV-associated tumors (13, 15) and EBV-transformed lymphoblastoid B cells (LCL) (13). Although the expression pattern of EBV-miRNAs in vivo is unexplored, and their targets are largely unknown, comprehensive studies in vitro indicate that their expression pattern is linked to viral latency stage (13, 14, 16). LCL display latency type III, phenotypically resemble proliferating activated B-cell blasts (12), and secrete large quantities of immunoregulatory exosomes (17, 18). LCL express both BHRF1 and BART miRNAs, albeit at different levels (11, 14, 16). Demonstration of functional transfer of BHRF1 and/or BART miRNAs to noninfected cells via exosomes may have implications specifically for EBV infection but also generically for miRNA-mediated biological processes including embryogenesis, tissue homeostasis, and immunomodulation or in patholo-gies such as oncogenesis.
Results
EBV-Infected Activated B Cells Secrete Exosomes that Contain EBV-miRNAs.
Exosomes are microvesicles 30–100 nm in size produced by reverse budding of the limiting membrane of multivesicular endosomes (MVEs) (Fig. 1 A and B). B-cell derived exosomes are secreted into the extracellular milieu upon fusion of the MVE with the plasma membrane (Fig. S1) and are characterized at the protein level by the presence of functional HLA-DR and the tetraspanin CD63 and the absence of cellular cytochrome C (Fig. 1C) (17, 19).
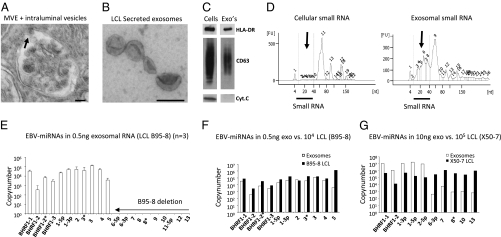
Fig. 1.
Exosomes from EBV-infected lymphoblasts (LCL) are enriched in small RNA and EBV-m iRNAs. (A) EM image of an MVE of an LCL with ongoing inward budding of the limiting membrane (arrow). (B) Purified exosomes isolated from LCL culture medium. (Scale bar, 100 nm,). (C) Western blots for HLA-DR, CD63, and cytochrome C of cell lysates and lysates from purified exosomes (CD63 under nonreducing conditions). (D) Bioanalyzer results of equal amounts of cellular (LCL) RNA compared with RNA isolated from purified LCL exosomes treated with RNase A (10 ng/μL). Small RNA species (indicated by arrows) are highly enriched in exosomes. (E) Detection of EBV-encoded mature miRNAs by multiplex quantitative RT-PCR using dilution series of chemically synthesized oligonucleotides (13). Shown are the average copy numbers measured in 500 pg RNA from three independent B95-8 LCL exosome purifications/isolations. One sample was treated with 10 ng/μL RNase A, one was treated with 400 ng/μL RNase A, and one sample was untreated. (F) Comparison between individual cellular EBV-miRNA copy-numbers in ∼104 LCL (B95-8) and relative abundance of exosomal EBV-miRNA copy numbers. (G) EBV-miRNA copy-numbers measured in 10 ng exosomal RNA from the X50-7 LCL compared with total cellular RNA. Cluster 2 BART EBV-miRNAs are ∼1,000-fold less abundant in exosomes than expected from their individual cellular expression levels. Error bars (SD) are derived from triplicate experiments.
We determined the possible presence of RNA molecules in purified exosomes from an LCL transformed by the B95-8 strain of EBV. Characterization of the RNA profile using a Bioanalyzer indicated that the exosomal RNA was smaller than 500 nucleotides and protected from exogenous RNase activity (Fig. S2 A–D). Of note, compared with cellular RNA, LCL exosomes are highly enriched in small RNA species, including the 19–22 nucleotide class of noncoding regulatory miRNAs (1) (Fig. 1D).
LCL express BHRF1 and BART miRNAs, albeit at variable levels (11, 14, 16). To determine whether EBV-miRNAs are part of exosomal (LCL) RNA, we used multiplex quantitative RT-PCR for mature EBV-miRNAs (13). Standard curves were generated from serial dilutions of chemically synthesized oligonucleotides (Fig. S2 E and F) allowing absolute quantification of individual miRNA copies. We detected from as few as 102 up to 105 of copies of BHRF1 and cluster 1 BART EBV-miRNA in 0.5 ng exosomal RNA (Fig. 1E) (cluster 2 BART miRNA are absent because of a genomic deletion in the B95-8 EBV strain). Generally, the abundance of individual EBV-miRNA in B95-8 exosomes correlated with cellular expression level, although BART5 appears to be excluded (Fig. 1F). We did not detect the noncoding primary BART transcripts by sensitive semiquantitative RT-PCR, although these transcripts were abundant in LCL cells. Thus mature miRNAs seem to be secreted selectively via the exosomal pathway. The finding that BART miRNAs were expressed in LCLs at relatively high levels compared with initial observations using Northern analysis (depicted are ∼60-cell equivalents) was surprising (14) but was comparable with other findings using a quantitative approach (16). We detected both cluster 1 and cluster 2 BART-miRNAs in exosomes secreted by a spontaneous LCL presumably carrying wild-type EBV (Fig. S3A). Surprisingly, in the X50-7 LCL that also expresses all BART miRNAs (20), cluster 2 BART miRNAs were ∼1,000-fold less abundant in exosomes (Fig. 1G). This difference was not related to the cellular expression level, because the copy numbers of EBV-miRNAs were highly similar, but may depend on the specific localization in the genome (i.e., cluster 1 vs. cluster 2; Fig S3 B and C). In conclusion, all three EBV-driven LCL express EBV-miRNAs that are secreted through exosomes, although some EBV-miRNAs may be excluded, depending on the cellular background.
EBV-miRNAs Are Delivered to and Internalized by Monocyte-Derived Dendritic Cells.
We proposed that EBV-miRNAs may function outside infected B cells by exosome-mediated delivery into physiologically relevant recipient cells. Dendritic cells (DC) control T-cell–mediated immunity of EBV infection and control EBV-driven transformation (21, 22). In addition, DC modulate adaptive immune responses by internalizing exosomes (23, 24). To monitor whether exosomes transfer to primary immature monocyte-derived DC (MoDC), we labeled purified LCL exosomes with a green fluorescent lipid dye (PKH67) that could be made visible with confocal microscopy through capture by HLA-DR–coated beads (Fig. 2A). When purified PKH-labeled exosomes were incubated with MoDC for 1–2 h, MoDC became fluorescent; increasing the amount of exosomes led to more and brighter fluorescent MoDC (Fig. 2B), indicating specific uptake. Flow cytometry confirmed the exosome uptake and revealed a linear relationship between the mean fluorescence index (MFI) and the amount of PKH67-labeled exosomes added (Fig. 2C). Internalization of exosomes was inhibited by disruption of actin filaments using cytochalasin-B and was reduced in cytokine matured as compared with immature MoDC (Fig. S4 B–D). Confocal imaging of MoDC that internalized fluorescent exosomes showed a patched intracellular fluorescence pattern, consistent with endocytosis into discrete intracellular compartments (Fig. 2D). Taken together, these data indicate that exosomes are actively internalized by MoDC.
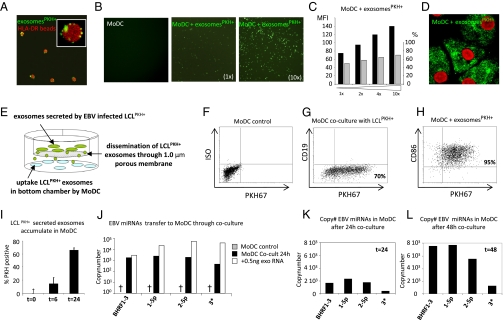
Fig. 2.
EBV-miRNAs are transferred from LCL to MoDC via exosomes. (A) Confocal image of purified PKH67-labeled LCL (RN) exosomes (green) captured by HLA-DR–specific Dynal beads (red). (Inset: Dynal bead with multiple LCL-derived exosomes captured on the surface.) (B) Primary MoDC incubated for 2 h with increasing amounts of PKH67-labeled purified exosomes. (C) Quantification of B by FACS showing an increase in MFI (black bars) and percentage (gray bars) of PKH67-positive cells. (D) Confocal image of primary (immature) MoDC incubated for 2 h with purified PKH67-labeled LCL exosomes. TO-PRO staining (red) indicates the nucleus. (E) Schematic of the transwell coculture model with PKH67-labeled LCL (producers) in the top well and primary MoDC (recipients) in the bottom well. A porous (1.0-μm) membrane allows transfer of fluorescent exosomes but precludes LCL migration. (F and G) FACS results representing CD86+/CD19− MoDC in the bottom chamber before and after 24-h coculture with PKH67-labeled LCL stimulated for 3 h with 10 μM monensin. (H) MoDC incubated for 2 h with purified PKH67-labeled exosomes added directly to the top chamber. (I) FACS results showing MFI of MoDC upon coculture with PKH67-labeled LCL. Error bars (SD) are derived from duplicate wells. (J) Quantitative RT-PCR for EBV-miRNAs in a subset (∼2 × 104) of EBV-negative primary MoDC cocultured with B95-8 LCL for 24 h, as shown in E (black bars), compared with the level of these miRNAs in 500 pg RNA from purified B95-8 exosomes (white bars). (K and L) Comparison of individual miRNA levels in a subset of MoDC cocultured for 24 (K) and 48 h (L) in the presence of B95-8 LCL.
We mimicked exosome transfer from LCL to MoDC using a coculture model as depicted in Fig. 2E. Prolonged coculture between PKH67-stained LCL and unstained MoDC led to in-creasing fluorescence in the MoDC, suggesting LCL continuously release PKH67-positive exosomes that are internalized by neighboring MoDC (Fig. 2 F and G), whereas CD19+ LCL do not pass the membrane. Transference of exosomes through the membrane was confirmed by adding purified PKH67-labeled exosomes in the top chamber (Fig. 2H). Internalized exosomes were measurable at 6 h, and after 24 h of coculture >70% MoDC were fluorescent (Fig. 2I). Pharmacological stimulation of exosome release increased MoDC fluorescence (Fig. S5). We detected multiple EBV-miRNAs after coculture with LCL (Fig. 2J), suggesting that exosomal miRNAs were transferred during LCL-MoDC coculture. Approximately 2 × 103 copies of EBV-miRNA BART1-5p were detected in a subset of the MoDC after 24 h coculture (Fig. 2K), and this level increased 4-fold after an additional 24 h of coculture (Fig. 2L). In conclusion, EBV-miRNAs are transferred to and accumulate in primary MoDC through continuous internalization of exosomes secreted by neighboring LCL.
Exosome-Mediated Transfer of EBV-miRNAs Leads to Repression of EBV Target Genes.
Exosomes internalized by DC localize to endosomes (24) that recently were determined to be sites of RNA-induced silencing complex (RISC)-dependent miRNA-mediated gene silencing (25). Virus-encoded miRNAs may exploit the host miRNA machinery as part of a viral strategy to avoid host immune responses by silencing endogenous immunoregulatory genes (26), as recently shown in EBV lymphomas where the EBV-miRNA BHRF1-3 (15) targets the immunostimulatory gene CXCL11 (11). We hypothesized that EBV-infected B cells secrete exosomes that may transport functional EBV-miRNAs to uninfected recipient cells. Because BHRF1-3 is secreted by LCL via exosomes, we reasoned that exosome transference could lead to altered CXCL11 expression in recipient cells.
We analyzed multiple model cell lines for efficiency of LCL exosome internalization and found that HeLa cells internalized LCL exosomes efficiently (Fig. 3B). To show that exosome-mediated transfer of EBV-miRNAs is functional, we constructed a firefly luciferase pMir-Report vector (Ambion) carrying the complete 3′-UTR-cDNA sequence of CXCL11. We incubated HeLa cells expressing the CXCL11-3′UTR with purified (BHRF1-3–positive) exosomes and detected 80% reduction (P < 0.001) in luciferase activity (Fig. 3C), whereas a CXCL11-3′UTR reporter construct with disrupted BHRF1-3 miRNA binding sites (11) (Fig. S6) showed significantly less reduction. In direct comparison we observed ∼60% attenuation of the inhibitory effects of exosomes that can specifically be attributed to BHRF1-3 transference (Fig. 3D). These results indicate that suppression of CXCL11 is specific and largely dependent on EBV-miRNA BHRF1-3 transfer through exosomes.
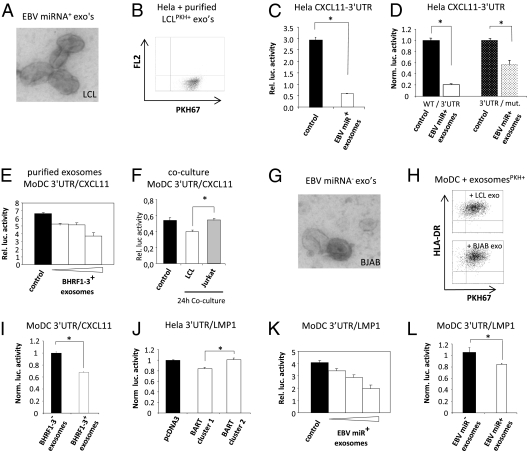
Fig. 3.
Internalization of EBV-miRNA containing exosomes leads to gene silencing in recipient primary DC. (A) EM image of purified secreted exosomes from EBV-infected B95-8 LCL. (B) Flow cytometry results of HeLa cells incubated for 2 h with PKH-labeled purified LCL exosomes. (C) Relative luciferase activity in HeLa cells after 8 h of cotransfection with a 3′UTR-CXCL11 reporter construct without (control) and with 50 μL of purified B95-8 LCL exosomes (white bar). (D) Normalized luciferase activity in HeLa cells transfected with wild-type (WT) 3′UTR-CXCL11 reporter construct or a mutated construct with disrupted BHRF1-3 target sites, incubated with and without EBV-positive exosomes. (E) Relative luciferase activity in primary MoDC transfected with 3′UTR-CXCL11 incubated for 24 h with 2-fold increasing (25, 50, and 100 μL) amounts of purified EBV-miRNA–positive LCL exosomes. (F) Relative luciferase activity measured in MoDC transfected with 3′UTR-CXCL11 and cocultured for 24 h with B95-8 LCL (white bar) or EBV-negative Jurkat cells (gray bar). (G) EM image of purified EBV-miRNA–negative BJAB exosomes. (H) Flow cytometry results of purified PKH67-labeled BJAB or LCL exosomes (HLA-DR+) incubated for 2 h with MoDC, indicating comparable internalization. (I) Normalized luciferase activity measured in MoDC transfected with a 3′UTR-CXCL11 reporter after 24 h incubation with 2 × 100 μL purified LCL (EBV-miRNA BHRF1-3+) and BJAB (EBV-miRNA BHRF1-3−) exosomes, (*, P < 0.025 in a two-tailed student t test). (J) Luciferase activity in HeLa cells with a 3′UTR-LMP1 luciferase and an empty vector or cluster 1 and cluster 2 BART miRNA expression vectors. (K) Dose–response as described in E with a 3′UTR-LMP1 reporter in MoDC. (L) As in I with 2 × 50 μL purified LCL exosomes and 2 × 100 μL BJAB-derived exosomes. *, P < 0.02, two-tailed student t test. Error bars (SD) in all graphs are derived from triplicates.
To demonstrate that suppression was not cell-type specific, we transfected the CXCL11-3′UTR in primary MoDC and added increasing amounts of LCL exosomes. This addition resulted in a dose-dependent suppression of luciferase activity (up to 50%) (Fig. 3E). Thus the repression of CXCL11 is dependent on the amount of LCL exosomes and is reproducible in multiple cell types. We further observed that continuous (24 h) EBV-miRNA transfer through coculture led to significant (∼20%; P < 0.01) repression of CXCL11 compared with EBV-negative control cells (Fig. 3F). To confirm that EBV-miRNAs in exosomes led to gene silencing, we purified exosomes from EBV-negative BJAB cells. Although LCL and BJAB exosomes are similar macroscopically (Fig. 3 A and G), in protein composition (Table S1), and in uptake by MoDC (Fig. 3H), LCL exosomes are superior in repressing of CXCL11-3′UTR–mediated luciferase activity (Fig. 3I). Collectively, these studies indicate that LCL exosomes internalized by MoDC led to silencing of ectopically expressed CXCL11 mRNA mediated through BHRF1-3.
We investigated the possibility that BART miRNAs in exosomes function similarly to BHRF1-3 and generated a reporter containing the complete 3′UTR cDNA sequence of EBV latent membrane protein 1 (LMP1), a confirmed target of BART miRNAs (27). An advantage of a viral target is the reduced risk of unforeseen repression by other introduced (cellular) exosomal miRNAs. We verified the functionality of the LMP1-3′UTR reporter in HeLa cells by cotransfection with a plasmid containing either cluster 1 or cluster 2 BART. As predicted, cluster 2BART had no effect on luciferase activity (27), confirming the specificity of the LMP1-3′UTR reporter construct (Fig. 3J). We next added increasing quantities of B95-8 LCL exosomes carrying predominantly cluster 1 BART miRNAs to primary MoDC transfected with the LMP1-3′UTR reporter. We observed a dose-dependent repression of luciferase activity (Fig. 3K), whereas EBV-negative exosomes had no such effect (Fig. 3L). We conclude that EBV-encoded BHRF1 and BART miRNAs are transferred through exosomes to recipient cells where they are directed to cellular sites of miRNA-mediated gene repression, causing functional translational repression of their target mRNAs.
EBV BART miRNAs Are Present in Peripheral B Lymphocytes and Non-B Cells.
EBV-infected cell lines in vitro express EBV-miRNAs at variable levels, but little is known about which EBV-miRNAs are expressed in EBV-infected circulating B cells. One complicating factor is the extremely low infection frequencies observed in healthy carriers (28), making detection very difficult.
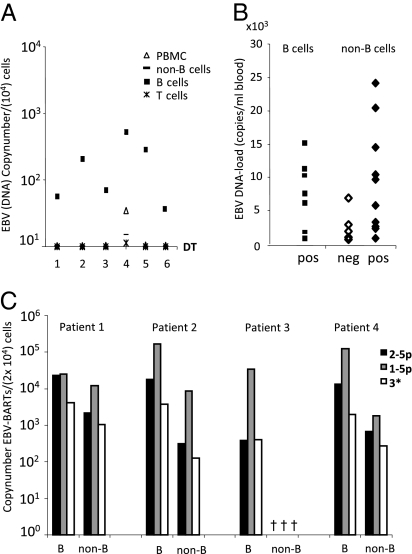
To overcome this difficulty, we investigated BART miRNA expression in circulating B cells from asymptomatic HIV-infected patients with increased EBV-DNA loads. These patients have a restricted EBV-gene-expression pattern similar to that seen in healthy individuals (29). To confirm the cellular tropism of EBV for B cells, we fractionated peripheral blood mononuclear cells (PBMCs) in T-, B-, and non-B-cell populations by cell sorting and measured the EBV-DNA load (DNA copies/mL blood) by quantitative DNA-PCR that targets a conserved region of the viral genome (29). As expected, EBV-DNA was restricted to the B-cell population and was not detectable in T cells or monocytes that make up the bulk of the non-B-cell population (Fig. 4A). In these B-cell fractions we readily detected a subset of mature BART miRNAs (Fig. 4B). Surprisingly, in ∼60% (n = 15) of the patients, we also detected these BART miRNAs in the non–EBV-carrying non-B cells (Fig. 4B). The likelihood of detecting BART miRNAs in non-B cells was dependent on viral load. However, when normalized to cell numbers, the relative abundance in B cells and non-B cells was strikingly similar, arguing against contamination (Fig. 4C). We determined the fractionated non-B-cell populations to be ∼97% pure, suggesting a maximum of ∼3% contaminating B cells. B-cell contamination therefore is unlikely to be the reason we detected BART miRNAs in the non-B-cell fractions, first because infection frequencies are very low, even in immune-suppressed patients with elevated EBV levels (29, 30), and second because the viral genome was undetectable in the non-B cells. Overall, our data suggest that in asymptomatic patients BART miRNAs are expressed by latently infected circulating B cells but also are present in noninfected non-B cells, suggesting miRNA transfer in vivo.
Fig. 4.
EBV-miRNAs are present in infected B cells and noninfected non-B cells in asymptomatic patients. (A) PBMCs from patients with various EBV loads were fractionated in non-B-cell, B-cell, and T-cell populations. Quantitative EBV-DNA PCR was performed. Shown are the number of EBV-DNA copies/104 cells. (B) Quantitative RT-PCR using RNA from fractioned B cells and non-B cells for BART1-5p, BART2-p, and BART3* EBV-miRNAs. Represented on the x axis are samples in which one or more EBV-miRNAs were detected in B cells (filled squares) or non-B cells (filled diamonds). The y axis shows the corresponding viral DNA loads (copies/mL blood). In 40% of the non-B-cell samples we were unable to detect any EBV-miRNAs (open diamonds). (C) Quantitative multiplex RT-PCR for three EBV-miRNAs in purified B-cell and non-B-cell fractions (2 × 104 cells) of four patients with varying EBV-DNA loads (copies/mL blood): 23,000 (patient 1), 10,000 (patient 2), 5,000 (patient 4), and 300 (patient 3). Shown are the copy numbers in log-scale for BART1-5p (black bars), BART2-p (gray bars), and BART3* (white bars) individually in each of the two cell fractions for each patient.
Discussion
We propose that miRNAs function in a paracrine-like fashion between immune cells through the intercellular exosomal pathway. In vitro studies revealed that the persistent human γ-herpes virus EBV induces gene repression in neighboring noninfected cells through exosomal transference of EBV-miRNAs. Such an adaptation to host cell biology is in agreement with the hypothesis that herpes viruses evolved to encode viral miRNAs and exploit the host cell miRNA machinery for their own benefit (2, 11).
Studies describing exosome physiology have been confined mostly to in vitro models using purified exosomes. The coculture method we describe here to demonstrate EBV-miRNA transfer may be more physiologically relevant than the use of purified exosome preparations alone. Nevertheless, we detected thousands of individual miRNA copy-numbers in as little as 0.5 ng of exosomal RNA. These levels are physiologically relevant, because the minimum threshold in mammalian cells to repress a target mRNA was estimated to be 100 miRNA copies (31). Indeed, the EBV-miRNAs secreted by LCL through exosomes were not inconsequential but, upon transfer, led to a reproducible and significant dose-dependent miRNA-mediated repression of mRNA targets in multiple cell types, including primary MoDC.
Whether EBV-miRNAs are secreted through exosomes and modulate gene expression in adjacent or distant cells in humans remains to be resolved. It perhaps is relevant that exosome miRNA transfer in vivo is likely to operate in the tumor or lymph node microenvironment, where much higher concentrations of exosomes may be present than described here for circulating cells and culture supernatants. This further emphassizes the potential role of miRNA and exosomal transfer in EBV biology. From the published literature, it is nevertheless clear that cellular miRNAs are present in exosomes from both cultured cells and human sera (5–7, 32). Interestingly, we detected significant numbers of BART-miRNAs in circulating noninfected non-B cells, suggesting that these miRNAs may have been transferred because the EBV genome was absent in these cells. Additionally, BART1-5p was consistently the most abundant miRNA in both circulating infected B cells and in the noninfected non-B cells, and we could not detect BHRF1-3 in either the B cells or the non-B cells. Taken together, these results suggest that EBV-miRNAs are transferred in vivo. The functional significance of these phenomena is the focus of further investigations.
Because multiple studies show that miRNAs are present in microvesicles/exosomes secreted by multiple cell types in culture and human sera (5–7, 32), a cellular miRNA-loading mechanism may exist that directs miRNAs to the intraluminal vesicles of MVEs. Indeed two independent reports showed that the RNA-induced silencing complex (RISC) is closely associated with MVEs that regulate miRNA-mediated gene silencing (25, 33). Because RISC proteins have been detected in exosomes (25, 33), it is tempting to speculate that miRNA loading into exosomes is controlled by RISC-MVE association. Interestingly, in one LCL cell-line (X50-7) some equally expressed EBV-miRNAs were 10,000-fold less frequent in exosomes, suggesting selective miRNAs were loaded into intraluminal vesicles of MVEs in these cells. Finally, the observation that MVEs are linked to miRNA physiology could also explain why exogenous exosome-associated miRNAs are capable of silencing genes in recipient cells. Exosomes internalized by DC localize to late endosomes (24), the newly identified subcellular compartments for mRNA recognition and miRNA-mediated gene silencing (25, 33).
In summary, we show in this report that viral (EBV) miRNAs are secreted from infected B cells and are functional upon transfer via exosomes in primary MoDC. EBV-miRNAs are present in circulating, noninfected non-B cells, suggesting that EBV-miRNAs transfer from infected to noninfected cells in vivo. Previous studies showed the presence of cellular miRNAs in vesicles isolated from human peripheral blood (5–7). Taken together these data are consistent with the notion of functional miRNA transfer through exosomes as a possible mechanism of intercellular communication and immune regulation (34), although alternative methods of transfer cannot be ruled out (8, 35).
Materials and Methods
Cell Culture Exosome Isolation, Purification, and RNA/DNA Isolation.
EBV-positive LCLs RN (B95-8, kindly provided by W. Stoorvogel), IM1 (spontaneous LCL), X50-7, and EBV-negative (BJAB) B-cell lines were cultured in RPMI-1640 (BioWitthaker), supplemented with 10%, exosome-depleted, FBS (HyClone; Perbio Sciences). Exosomes were isolated and purified from the supernatants of B-cell cultures using the differential centrifugation protocol as described previously (17). Exosomes were pelleted and washed at 70,000 × g for 1 h (2×) and finally were dissolved in 200 μL PBS and analyzed by EM and Western analysis to confirm the presence and purity of exosomes. Exosome purifications did not contain EBV virus as judged by negative findings in a virus capsid antigen-p40 and gp350/220 Western analysis. Total RNA from exosomes/cultured cells and clinical samples was isolated using TRIzol (Invitrogen). When low yields were expected , 5 μL glycogen (Roche) was added to the isopropanol precipitation step. Intact exosomes preps were pretreated with 10–400 μg/mL RNase A (Sigma) before TRIzol RNA isolation when indicated. The amount, quality, and composition of isolated RNA was analyzed by the NanoDrop 1000 spectrophotometer (Thermo Scientific) for total RNA and an Agilent 2100 Bioanalyzer for small RNA profiles. DNA isolation was performed using a silica-based method described previously (29). The generation of primary (immature) MoDC is explained in detail in SI Materials and Methods.
Patients and Clinical Specimens.
Random asymptomatic HIV carriers (n = 197) visiting the Slotervaart Hospital (Amsterdam, the Netherlands) for routine check-up between 2004 and 2006 were enrolled in a previously published study (29). Whole blood (≈10 mL) was collected from these individuals for routine diagnostic testing for plasma HIV-RNA load and CD4 T-cell counts . Blood not used for these purposes was used for EBV research purposes as described herein.
EBV-DNA, -RNA, and -miRNA Detection by Quantitative PCR and Multiplex RT-PCR.
Multiplex EBV-miRNA RT-PCR was performed as previously described using stem-loop RT primers for maximal 10 EBV-miRNAs in one RT reaction (13). A list of all primers and probes used in this study is provided in Table S2.
Exosome Transfer Studies.
LCL were labeled with PKH67 dye (Sigma) according to manufacturer's protocol and seeded into porous 1-μm 24-well Transwell devices (Corning-Costar). Six-day-old differentiated MoDC (CD14−, CD1a+ as determined by FACS) were placed in the bottom well and analyzed by FACS for PKH67 uptake in a 6- to 24-h period after EDTA treatment. Total MoDC RNA at 24 h and 48 h was isolated with the TRIzol method for detection of EBV-miRNAs by quantitative RT-PCR.
Cloning, Transfection, and Luciferase Reporter Assays.
Total cDNA from RN and BJAB cells was used to isolate the 3′-UTRs of LMP1 and CXCL11, respectively, by PCR. Forward primer 5′-ACGTACTAGTGCCTTCTAGGCATTACCATGTC-3′ and reverse primer 5′-ACGTAAGCTTGCTGCATCACAAGTCACATCAA-3′ were used for the 3′-UTR of LMP1 (GenBank accession number X01995), and forward primer 5′-ACGTACTAGTGCATATGAAGTCCTGGAAAAGG-3′ and reverse primer 5′-ACGTAAGCTTGCGAAAGGTTGTGGTAGTTTAT-3′ were used for the 3′-UTR of CXCL11 (GenBank accession number NM_005409). PCR products were cloned into the SpeI and HindIII sites of pMir-Report vector (Applied Biosystems). The mutated 3′-UTR-CXCL11 was purchased from Geneart. Transfection of pMir-3′-UTR-LMP1 or pMir-3′-UTR-CXCL11 into differentiated MoDC was achieved by Nucleofection (Amaxa). Cells always were cotransfected with a plasmid containing an expression cassette for Gaussia luciferase for normalization.
Supplementary Material
Acknowledgments
The authors thank Drs. W. Stoorvogel for the B95-8 RN LCL cell line and J. Neefjes for the CD63 and anti-HLA-DR antibodies, Tineke Vendrig and H. Janssen for assisting with EM analysis, Dr. D. Hayward for providing EBV-miRNA BART cluster constructs, Drs. C. Jimenez and S. Piersma for helpful assistance with proteomic analysis, and A. Zomer, A. Muggen, J. Oldenburg, and B. van Thiel for their contributions to the LCL-MoDC co-culture model. D.M.P. is funded by the Netherlands Organization for Scientific Research (NWO-Veni). The work was funded in part by Grant KWF2007-3775 from the Dutch Cancer Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914843107/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.Dolken L, et al. Mouse cytomegalovirus microRNAs dominate the cellular small RNAs profile during lytic infection and show features of post-transcriptional regulation. J Virol. 2007;81(24):13771–13782. doi: 10.1128/JVI.01313-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 8.Belting M, Wittrup A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: Implications in health and disease. J Cell Biol. 2008;183:1187–1191. doi: 10.1083/jcb.200810038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 10.Cullen BR. Viral RNAs: Lessons from the enemy. Cell. 2009;136:592–597. doi: 10.1016/j.cell.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 12.Thorley-Lawson DA. Epstein-Barr virus: Exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 13.Cosmopoulos K, et al. Comprehensive profiling of EBV microRNAs in nasopharyngeal carcinoma. J Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia T, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by EBV-MIR-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–397. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raposo G, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middeldorp JM, Pegtel DM. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol. 2008;18:388–396. doi: 10.1016/j.semcancer.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Wubbolts R, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 20.Cameron JE, et al. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–266. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bickham K, et al. Dendritic cells initiate immune control of Epstein-Barr virus transformation of B lymphocytes in vitro. J Exp Med. 2003;198:1653–1663. doi: 10.1084/jem.20030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim WH, Kireta S, Russ GR, Coates PT. Human plasmacytoid dendritic cells regulate immune responses to Epstein-Barr virus (EBV) infection and delay EBV-related mortality in humanized NOD-SCID mice. Blood. 2007;109:1043–1050. doi: 10.1182/blood-2005-12-024802. [DOI] [PubMed] [Google Scholar]
- 23.Montecalvo A, et al. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during T cell allorecognition. J Immunol. 2008;180:3081–3090. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- 24.Morelli AE, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 25.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 26.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo AK, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 29.Stevens SJ, et al. Aberrant Epstein-Barr virus persistence in HIV carriers is characterized by anti-Epstein-Barr virus IgA and high cellular viral loads with restricted transcription. AIDS. 2007;21:2141–2149. doi: 10.1097/QAD.0b013e3282eeeba0. [DOI] [PubMed] [Google Scholar]
- 30.Gross AJ, Hochberg D, Rand WM, Thorley-Lawson DA. EBV and systemic lupus erythematosus: a new perspective. J Immunol. 2005;174:6599–6607. doi: 10.4049/jimmunol.174.11.6599. [DOI] [PubMed] [Google Scholar]
- 31.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 32.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 35.Rechavi O, et al. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971–1979. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.