Abstract
The development of hippocampal circuitry depends on the proper assembly of correctly specified and fully differentiated hippocampal neurons. Little is known about factors that control the hippocampal specification. Here, we show that zinc finger protein Zbtb20 is essential for the specification of hippocampal CA1 field identity. We found that Zbtb20 expression was initially activated in the hippocampal anlage at the onset of corticogenesis, and persisted in immature hippocampal neurons. Targeted deletion of Zbtb20 in mice did not compromise the progenitor proliferation in the hippocampal and adjacent transitional ventricular zone, but led to the transformation of the hippocampal CA1 field into a transitional neocortex-like structure, as evidenced by cytoarchitectural, neuronal migration, and gene expression phenotypes. Correspondingly, the subiculum was ectopically located adjacent to the CA3 in mutant. Although the field identities of the mutant CA3 and dentate gyrus (DG) were largely maintained, their projections were severely impaired. The hippocampus of Zbtb20 null mice was reduced in size, and exhibited increased apoptotic cell death during postnatal development. Our data establish an essential role of Zbtb20 in the specification of CA1 field identity by repressing adjacent transitional neocortex-specific fate determination.
Keywords: cell fate, neocortex, zinc finger transcription factor
The hippocampus is an archicortical structure located at the medial-temporal edge of the neocortex. It can be divided into distinct cytoarchitectonic regions: the CA1 and CA3 of Ammon’s horn, and the dentate gyrus (DG) (1). All three regions consist of a principal layer of densely packed neurons—pyramidal cells in the CA1 and CA3, and granule cells in the DG. The subiculum and the transitional neocortex (also known as mesocortex) lie between Ammon’s horn and the neocortex. The subiculum comprises a principal cell layer of large, less-compact pyramidal neurons, whereas the cytoarchitecture of the transitional neocortex is distinct from the archicortex in that the principal laminae comprise several subsets of relatively less compact neurons organized into deep and upper layers (2).
The complex developmental cell fate decisions that orchestrate the partitioning of the cerebral cortex into neo- and archicortices remain poorly understood. The hippocampal primordium develops at the caudomedial edge of the dorsal telencephalon and is flanked dorsally by the transitional neocortex primordium. During embryonic stages, it consists of three distinct germinative zones that give rise to CA pyramidal neurons and DG granule neurons (3). The cortical hem, an embryonic structure located adjacent to the ventral edge of the hippocampal anlage, releases patterning cues and functions as a hippocampal organizer (4). In the absence of Wnt3a from the hem, or loss of Wnt signaling component LEF1 or β-catenin in the presumptive anlage, the hippocampal primordium fails to develop (5, 6). Furthermore, a number of other factors have been found to participate in the initial phase of hippocampal development by regulating Wnt signaling, including FOXG1, FGF, BMPs, Gli3, and EMX1/2 (7, 8). However, little is known about how hippocampal field identities are specified and how differential cell fate decisions in the hippocampal and transitional neocortex primordia are developmentally controlled.
Zbtb20, a new member of the BTB/POZ zinc finger family, plays a variety of important roles in multiple systems, as suggested by the severe phenotypes in the mice lacking Zbtb20 (9). In liver, Zbtb20 acts as a transcriptional repressor of alpha-fetoprotein gene (10). It has been implied that Zbtb20 may also be involved in corticogenesis (11), but its physiological role in hippocampal development has not been defined. In this study, we present evidence that Zbtb20 is essential for the specification of CA1 field identity by repressing the acquisition of transitional neocortex cell fates.
Results
Zbtb20 Is Expressed in the Developing Hippocampal Anlage.
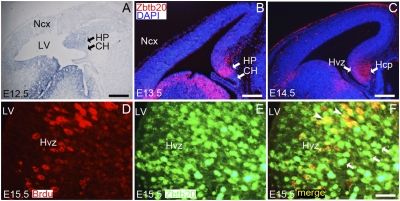
Zbtb20 mRNA was first detected by in situ hybridization in the hippocampal primordium as early as embryonic day (E) 12.5, and was greatly increased by E13.5 (Fig. 1Aand Fig. S1A). Anti-Zbtb20 immunostaining revealed that Zbtb20 protein was expressed within the E13.5 hippocampal primordium, but was excluded from the cortical hem and neocortical region (Fig. 1B). By E14.5, when immature pyramidal cells from Ammon’s horn start to migrate out of the proliferative ventricular zone (VZ) to form the cortical plate (CP) (12), Zbtb20 expression was observed in presumptive postmitotic pyramidal cells in the CP and, to a much less extent, in the VZ (Fig. 1C). In addition, robust Zbtb20 expression was detected in the dentate migratory path and the DG anlage at E16.5 (Fig. S1B). Zbtb20 expression was maintained in the hippocampus throughout late embryonic stages and early postnatal development (Fig. S1 B and C), but was absent in subiculum, which is consistent with the previous report (11).
Fig. 1.
Zbtb20 expression in the developing hippocampus. (A) Zbtb20 expression was examined by in situ hybridization on the sagittal section of E12.5 forebrain. (B and C) Zbtb20 expression was detected by immunohistochemistry using anti-Zbtb20 antibody 9A10 on the coronal sections through the forebrain at E13.5 (B) or E14.5 (C). (D–F) Double-immunostaining of anti-BrdU and anti-Zbtb20 on the E15.5 coronal brain sections. (F) The majority of BrdU+ cells in the hippocampal VZ (Hvz) were Zbtb20+ (yellow; indicated by arrowheads), and a large number of Zbtb20+ cells outside the Hvz were BrdU− (green; indicated by arrows). HP, hippocampal primordium; Ncx, neocortex; Hcp, hippocampal cortical plate; LV, lateral ventricle. (Scale bars: 250 μm for A and C; 175 μm for B; 30 μm for D–F.)
To further define the expression distribution of Zbtb20, we pulse labeled with bromodeoxyuridine (BrdU) at E15.5 for 1 h, the stage at which the generation of pyramidal cells in Ammon’s horn is most active (4). We found that the majority of BrdU-positive cells in the hippocampal VZ were colabeled with anti-Zbtb20, and the majority of Zbtb20-positive cells located outside the VZ were not labeled with BrdU, indicating that these were postmitotic cells (Fig. 1 D–F). In summary, Zbtb20 transcripts are detected in the hippocampal primordium as early as E12.5, and thereafter Zbtb20 is expressed specifically in progenitor cells and postmitotic neurons of the developing hippocampus.
Cytoarchitectonical Transformation in Zbtb20−/− CA1.
To investigate the role of Zbtb20 in hippocampal development, we analyzed the hippocampal anlage of Zbtb20 null mice and found that, though it did not differ noticeably from wild-type or heterozygous littermate controls with respect to its size, cell density, and cellular distribution pattern before E16.5 (Fig. S2 A and B), morphological defects became apparent by E18.5. At this developmental time point, the wild-type hippocampal anlage has a thin and compact CP that is clearly distinguishable from the intermediate zone (IZ) and the CP of the adjacent transitional neocortex. In contrast, the E18.5 CA1 region of Zbtb20−/− mice was wider and less dense, and the reticular IZ was thinner than that in control and thus appeared very similar to the adjacent transitional neocortex (Fig. S2 C and D).
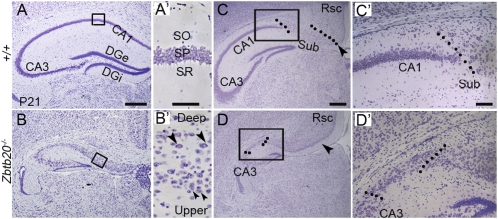
This transformation became progressively more conspicuous during early postnatal development, when both the hippocampus and transitional neocortex gradually acquire their mature features (Fig. S2 E–H). By postnatal day 21 (P21), the pyramidal cell layer in the CA1 region of Zbtb20 null mice was divided into two principal layers: a compact upper layer comprising small neurons and a deep layer with relatively large, loosely packed neurons. This formation of deep and upper lamination is highly reminiscent of the dorsal transitional neocortex (13). In addition, the stratum radiatum (SR) and the stratum oriens (SO), normally located on either side of the pyramidal layer in wild type, were unidentifiable in Zbtb20 null mice (Fig. 2 A and B and A′ and B′). At more caudal levels, the subiculum lies between the CA1 and the retrosplenial cortex (Rsc) in wild-type mice. However, the subiculum appeared to be located adjacent to CA3 region in mutant, as shown by the appearance of a single principal cell layer with relatively large, loosely packed neurons (2) (Fig. 2 C and D and C′ and D′). Taken together, these findings suggest that Zbtb20 ablation leads to the transformation of the CA1 field into a transitional neocortex-like structure and the corresponding translocation of subiculum adjacent to the CA3.
Fig. 2.
Abnormal cytoarchitecture of the Zbtb20−/− hippocampus. (A–D) Nissl-stained coronal forebrain sections at P21. (A′ and B′) High-magnification views of the boxed areas in A and B, respectively. The normally single layer of the stratum pyramidale (SP; A′) was transformed into two principal layers in the mutant CA1 field (B′): an upper layer (small arrowheads) and a deep layer (large arrowheads). (C and D) At a more caudal level, a subiculum-like structure appeared adjacent to the mutant CA3. Dotted lines indicate boundaries of subiculum. (C′ and D′) High-magnification views of the boxed areas in C and D, respectively. Sub, subiculum. (Scale bars: 400 μm for A–D; 50 μm for A′ and B′; 100 μm for C′ and D′.)
In addition to the CA1, the CA3 and DG also exhibited severe morphological defects in Zbtb20 knockout mice. At E18.5, the mutant CA3 began to exhibit a wider and more loosely packed CP than in wild type (Fig. S2 C and D). In contrast to the mutant CA1, the mutant CA3 maintained a single pyramidal layer comprising morphologically homogenous neurons during postnatal development (Fig. 2 A and B and Fig. S2 E–H). However, compared with the wild-type control, the mutant DG was obviously smaller at E18.5 (Fig. S2 C and D), and exhibited shorter bladelike structure during postnatal development, with the external blade (DGe) even much shorter (Fig. S2 E–H). At P7, many DG neurons in mutant were still present in the DG migratory path from dentate VZ to DG anlage, whereas few were observed in the wild-type counterpart. At P21, only a residual of DG was left in mutant (Fig. 2 A and B). These data indicate the severe defects of DG development in the absence of Zbtb20.
Increased Apoptosis in the Postnatal Hippocampus of Zbtb20 Null Mice.
Beginning at P3, the hippocampus of Zbtb20 null mice was smaller and less compact than in wild-type and heterozygous littermates, and the density of cells progressively decreased during postnatal development (Fig. 2 A and B and Fig. S2 E–H). This reduction could be due to a decrease in cell proliferation, an increase in cell death, and/or neuronal disorganization. To evaluate the cell proliferation of hippocampal precursors, we conducted BrdU incorporation assays at E14.5, the peak period of pyramidal neuron generation in Ammon’s horn (4), and at E18.5, when DG granule neurogenesis is most active (14). After 1 h of pulse, the frequency of BrdU-labeled cells both in the E14.5 hippocampal and transitional VZ and in the E18.5 DG were comparable between Zbtb20 mutant and wild-type embryos (Fig. S3 A–F), which indicate that Zbtb20 ablation does not alter the precursor proliferation in the hippocampal or adjacent transitional VZ.
To determine the apoptotic cell death of hippocampal neurons, we performed TUNEL labeling. At E18.5, a few TUNEL-positive cells were detected in the mutant hippocampus, with no significant difference from the wild-type control (Fig. S3 G–I). From P0 through P21, however, there was a remarkable increase in the number of apoptotic cells in the mutant hippocampus (Fig. S3 I–K). At P7, for example, there was a 10-fold increase in TUNEL-positive cells in mutant hippocampus compared with control. These data suggest that the reduced hippocampus in Zbtb20−/− mice is mainly caused by the increased postnatal apoptosis of hippocampal neurons.
Ectopic Expression of Transitional Neocortex-Specific Genes in Zbtb20−/− CA1.
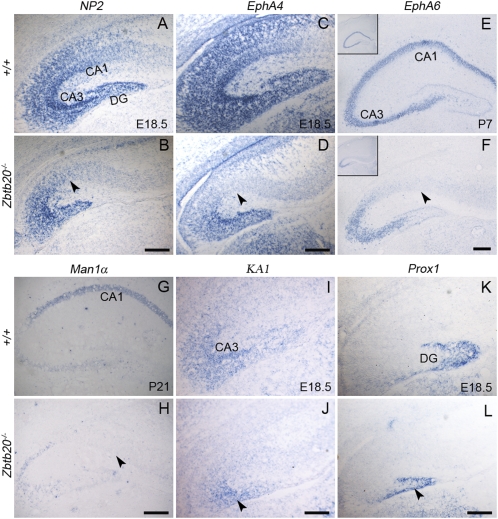
To determine its molecular identity, we then evaluated the expression of hippocampal-specific genes in Zbtb20−/− mice. NP2 and EphA4 are normally expressed in the entire hippocampus from early embryonic development (15, 16), but were greatly reduced in the mutant CA1 field at E16.5, 2 days before the onset of the cytoarchitectonical abnormalities we observed (Fig. S4 A–D). By E18.5, both factors were almost undetectable in the mutant CA1, although they were expressed in the CA3 and DG fields of knockouts (Fig. 3 A–D). Similarly, EphA6 is strongly expressed by postnatal wild-type CA1 and CA3 (http://www.brain-map.org/), but was almost lost in the mutant CA1 (Fig. 3 E and F). Furthermore, Mannosidase 1 alpha (Man1α), a specific marker of mature CA1 neurons (17), was totally absent from the Zbtb20−/− CA1 region at P21 (Fig. 3 G and H). In contrast to its homogenous expression in wild-type CA1, Oct6 was detected in some neurons scattered in the mutant CA1 region, with a pattern reminiscent of that in transitional neocortex (Fig. S4 E and F) (18). These findings strongly indicate that the neurons in the mutant CA1 region have lost their CA1 identity.
Fig. 3.
Loss of hippocampus-specific markers in the Zbtb20−/− CA1 region. Coronal forebrain sections were detected for the indicated genes by in situ hybridization at the indicated stages. (A–F) NP2, EphA4, and EphA6 expression was present in the mutant CA3 and DG, but undetectable in the mutant CA1 (arrowheads in B, D, and F). (G and H) CA1-specific Man1α expression was totally lost in the mutant CA1 (arrowhead in H). (I–L) Decreased KA1 and Prox1 expression in the mutant CA3 (arrowhead in J) and DG (arrowhead in L), respectively. (Scale bars: 200 μm for A–F and I–L; 350 μm for G and H.)
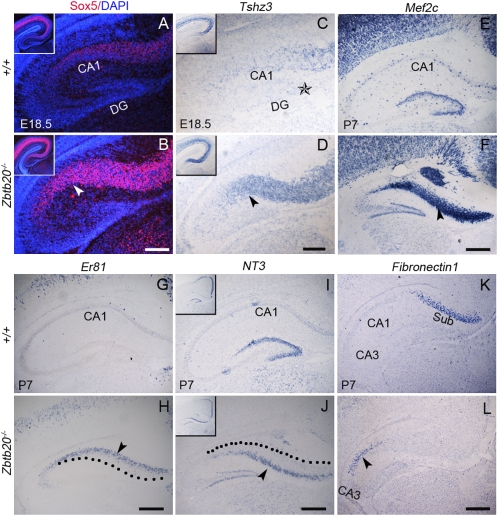
We next analyzed the expression of various transitional neocortex-specific genes in the mutant CA1 region. At E16.5, the expression domains of Sox5 (a layer V, VI, and subplate marker) (19), Tshz3 (a marker of all neocortical layers) (20), and Id2 (a layer II–III, V–VI, and subplate marker) (21) were found to expand ventrally in mutants (Fig. S5 A–F) and were present in the E18.5 CA1 (Fig. 4 A–D and Fig. S5 G and H). Moreover, the expression domain of neurotrophin 3 (NT3, a transient marker for upper layer in cingulate and retrosplenial cortices) (5), was also expanded ventrally in mutants at E18.5 (Fig. S5 I and L), and NT3-positive neurons were distributed throughout the upper layer of the mutant CA1 region at P7 (Fig. 4 I and J). ER81 (a layer V marker) (22), Cux2 (a layer II–IV marker) (23), and Mef2c (a layer I–VI marker) (24) were ectopically observed in the mutant CA1 region (Fig. 4 E–H and Fig. S5 K and L). Furthermore, at more caudal levels, the expression domain of Fibronectin1 (a marker for subiculum), normally located between CA1 and retrospenial cortex in wild type (http://www.brain-map.org/), was ectopically present adjacent to the lateral border of the CA3 (Fig. 4 K and L). These observations, together with the loss of CA1-specific markers, suggest that the neurons in mutant CA1 region have acquired a transitional neocortex-like identity. Interestingly, none of these genes were ectopically expressed in the CA3 and DG fields. Thus, both histological and genetic evidence strongly indicates that Zbtb20 ablation leads to the ventral expansion of transitional neocortex and a shift of subiculum position at the expense of the CA1 region. Given the apparently normal cell proliferation of the progenitors in hippocampal and transitional VZ, the cytoarchitectonical transformation is most likely caused by the misspecification of the mutant CA1 precursors.
Fig. 4.
Ectopic expression of transitional neocortex-specific genes in the Zbtb20−/− CA1 region. Coronal forebrain sections were detected for indicated genes by immunohistochemistry (A and B) or in situ hybridization (C–L) at the indicated stages. The arrowheads in B and D indicate the mutant CA1 field. The star in C indicates the notch at the DG crest. The arrowheads in H and J indicate the deep and upper layer of the mutant CA1, respectively. The dotted lines in H and J outline the upper and deep boundary of mutant CA1, respectively. (Insets) Low-magnification views of the hippocampus (A–D and I–J). (K and L) At caudal level, the expression domain of Fibronectin1, a marker for subiculum (K), was shifted adjacent to the mutant CA3 (L). (Scale bars: 200 μm for A–D; 250 μm for E–L.)
Although CA3 neurons in mutants still expressed NP2 and EphA4, in situ signals appeared to be somewhat weak in comparison with wild-type controls (Fig. 3 A–D and Fig. S4 A–D). Similarly, KA1 expression in the CA3, and Prox1 in the DG was reduced in mutant mice from E16.5 onward (Fig. 3 I–L and Fig. S4 G–L). These suggest that in the absence of Zbtb20, the specification of CA3 and DG neurons is largely maintained, although their differentiation is impaired.
Transitional Neocortex-Like Migratory Behavior of the Zbtb20−/− CA1 Neurons.
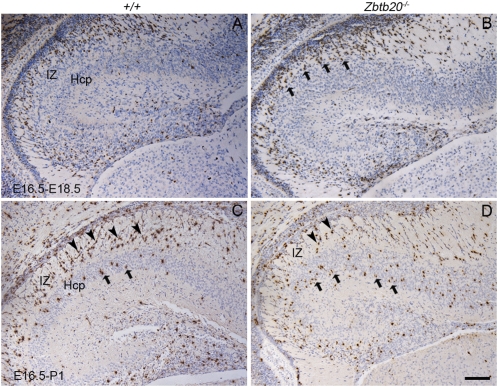
Hippocampal progenitors exhibit distinct migratory behaviors from their counterparts of transitional neocortex. To examine the migration of hippocampal neurons, we pulse labeled the neurons born at E16.5 with BrdU and analyzed their migration at E18.5 and P1. Consistent with previous reports (12), most wild-type CA1 neurons born at E16.5 still remained within the IZ at E18.5 and P1, which resulted in a compact IZ (Fig. 5 A and C). In contrast, some of the mutant CA1 neurons born at E16.5 had crossed the IZ and reached the CP by E18.5 (Fig. 5B), and the majority of them had penetrated the CP by P1 (Fig. 5D). These findings revealed that the mutant CA1 neurons migrated more rapidly through the IZ to the CP than their wild-type counterparts, at a rate similar to transitional neocortex neurons (12). Therefore, the altered migratory behavior also supports the notion that the mutant CA1 neurons have acquired the identity of transitional neocortex neurons. In addition, the accelerated migration may partly explain the appearance of a less-compact mutant CA1 IZ at E18.5 (Fig. S2 C and D).
Fig. 5.
Accelerated migratory behavior of the Zbtb20−/− CA1 neurons. Hippocampal progenitors were labeled by BrdU incorporation at E16.5, and subjected to BrdU immunostaining at E18.5 or P1. The majority of wild-type CA1 neurons born at E16.5 (BrdU+ cells, dark brown) were still in the IZ (arrowheads in C) by P1 (A and C), whereas some of BrdU+ cells in the mutant CA1 had crossed the IZ and reached the CP by E18.5 (arrows in B), with the majority of them penetrating into the CP by P1 (arrows in D). (Scale bars: 175 μm.)
Impaired Neuronal Connections in Zbtb20−/− Hippocampus.
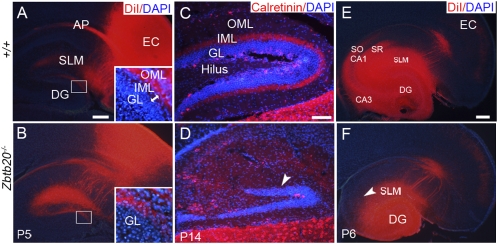
To examine the entorhinal projections to DG, we placed small crystals of DiI into entorhinal cortex (EC). Consistent with the previous report (25), the entorhino-hippocampal axons terminated in the stratum lacunosum-moleculare (SLM) and the outer molecular layer (OML) of DG in P5 wild-type mice. In Zbtb20 null mice, DiI-labeled axons in the SLM were increased, but those in the OML were comparable to wild-type DG (Fig. 6 A and B). The inner molecular laye (IML) could be clearly identified in wild-type DG with no entorhino-hippocampal axons, but it was invisible in mutant DG. To distinguish two possibilities of either abnormal penetration of entorhino-hippocampal projections into the IML or loss of the IML in Zbtb20 mutant DG, we examined expression of Calretinin, which is indicative for the axons of mossy cells in the IML (7). Calretinin-positive fibers were densely distributed in wild-type IML, but absent in mutant DG (Fig. 6 C and D), indicating the loss of IML in mutant DG. After placing DiI in the DG of P14 mice, the mossy fibers were clearly visualized in the stratum lucidum of wild-type CA3, but absent in the mutant (Fig. S6 A and B). Similarly, Schaffer collaterals, which extend from CA3 to CA1, were present in the wild types rather than the mutants after placing DiI in the CA3 and DG (Fig. 6 E and F). Taken together, the tracing results show the remarkable defects in hippocampal neuronal network in Zbtb20 mutant mice.
Fig. 6.
Impaired connections in Zbtb20−/− hippocampus. (A and B) Horizontal brain sections taken after the placement of DiI in the collateral EC at P5. (Insets) High-magnification views of the boxed areas in A and B, respectively. (C and D) Immunohistochemistry study for Calretinin-positive fibers on P14 saggital sections. (E and F) Schaffer collaterals projecting from the CA3 to the SO and SR of CA1 were detectable in wild-type hippocampus (E) but not in the mutant (F) after placing DiI in the P6 CA3 and DG. (Scale bars: 200 μm for A, B, E, and F; 100 μm for C and D.)
Apparently Normal Wnt Signaling in the Zbtb20−/− Hippocampal Primordium.
To determine whether the loss of Zbtb20 affects Wnt signaling in the CA region, we examined Wnt pathway components at E13.5–14.5, when Zbtb20 is remarkably expressed with the occurrence of active hippocampal neurogenesis. Consistent with previous reports (26), we found that in wild-type mice the Wnt receptor Frizzled 5 (Fzd5) and secreted frizzled-related protein 1 (SFRP1) were expressed in the transitional/neocortical VZ, whereas SFRP3 was expressed in the hippocampal and transitional VZ, but excluded from the neocortex (Fig. S7 A–F). Wnt inhibitory factor (WIF) expression was restricted to the hippocampal VZ just dorsal to the cortical hem, whereas Axin2, an intracellular negative regulator of Wnt signal transduction and a known downstream target of canonical Wnt signaling, was highly expressed in the hem and the adjacent hippocampal VZ (Fig. S7 G–J). In Zbtb20 mutant mice, the pattern of expression of each of these Wnt pathway members was not notably different from wild type. Furthermore, the β-catenin activity at E13.5, detected broadly in the telencephalic VZ and the cortical hem, was similar between Zbtb20 knockouts and wild-type mice (Fig. S7 K and L). These data suggest that loss of Zbtb20 does not lead to significant alterations of canonical Wnt signaling in hippocampal primordium. In addition, by immunohistochemistry analysis, we did not find any remarkable change of the activating status of the signaling components of TGF-β, BMP, or FGF in the mutant hippocampal and adjacent transitional VZ (Fig. S7 M–R).
Altered Gene Expression Profile in Zbtb20−/− Hippocampus.
To define the transcriptional signature of the mutant hippocampus, we performed transcript profiling experiments. It turned out that there were a total 153 up-regulated and 192 down-regulated unique probe sets that exhibited a minimum 2-fold change and reached statistical significance. Among of them, some were indicated by bioinformatic analysis to participate in neuron development, migration, apoptosis, and axon guidance (Table S1). Notably, the increased expression of Tshz3, NT3, Mef2c, and Id2 in knockout hippocampus is consistent with their ectopic expression in mutant CA1 (Fig. 4 C–F, I, and J and Fig. S5 C–J). The up-regulation of Tbr1 and the orphan nuclear receptor Nr4a2 (a marker for subiculum and deep layer projection neurons) (27) and down-regulation of the trophic factor BDNF (a survival factor of hippocampal progenitors) (28) in mutant hippocampus were confirmed by RT-PCR (Fig. S6C), the latter may partly account for the increased apoptosis of the mutant hippocampal neurons. Taken together, these findings indicate that Zbtb20 orchestrates the expression of key factors for hippocampal development.
Discussion
In this study, we examined the role of Zbtb20 in hippocampal development. Our findings provide compelling evidence that Zbtb20 is essential for the specification of CA1 field identity and the postnatal survival of hippocampal neurons.
Our findings suggest that the hippocampal CA1 field from the mice lacking Zbtb20 is transformed into a transitional neocortex-like structure and the subiculum is correspondingly shifted into the lateral border of mutant CA3. Although Zbtb20−/− CA1 progenitor cells seemed to proliferate normally in the hippocampal primordium, they migrated rapidly to the CP, pausing for an abnormally short period in the IZ, in a manner similar to that of transitional neocortex progenitors. The CA1 fields of Zbtb20−/− mice lacked the typical compact and homogeneous composition of the pyramidal cell layer and instead developed deep and upper cortical layers reminiscent of the transitional neocortex. Remarkably, Zbtb20−/− CA1 neurons failed to express or expressed reduced levels of CA1 markers, such as NP2, EphA4, EphA6, and Man1α, but maintained Oct6 expression, which is normally expressed in CA1 and transitional neocortex. Furthermore, the mutant CA1 field ectopically expressed many transitional neocortex-specific markers, including the transcription factors Sox5, Tshz3, Id2, Mef2c, ER81, Cux2, and the trophic factor NT3. In addition, the expression domain of Fibronectin1, a subiculum-specific marker, was shifted laterally and located adjacent to mutant CA3. When the results of this study are placed in context with previous work that misexpression of Zbtb20 induces a CA1-like transformation of the transitional neocortex and subiculum (11, 29), we conclude that Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Whether Zbtb20 accordingly plays a role in the projections of CA1 neurons remains to be defined.
Explant culture experiments have suggested that the hippocampal primordium is well established by E12.5 (3), supporting a “protomap” model, which posits that embryonic patterning cues act on cortical progenitor cells to specify the different areas of the mature cortex (8). During the initial phase of hippocampal development (before E12.5), Wnts from the cortical hem are critical for the expansion of, and most likely the selection of, a pool of hippocampal progenitor cells (5, 6). Our study showed that the loss of Zbtb20 did not cause any notable alterations to the expression and activity of both canonical and noncanonical Wnt signaling components in the hippocampal region at embryonic stages. These observations suggest that Wnt signaling alone is not sufficient to induce archicortex differentiation. This raises the possibility that Wnt signaling directs the expansion of hippocampal progenitors, and Zbtb20 thereafter directs the specification of presumptive hippocampal, particularly CA1, progenitors. Alternatively, Zbtb20 may be an obligate downstream target of Wnt signaling in the specification of CA1 cell fates. At present, we do not have data to distinguish between these two possibilities. Given that Zbtb20 is activated in the hippocampal anlage at E12.5, before the onset of CA1 differentiation (which begins at E15.5), and is expressed by both proliferating and postmitotic hippocampal progenitor cells, we reason that Zbtb20 is necessary to maintain archicortical potential in the presumptive CA1 subpopulation of multipotent cortical progenitors, and/or to maintain CA1 area identities in postmitotic neurons by suppressing transitional neocortex and subiculum differentiation regimes. However, to determine the exact roles of Zbtb20 in progenitor cells and postmitotic neurons, experiments would need to be conducted wherein Zbtb20 expression is temporally controlled in hippocampal precursors and postmitotic neurons, respectively. Notably, it was recently reported that down-regulation of Zbtb20 expression by RNA interference from E15 results in the loss of Calbindin-D28k expression by CA1 neurons (29), although their cell fate was not elaborately determined.
Another intriguing finding in this study is the field-dependent manner in which Zbtb20 controlled hippocampal development. In contrast to its essential role in the specification of CA1 neurons, Zbtb20 appeared to be dispensable for the specification of CA3 pyramidal neurons and DG granule cells, whose identities were largely maintained in Zbtb20 knockout mice. This CA1-specific effect of Zbtb20 was also observed when Zbtb20 was ectopically expressed in transitional neocortex (29). This supports the hypothesis that hippocampal subfields are patterned by unique mechanisms (18). Early differentiation of the CA1 and CA3 regions begins at the two hippocampal poles respectively, and progresses inwards, suggesting that the signals that specify the two CA field identities are derived from organizers close to each pole (18). Interestingly, we observed that the expression domain of transitional neocortex markers encroached into the CA1 region of Zbtb20 mutants gradually, from dorsal to ventral and eventually expanding to, but not beyond, the CA1/CA3 boundary. This phenomenon suggests that, although Zbtb20 is expressed in the CA3 region from E12.5 and into postnatal development, this subfield relies on separate mechanisms to suppress the expression of transitional neocortex patterning genes.
Our study also indicates that Zbtb20 is required for the postnatal survival of hippocampal neurons. We found that increased apoptosis occurred throughout the entire region of Zbtb20 null hippocampus during postnatal development. This increase was not likely to be an indirect effect of global metabolic dysfunction (9), because the majority of Zbtb20−/− mice appeared normal and indistinguishable from wild-type controls before P10, although they later developed hypoglycemia. Therefore, we reason that the high levels of postnatal cell death in hippocampal regions may result from cell-autonomous defects, probably including the decreased BDNF expression. It is also possible that nonautonomous factors contributed to the occurrence of increased cell death. In Zbtb20 knockout mice, the disruption of appropriate network connections would diminish synaptic activity and trophic support.
Taken together, this study establishes a critical role for Zbtb20 in the specification of CA1 field identity and in the maintenance of neuronal survival in the developing hippocampus, and provides insight into the regulation of corticogenesis.
Materials and Methods
Animals.
Zbtb20 null mice were described previously (9). The wild-type or heterozygous littermates were used as control. Mice were kept in a specific pathogen-free (SPF) facility, and all animal experiments were preformed according to institutional guidelines. Tissues were harvest as described in SI Text.
In Situ Hybridization.
Antisense riboprobes labeled with digoxygenin-UTP were transcribed from cDNA clones (SI Text and Table S2). After overnight hybrization with riboprobes, coronal forebrain cryosections were detected with anti-digoxygenin (Roche) antibody conjugated to alkaline phosphatase, and developed by nitro blue tetrazolium.
Immunohistochemistry.
Tissue sections were incubated with the appropriate primary antibody (SI Text) and visualized with the indirectly coupled Alexa Fluor 594 (SI Text). DAPI was used for nuclear counterstaining. For Zbtb20 and BrdU colabeling, embryos were pulse labeled with BrdU for 1 h by i.p. injection into timed-pregnant female. Then BrdU was detected by Alexa Fluor 594, and Zbtb20 visualized with Alexa Fluor 488.
Cell Proliferation and Apoptosis Assays.
For proliferation assays, the embryos were labeled with BrdU as described, and the forebrain sections analyzed for BrdU incorporation. TUNEL staining was performed for apoptosis assay. The frequency of labeled cells was measured by cells/100 μm2 area before statistical test using Student’s t test. See also SI Text for details.
Neuronal Migration Assay in Vivo.
Timed pregnant mice were injected i.p. with BrdU (50 mg/kg body weight) at E16.5, and brains were harvested at E18.5 or P1. Coronal forebrain sections were subjected to BrdU immunostaining.
DiI Tracing Study.
Small crystals of FAST DiI were inserted into collateral EC or DG and CA3, brains were incubated in 4% paraformaldehyde for 1∼10 days, sectioned into 60 μm thickness, counterstained with DAPI, and photographed under fluorescence light (SI Text).
Transcript Profiling.
P2 hippocampi were pooled for RNA extraction. Microarray analysis was performed using Affymetrix mouse 430 2.0 chip (SI Text). All data sets are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). Some of the differentially expressed transcripts were confirmed by in situ hybridization or real-time RT-PCR (SI Text).
Supplementary Material
Acknowledgments
We thank Drs. F. Guillemot and Q. Ma for providing plasmids, and Prof. Yizhang Chen for his critical review of the manuscript. This work was supported by National Natural Science Foundation of China Grant 30770456 (to W.Z.) and 30970589 (to Z. Xie), National “973” Program of China Grant 2006CB503910 (to W.Z.), and National “863” Program of China Grant 2007AA02Z173 (to W.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.R.R. is a guest editor invited by the Editorial Board.
Data deposition: All data sets have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no GSE19513).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912315107/DCSupplemental.
References
- 1.Witter M, Amaral DG. In: The Rat Nervous System. Paxinos G, editor. San Diego: Elsevier Academic; 2004. pp. 635–704. [Google Scholar]
- 2.Ishizuka N. Laminar organization of the pyramidal cell layer of the subiculum in the rat. J Comp Neurol. 2001;435:89–110. doi: 10.1002/cne.1195. [DOI] [PubMed] [Google Scholar]
- 3.Tole S, Grove EA. Detailed field pattern is intrinsic to the embryonic mouse hippocampus early in neurogenesis. J Neurosci. 2001;21:1580–1589. doi: 10.1523/JNEUROSCI.21-05-01580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 5.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 6.Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122:129–143. doi: 10.1016/s0306-4522(03)00519-0. [DOI] [PubMed] [Google Scholar]
- 7.Förster E, Zhao S, Frotscher M. Laminating the hippocampus. Nat Rev Neurosci. 2006;7:259–267. doi: 10.1038/nrn1882. [DOI] [PubMed] [Google Scholar]
- 8.Hébert JM, Fishell G. The genetics of early telencephalon patterning: Some assembly required. Nat Rev Neurosci. 2008;9:678–685. doi: 10.1038/nrn2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutherland AP, et al. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol. 2009;29:2804–2815. doi: 10.1128/MCB.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z, et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105:10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen JV, Nielsen FH, Ismail R, Noraberg J, Jensen NA. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development. 2007;134:1133–1140. doi: 10.1242/dev.000265. [DOI] [PubMed] [Google Scholar]
- 12.Nakahira E, Yuasa S. Neuronal generation, migration, and differentiation in the mouse hippocampal primoridium as revealed by enhanced green fluorescent protein gene transfer by means of in utero electroporation. J Comp Neurol. 2005;483:329–340. doi: 10.1002/cne.20441. [DOI] [PubMed] [Google Scholar]
- 13.Lopes da Silva FH, Witter MP, Boeijinga PH, Lohman AH. Anatomic organization and physiology of the limbic cortex. Physiol Rev. 1990;70:453–511. doi: 10.1152/physrev.1990.70.2.453. [DOI] [PubMed] [Google Scholar]
- 14.Reznikov KY. Cell proliferation and cytogenesis in the mouse hippocampus. Adv Anat Embryol Cell Biol. 1991;122:1–74. doi: 10.1007/978-3-642-76447-9. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Chédotal A, He Z, Goodman CS, Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 16.Liebl DJ, Morris CJ, Henkemeyer M, Parada LF. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J Neurosci Res. 2003;71:7–22. doi: 10.1002/jnr.10457. [DOI] [PubMed] [Google Scholar]
- 17.Lein ES, Callaway EM, Albright TD, Gage FH. Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. J Comp Neurol. 2005;485:1–10. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- 18.Tole S, Christian C, Grove EA. Early specification and autonomous development of cortical fields in the mouse hippocampus. Development. 1997;124:4959–4970. doi: 10.1242/dev.124.24.4959. [DOI] [PubMed] [Google Scholar]
- 19.Kwan KY, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caubit X, Tiveron MC, Cremer H, Fasano L. Expression patterns of the three Teashirt-related genes define specific boundaries in the developing and postnatal mouse forebrain. J Comp Neurol. 2005;486:76–88. doi: 10.1002/cne.20500. [DOI] [PubMed] [Google Scholar]
- 21.Neuman T, et al. Neuronal expression of regulatory helix-loop-helix factor Id2 gene in mouse. Dev Biol. 1993;160:186–195. doi: 10.1006/dbio.1993.1297. [DOI] [PubMed] [Google Scholar]
- 22.Yoneshima H, et al. Er81 is expressed in a subpopulation of layer 5 neurons in rodent and primate neocortices. Neuroscience. 2006;137:401–412. doi: 10.1016/j.neuroscience.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 23.Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
- 24.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng JB, Yu DM, Wu P, Li MS. The tracing study of developing entorhino-hippocampal pathway. Int J Dev Neurosci. 2007;25:251–258. doi: 10.1016/j.ijdevneu.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Diep DB, Hoen N, Backman M, Machon O, Krauss S. Characterisation of the Wnt antagonists and their response to conditionally activated Wnt signalling in the developing mouse forebrain. Brain Res Dev Brain Res. 2004;153:261–270. doi: 10.1016/j.devbrainres.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Arimatsu Y, Ishida M, Kaneko T, Ichinose S, Omori A. Organization and development of corticocortical associative neurons expressing the orphan nuclear receptor Nurr1. J Comp Neurol. 2003;466:180–196. doi: 10.1002/cne.10875. [DOI] [PubMed] [Google Scholar]
- 28.Choi SH, Li Y, Parada LF, Sisodia SS. Regulation of hippocampal progenitor cell survival, proliferation and dendritic development by BDNF. Mol Neurodegener. 2009;4:52. doi: 10.1186/1750-1326-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen JV, Blom JB, Noraberg J, Jensen NA. Zbtb20-induced CA1 pyramidal neuron development and area enlargement in the cerebral midline cortex of mice. Cereb Cortex. 2009 doi: 10.1093/cercor/bhp261. 10.1093/cercor/bhp261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.