Abstract
A systematic approach to the discovery of conformation-specific antibodies or those that recognize activation-induced neoepitopes in signaling molecules and enzymes will be a powerful tool in developing antibodies for basic science and therapy. Here, we report the isolation of antibody antagonists that preferentially bind activated integrin Mac-1 (αMβ2) and are potent in blocking neutrophil adhesion and migration. A novel strategy was developed for this task, consisting of yeast surface display of Mac-1 inserted (I) domain library, directed evolution to isolate active mutants of the I domain, and screening of phage display of human antibody library against the active I domain in yeast. Enriched antibody library was then introduced into yeast surface two-hybrid system for final quantitative selection of antibodies from monomeric antigen–antibody interaction. This led to highly efficient isolation of intermediate to high affinity antibodies, which preferentially reacted with the active I domain, antagonized the I domain binding to intercellular adhesion molecule (ICAM)-1, complement C3 fragment iC3b, and fibronectin, and potently inhibited neutrophil migration on fibrinogen. The strategy demonstrated herein can be broadly applicable to developing antibodies against modular domains that switch between inactive and active conformations, particularly toward the discovery of antibody antagonists in therapeutic and diagnostic applications.
Keywords: antibody antagonist, mac-1 integrin, phage display, yeast display
Conformational change of proteins is essential to affinity regulation in cell signaling molecules, interactions of viral proteins with host receptors, and proteins with enzymatic activity (1). Antibodies that can recognize the activation-induced epitopes (“neoepitope”) may in turn cause activation and/or probe different activation states of the molecules. In contrast to conformation-insensitive antibodies, neoepitope-specific antibodies may be used to diagnose activation state of cells and to deliver therapeutic agents to specific cells (2, 3). Some antibodies produced by animal immunization are specific to active conformation of antigens (4–6). Alternative to screening hybridoma antibodies for conformation- or activation-specific properties, in vitro methods can be used to isolate antibodies by repeated cycles of selection and depletion (subtractive panning) against wanted and unwanted antigens, respectively (7–9). In this regard, phage display systems of naïve, immunized, or synthetic antibody libraries (10, 11) have been most successful in selection of human antibodies against purified proteins, surface molecules of mammalian cells, or a library of antigens in yeast cells.
Complex linkage between conformation and affinity regulation exists in integrins, which are transmembrane heterodimers consisting of noncovalently associated α and β subunits (12). As leukocyte infiltration into the tissue requires activation of integrins, integrins expressed in leukocytes are important therapeutic targets in autoimmune and inflammation-related diseases. This is evidenced by the fact that antibody antagonists targeting leukocyte-specific integrins such as lymphocyte function-associated antigen (LFA)-1 (αLβ2), Mac-1 (αMβ2), αIIbβ3, and α4β1/α4β7 have been approved or are under preclinical and clinical studies (13–15). To reduce potential unwanted side effects caused by indiscriminate binding of antibodies to integrins, systematic in vitro approaches have been utilized to produce antibodies that are more specific to activated integrins (8, 9, 16). For example, a series of subtractive panning of phage library against different forms of Mac-1 expressed in mammalian cells (9) or the inserted (I) domain of LFA-1 (2, 8) have been used to isolate single chain antibodies that preferentially react with activated integrins.
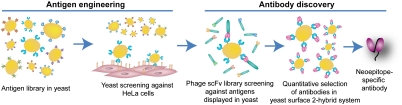
The I domain, a major ligand binding site in the I domain-containing integrins, exists in multiple conformations of low to high affinity to ligands, which is coupled to the transition from an inactive to an active state of integrins (17–19). Antibodies specific to the high affinity conformation of the I domain, therefore, may also be specific to active integrins in cells. Due to the fact that I domain is the ligand binding site, such antibodies will likely be activation-specific antagonists. Similar to the previous approach of selecting antibody library against antigen library (11), we examined if protein domains displayed on the surface of yeast and engineered for high affinity by directed evolution can be used to screen phage clones. We first isolated activating mutations by screening a yeast library of Mac-1 I domains against HeLa cells expressing ligands for Mac-1. Then phage library were panned against the active I domain in yeast, where the enrichment of reactive phage clones was monitored by flow cytometry. From enriched phage libraries, cDNAs encoding single chain fragment variable (scFv) antibody and active Mac-1 I domain were introduced as a pair of the bait and prey proteins into the yeast surface two-hybrid system (YS2H) (20) that was designed for quantitative estimation of protein–protein interactions. Antibodies selected from two steps of phage binding to yeast and antigen–antibody interactions in YS2H were found to react with the active Mac-1 I domain and be potent in blocking neutrophil adhesion and migration. The streamlined process of antigen display and engineering in yeast display system (21), phage panning directly with antigens displayed in yeast, and quantitative selection of antibodies in YS2H can be adapted to the discovery of neoepitope-specific antibodies against other mammalian proteins.
Results
Overview of Antibody Selection Strategy.
Various mammalian proteins have been expressed functionally in yeast as a fusion to a cell wall protein called agglutinin (22–26). To isolate mutations that induce active conformation, a mutagenesis library of the Mac-1 I domain was sorted against HeLa cells, which express ICAM-1 and other ligands for Mac-1 (Fig 1). Against yeast cells expressing active mutant of Mac-1 I domain, a phage library of human single chain antibody (scFv) was panned to select for activation-specific antibodies. Using immunofluorescence flow cytometry, phage binding to yeast cells was monitored by antibody binding to the His tag, which is placed between the scFv and the phage coat protein, pIII. With successive sorting, the enrichment of reactive phage clones led to an increase in antiHis antibody binding. Next, the scFv cDNA library amplified from enriched phage clones was then introduced into the YS2H vector, where the affinity between antigen and antibody can be estimated by the flow cytometry measurement of antitag antibody binding. Final selection of antibodies in the YS2H enabled quantitative assessment of candidate antibodies according to their 1∶1 binding affinity to the antigen.
Fig. 1.
Streamlined antibody selection strategy. A mutagenesis library of antigens expressed in yeast display system (21) is subjected to directed evolution to isolate mutations to induce active conformation, to which a phage library of human single-chain antibody is screened. Final selection of specific antibodies is made in yeast surface 2-hybrid system (20) according to quantitative assessment of monomeric antigen–antibody interactions.
Selection of Active Mac-1 I Domain by Directed Evolution.
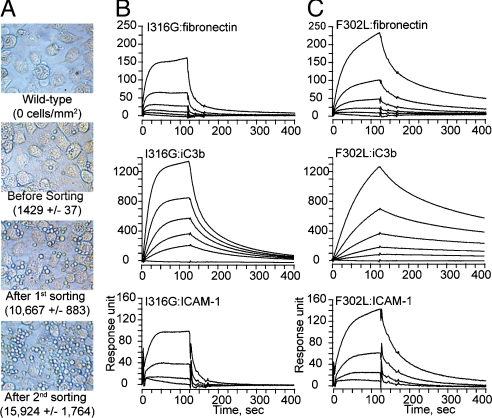
The I domain in integrin Mac-1 exists at low and high affinity conformation, which is stabilized in inactive and active state of full-length Mac-1, respectively. To isolate mutations that would induce high affinity conformation of the I domain, we constructed a mutagenesis library of the I domain (Asp132—Thr322) in yeast display and sorted yeast cells by their binding to HeLa cells. Whereas yeast cells expressing the wild-type I domain were easily washed out from HeLa, the number of bound yeast cells progressively increased with successive sorting (Fig 2A). High affinity binders from the second sort library were collected and grown in SDCAA plate. Twenty-four clones from the plate were individually tested for binding to HeLa, from which five unique clones containing one or two mutations were isolated. These five clones bound to HeLa better than the second sort library (Fig S1). One highest binder was found to contain Leu substitution for Phe-302. Phe-302 is located in the loop between the β6-strand and the α7-helix (Fig S1B), which was identified as an activation hot spot in Mac-1 (F302W) (18) and its equivalent position as a hot spot in LFA-1 (F292G) (22). To confirm that the mutation of F302L induced high affinity conformation of the I domain, the I domains with the mutations of F302L and the previously identified I316G (27) were produced in bacteria and tested for binding to ICAM-1, iC3b, and fibronectin using surface plasmon resonance (Fig 2 B and C). The mutation of I316G would disrupt van der Waals interactions of Ile-316 with the neighboring residues in a low affinity conformation (Fig S1E), inducing high affinity conformation (27). Whereas the wild-type I domain showed little binding to fibronectin and ICAM-1, and low-level binding to iC3b (KD = 1.8 μM) (Table S1), the active mutants of F302L and I316G bound much stronger to all three ligands (Fig 2 B and C). Overall, the F302L exhibited slower kinetics in both association (kon) and dissociation rates (koff) than the I316G, and bound with higher affinity to the ligands (Fig 2 B and C and Table S1).
Fig. 2.
Selection of high affinity Mac-1 I domains. (A) A mutagenesis library of Mac-1 I domain in yeast was screened with HeLa cells to isolate high affinity mutants. Yeast cells appear as small bright spheres in the images. (B and C) SPR measurement of the binding of active I domains (I316G and F302L) to ICAM-1, iC3b, and fibronection. I domains were injected at a series of two-fold serial dilutions starting at 250 nM.
Selection of Antibody Library Against Yeast Cells Expressing Active Mac-1 I Domain.
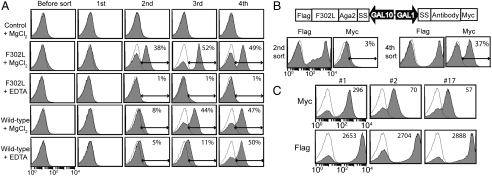
Nonspecific binders of phage clones were first depleted by incubation with yeast cells expressing unrelated proteins. The remaining phage clones were then panned against the yeast cells expressing Mac-1 I domain with F302L. The binding of the enriched phage clones to the F302L after each round of sorting was examined by flow cytometry (Fig 3A). The binding of the second sort phage library to F302L was metal-ion dependent (38% vs. 1% with EDTA), indicating that the metal-ion dependent adhesion site (MIDAS) is the key epitope for antibody binding. Positive binding to the wild-type I domain was also observed with the third and fourth round sort. The fourth sort library contained phage clones that reacted with the wild-type I domain with or without the metal-ion. For the final selection of antibodies, we constructed the YS2H vector with the F302L cDNA as the bait and the scFv cDNA as the prey proteins that were cloned from the second and the fourth sort libraries (Fig 3B). In YS2H, the expression of Myc and Flag tag indicates, respectively, the amount of antibody captured by the antigen and the level of antigen expression. Although the second and the fourth sort phage libraries bound comparably to yeast cells (Fig 3A), when the pair of antigen and antibody were expressed in yeast, yeast expressing the second sort antibody library exhibited much lower Myc expression than the fourth sort library (3% vs. 37%) (Fig 3B). This discrepancy may arise from the excess of phage clones used in flow cytometry, which resulted in comparable binding to F302L irrespective of the difference in the percentage of reactive clones in the second vs. the fourth sort library. This is in contrast to Myc expression in YS2H, which reports the percentage of positive clones with mean fluorescence intensity correlating with the antigen–antibody interaction strength. Yeast cells expressing the fourth sort scFv library were grown in SDCAA plate, from which twenty-four clones were individually tested for tag expression. Nine clones among the twenty-four clones displayed positive Myc tag expression; from sequencing, three unique clones were identified. Whereas Flag tag expression was comparable in all three clones (clones 1, 2, and 17 were designated as AM01, AM2, and AM17), the level of Myc expression varied considerably with highest expression seen in clone AM01. Using the first-order Langmuir binding isotherm (SI Methods), the affinity (KD) for AM01 and AM17 was determined at 34 nM and 241 nM, respectively. Sequencing analysis revealed that these three clones were identical in the framework regions, differing only in the complementarity determining regions 2 and 3 (CDR2 & CDR3) (Fig S2).
Fig. 3.
Selection of phage library against the F302L displayed in yeast. (A) A phage library after each round of panning was examined for binding to the F302L and the wild-type I domain using flow cytometry. Unless noted otherwise, open and filled histograms correspond to antibody binding to uninduced and induced cells, respectively. Control indicates yeast cells expressing unrelated proteins. (B) The cDNAs amplified from the second and fourth round sort were introduced into the YS2H vector along with the F302L for final selection of antibodies. (C) Selected individual clones exhibit differential level of Myc expression, correlating with antibody-F302L binding affinity.
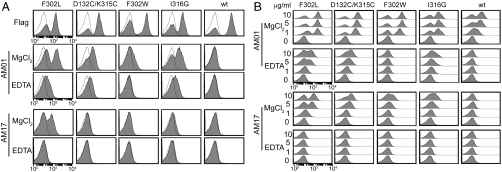
Soluble Antibody Binding to Mac-1 I Domain Expressed in Yeast.
In YS2H, the prey proteins exist on cell surface due to their binding to the bait as well as in culture media; the level of soluble protein in the media was estimated to be around 10 nM for scFv (Fig S3) (20). Antibody concentration in the media was high enough for flow cytometry application without the need for purification. These antibodies were examined for binding to the active I domain mutants that were induced by different mutations [D132C/K315C (28), F302W, and I316G] to confirm that selected antibodies are activation-specific but not specific to the mutation itself (Fig 4A). Single chain antibodies AM01 and AM17 both bound the F302L in a metal-ion dependent manner (10 mM MgCl2 vs. 10 mM EDTA). AM01 exhibited higher affinity binding than AM17, and bound to other active mutants. To further examine antigen–antibody binding affinity, AM01 and AM17 were expressed from bacteria and used at 1, 5, and 10 μg/ml to label the wild type and the active I domain mutants (Fig 4B). Overall, the binding of purified antibodies to I domains was in good agreement with the antibodies produced in yeast: Whereas AM01 bound the active I domains much higher than the wild type, the binding of AM17 was more specific to the F302L, displaying low-level binding to D132C/K315C, F302W, and I316G and no binding to the wild-type I domain. Without the metal ions, AM01 exhibited much lower binding to the active I domains, whereas binding was undetectable with AM17. It can be hypothesized that although the metal ion in the MIDAS in the active conformation is a critical epitope for both antibodies, other activation-independent epitopes may contribute to AM01 binding.
Fig. 4.
The measurement of antibody binding to the Mac-1 I domains. (A) Yeast culture supernatant containing antibodies were used without purification for binding to I domain expressing yeast cells. (B) To further confirm antibody specificity, AM01 and AM17 were produced from bacteria and used at 0–10 μg/mL concentrations for binding to yeast cells expressing I domain variants.
Surface Plasmon Resonance (SPR) Measurement of Mac-1 Binding to Activation-Specific or Activiation-Insensitive Antibodies.
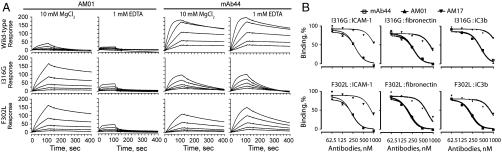
A number of antibodies against the Mac-1 I domain have previously been isolated, some of which react differentially with the activation state of Mac-1. mAb44 was found to bind Mac-1 irrespective of its activation state in a metal-ion independent manner (27). To compare newly selected AM01 with activation-insensitive mAb44, antibody binding to the wild type and the active I domains (I316G and F302L) was examined by flowing I domains over the SPR chip immobilized with AM01 and mAb44 (Fig 5A). The configuration of antibodies immobilized to the surface and the I domains as analytes was chosen to examine monomeric interaction, which was necessary as AM01 is monovalent whereas mAb44 is divalent. Consistent with AM01 binding to the I domains expressed in yeast cells (Fig 4), AM01 bound the active mutants much higher affinity (KD = 2 nM) than the wild-type I domain (KD = 66 nM) with strong dependence on the metal ion (Fig 5A). In contrast to preferential binding of AM01 to active I domains, mAb44 bound better to the wild-type I domain (KD = 0.4 nM) with little influence by the metal ions (Fig 5A and Table S1).
Fig. 5.
SPR measurement of the affinity of antibody to I domains and inhibition of I domain-ligand interactions. (A) Antibodies were immobilized to a Biacore chip, and the I domains were injected at a series of two-fold serial dilutions starting from 12.5 nM in buffer containing either 10 mM MgCl2 or 1 mM EDTA. (B) The I domains at 200 nM were preincubated with the antibodies, ranging from 62.5 to 1000 nM, and this complex was injected into a chip coated with Mac-1 ligands. The solid lines represent the best fit of the model to the data, from which the values of a half maximal inhibitory concentration (IC50) of antibodies were derived.
Inhibition of Mac-1 Binding to Ligands by Antibodies.
Therapeutic potency of antibody antagonists can be quantified as the concentration of antibody that produces half maximal inhibition of receptor binding to ligands or the half maximal inhibitory concentration (IC50). To measure the potency of inhibition of Mac-1 binding to ligands, Mac-1 I domains (I316G and F302L at 200 nM) mixed with scFv or antibodies (1000 to 62.5 nM) were injected over the SPR chip coated with ICAM-1, iC3b, and fibronectin (Fig 5B). The observations that AM01 and AM17 binding to the I domains was strongly dependent on the metal ions (Fig 4) suggest that the MIDAS is a critical epitope and antibodies should compete with the ligands for binding to the I domain. Under this condition, the IC50 of the antibodies should be proportional to the affinity of the antibody (or inversely proportional to the dissociation constant, KD) to antigen. The mAb44 and AM01 exhibited comparable inhibitory potency (IC50), whereas 3- to 4-fold higher concentration of AM17 was required to produce comparable inhibition (Fig 4 and Table S2). Higher potency of AM01 and mAb44 than AM17 agreed with their higher affinity to the active I domains.
Inhibition of Neutrophil Adhesion and Migration by Antibodies.
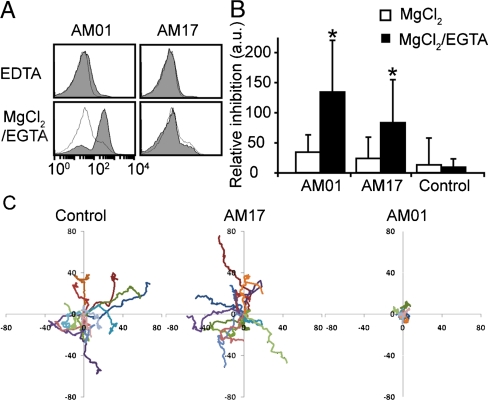
Next, we analyzed the binding of antibodies to neutrophils and inhibition of neutrophil adhesion and migration on fibrinogen-coated surface. To activate integrins without inducing cellular activation (29), neutrophils were treated with 1 mM MgCl2 and 1 mM EGTA to deplete free calcium ions and labeled with AM01 or AM17. Whereas little binding of AM01 and AM17 to neutrophils was observed without EGTA or with EDTA, AM01 binding increased significantly when cells were treated with 1 mM MgCl2 and 1 mM EGTA (Fig 6A). In contrast to its binding to the F302L expressed in yeast, the binding of AM17 to neutrophils was not detected with MgCl2/EGTA activation. This discrepancy was presumably due to the difference in the degree of activation by F302L vs. by the depletion of calcium ions in Mac-1. To evaluate the potency of antibodies in inhibiting neutrophil binding to fibrinogen-coated surface, 96-well V-bottom plate assay was used to measure the amount of detached cells by centrifugal force. In this assay, more cells would accumulate to the V-bottom tip if the affinity between antibody antagonist and Mac-1 is higher. Neutrophils were incubated for 15 min at RT in 20 μg/mL of antibodies with or without activation by EGTA, washed, and added to the wells coated with fibrinogen. Whereas the inhibition of neutrophil adhesion to fibrinogen without EGTA activation was not statistically significant, after EGTA activation significant inhibition of neutrophil adhesion was observed by AM01 and AM17 at 40% and 25% (the percent relative inhibition as defined in Methods), respectively (Fig 6B). Incomplete inhibition by antibodies may be due to the activation of Mac-1 after washing out unbound antibodies caused by the force applied to neutrophils during centrifugation of the plate, ligand-induced activation, and nonspecific neutrophil adhesion to the plate. To examine if these antibodies are capable of blocking neutrophil migration after stimulation with 10 nM formyl-Met-Leu-Pro (fMLP), which is the condition that would not only activate integrins but also upregulate Mac-1 expression (4), the migration of activated neutrophils on fibrinogen-coated surface was analyzed. Compared with no antibody control, preincubation of neutrophils with AM01 resulted in highly restricted migration within 15 μm from an initial attachment site. In contrast, AM17 did not significantly inhibit neutrophil migration over control.
Fig. 6.
Evaluation of antibodies in inhibition of neutrophil adhesion and migration. (A) Activation-dependent binding of antibodies to neutrophils was examined with immunofluorescence flow cytometry. The histograms represent antiHis antibody binding with (Filled) and without (Open) antibodies against Mac-1. (B) The inhibition of neutrophil adhesion to fibrinogen by antibodies was measured using 96-well V-bottom plate assays. Student’s t tests were used to determine statistical significance (p < 0.05 for * vs. control). (C) The migration of neutrophils on fibrinogen-coated surface after10 nM fMLP activation was recorded and analyzed. Each line represents the movement of single neutrophils.
Discussion
Antibodies specific to activated cell surface molecules may be used to probe cellular state as well as to target activated cells for the delivery of therapeutic agent (3). With protein domains expressed on the surface of yeast as an antigen, we demonstrate a streamlined approach applicable to developing activation-specific antibodies against the I domain-containing integrins. Compared to the previous approaches using soluble proteins as antigens (7, 8), the use of yeast cells expressing antigen offers several advantages such as obviating the need to purify soluble proteins and direct estimation of antibody affinity using immunofluorescence flow cytometry. More importantly, antigens displayed in yeast display system can be engineered by directed evolution approach to induce active conformation, which would mimic the conformation induced by activation signals in cells (22). Compared to rationally designed mutations, directed evolution approach led to mutations more potent in inducing active conformation of Mac-1 I domain, consistent with our previous studies with LFA-1 I domain (22). Compared to the diversity in cell surface molecules in mammalian cells, the protein antigens expressed in yeast as a fusion to agglutinin would likely represent one of the most abundant cell surface proteins (30). This can lead to highly efficient selection of specific phage clones, as evidenced by the isolation of positive clones after as few as two rounds of sorting. The use of YS2H for the final stage of antibody selection by quantitative estimation of antigen-antibody affinity was proven effective in identifying single chain antibodies of differing affinity. This method can overcome the problem of selecting phage clones biased toward those expressing multimeric scFv or with higher titer (31).
In contrast to depleting antibody library against the inactive I domains to select for activation-specific antibodies (8), we used yeast cells expressing unrelated proteins to deplete nonspecific binders. Even without the depletion against the inactive I domains, antibodies selected against the active I domain (F302L) (e.g., AM01 and AM17) preferentially bound or were specific to the active I domains induced by various mutations. We also found that a subset of enriched phage clones reacted with the wild-type I domain as well, which are likely to be activation-insensitive. Notably, all the reactive phage clones against the F302L were metal-ion dependent. Structural changes of the integrin I domains coupled to different affinity states have been studied extensively (18, 22, 32): It involves the rearrangement of the metal-ion coordinating and proximal residues in the MIDAS and the downward displacement of the C-terminal α7-helix. Therefore, some of the activation-specific antibodies may be specific to the residues in the MIDAS or in the α7-helix, not necessarily requiring the metal ions. The dominance of the metal ion in the MIDAS as an activation-specific epitope may be attributed to unstructured nature of the C-terminal region containing the α7-helix (19, 33) and may also reflect the nature of Mac-1 I domain in recognition of diverse molecules largely dependent on the electrostatic potential in the MIDAS.
The use of YS2H in the final selection of antibodies led to a number antibodies varying in affinities to the active I domain. We have chosen AM01 and AM17 that represented Myc expression highest to lowest among the selected yeast clones. Previously we have demonstrated that the affinity between two interacting proteins in YS2H can be quantitatively estimated from flow cytometry measurement of tag expression. Using the Langmuir isotherm equation, the predicted affinities (KD) were 34 nM for AM01 vs. 241 nM for AM17 to the F302L. These values closely approximate the SPR data indicating 3- to 4-fold higher concentration of AM17 than AM01 to induce comparable inhibition of the I domain binding to the ligands and the solution affinity of AM01 binding to F302L (KD = 2 nM). Quantitative selection of antibodies in YS2H based on 1∶1 interaction of antigen-antibody can be a powerful method in selecting antibodies of desired affinities to achieve an optimal level of antibody binding to cells and antigens.
The antibodies isolated in this study, such as those that preferentially bind activated Mac-1 can be developed into antagonists targeting aberrantly active leukocytes or to diagnose inflammatory diseases associated with inflamed leukocytes. It is hypothesized that aberrant or excessive infiltration of immune cells, wherein integrin activation may be dysregulated, lead to direct damage to the vasculature and the underlying tissue, and the diseases such as sepsis, cardiovascular disease, and other inflammation-related diseases (34, 35). Among leukocyte-specific integrins, Mac-1 (αMβ2) is expressed predominantly in phagocytic, myeloid cells such as neutrophils, monocytes, and macrophages, and is distinct in its ability to interact with a wide-range of ligands including ICAMs, fibrinogen, fibronectin, heparin, and iC3b (36). Due to this reason, the diseases associated with myeloid cells such as ischemia-reperfusion injuries in cerebral and myocardial infarction and sepsis have been targeted with antagonists to β2 integrins or to Mac-1 (15, 37, 38). To assess potential use of activation-specific antibodies developed in this study, we examined the ability of antibodies to block soluble Mac-1 I domain binding to ICAM-1, fibronectin, and iC3b, and to inhibit neutrophil adhesion and migration over fibrinogen-coated surface. The degree of reduction in neutrophil adhesion and migration was in agreement with the affinity of antibody binding to Mac-1 I domain. Although AM17 was less potent than AM01 due to lower affinity to Mac-1, conversion of scFv AM17 into bivalent immunoglobulins (IgG) may induce sufficiently potent affinity to Mac-1 with little interaction with inactive Mac-1.
Many mammalian proteins consist of multidomains, some of which can be expressed separately in a native conformation retaining modular functions. The use of modular domains expressed in yeast for antibody selection not only overcomes the difficulty with the functional expression of large mammalian molecules but also offers the advantage of narrowing antibody epitopes into specific region to function as agonists or antagonists. Some of the agonistic antibodies against integrins [such as mAbs KIM127 (39) and CBR LFA-1/2 (40)] map to a region in the β2 subunit that is buried in an inactive conformation but exposed in an active conformation. Therefore, agonistic antibodies can be developed by screening phage library against the domains or the regions that become exposed only in an active conformation. Likewise, antibodies screened against the domains that interact with the ligands may become antagonist antibodies. Furthermore, by tapping into the power of directed evolution implemented by the yeast display system, the protein domains that undergo conformational change or exhibit allostery can be stabilized into one conformation for selection of conformation-specific antibodies. In summary, the strategy demonstrated in this study can be applicable to the selection of conformation-specific or activation-dependent antibodies against human antigens that are hard to express in solution, highly homologous to those in other mammals for immunization, or that undergo conformational change that needs to be locked into one conformation by mutations.
Methods
Directed Evolution of Mac-1 I Domain for Active Conformation.
Random mutagenesis library of Mac-1 I domain was constructed according to the published protocol (SI Methods) (22).
Selection of Reactive Phage Clones.
A phage library (2 × 1013 colony forming unit) of human single chain (scFv) antibody [Tomlison I/J phage libraries (41)] was incubated in 3 mL PBS containing 1 mM MgCl2 and 2% instant nonfat dry milk (Carnation) for 30 min at RT, followed by 30 min incubation at RT with 107 yeast cells expressing unrelated proteins (heat shock factor 1) in 3 mL PBS containing 1 mM MgCl2. After incubation, cells were spun down and the supernatant containing unbound phage clones was adjusted with 10 mM MgCl2, to which 107 yeast cells expressing the active Mac-1 I domain (F302L) were added. After 30 min incubation at RT, yeast cells were spun down, washed with PBS with 10 mM MgCl2, and the pellet was treated with 1 mg/mL trypsin for 10 min at RT to release bound phage from yeast cells. Phage clones eluted into the supernatant were used to infect Escherichia coli (TG1) to produce the next round of phage library. The binding of phage clones to yeast cells was monitored by immunofluorescence flow cytometry using antibodies against His-tag placed between single chain antibody and pIII coat protein.
Neutrophil Adhesion and Migration Inhibition Assay.
The 96-well V-bottom plate (Greiner) assay (22) was used to measure the potency of antibodies in blocking neutrophil binding to fibrinogen (100 μg/mL). The percent relative inhibition by antibodies was calculated as 100× (F_antibody - F_BSA) / (F_max - F_BSA), where F_antibody, F_BSA, and F_max correspond to the level of BCECF-AM fluorescence from the neutrophils incubated with antibody and BSA, and with maximum centrifugation (873 g), respectively. Cell migration assay was carried out as previously described (SI Methods) (42).
See SI Text for additional methods, figures, and tables.
Supplementary Material
Acknowledgments.
This work was supported by an American Heart Association Scientist Development Grant and Northeast Biodefense Center Grant (M.M.J.). We thank Jeffrey C. Mattison and Carissa J. Ball (Biomedical Engineering Department, Cornell University) for their assistance with neutrophil preparation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914358107/DCSupplemental.
References
- 1.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimaoka M, et al. AL-57, a ligand-mimetic antibody to integrin LFA-1, reveals chemokine-induced affinity up-regulation in lymphocytes. Proc Natl Acad Sci USA. 2006;103:13991–13996. doi: 10.1073/pnas.0605716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci USA. 2007;104:4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond MS, Springer TA. A subpopulation of Mac-1 (CD11b/CD18) molecules mediates neutrophil adhesion to ICAM-1 and fibrinogen. J Cell Biol. 1993;120:545–556. doi: 10.1083/jcb.120.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elemer GS, Edgington TS. Monoclonal antibody to an activation neoepitope of alpha M beta 2 inhibits multiple alpha M beta 2 functions. J Immunol. 1994;152:5836–5844. [PubMed] [Google Scholar]
- 6.Lu C, Ferzly M, Takagi J, Springer TA. Epitope mapping of antibodies to the C-terminal region of the integrin beta 2 subunit reveals regions that become exposed upon receptor activation. J Immunol. 2001;166:5629–5637. doi: 10.4049/jimmunol.166.9.5629. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc Natl Acad Sci USA. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, et al. Identification and characterization of a human monoclonal antagonistic antibody AL-57 that preferentially binds the high-affinity form of lymphocyte function-associated antigen-1. J Leukocyte Biol. 2006;80:905–914. doi: 10.1189/jlb.1105649.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenhardt SU, et al. Generation of activation-specific human anti-alphaMbeta2 single-chain antibodies as potential diagnostic tools and therapeutic agents. Blood. 2007;109:3521–3528. doi: 10.1182/blood-2006-03-007179. [DOI] [PubMed] [Google Scholar]
- 10.Kretzschmar T, von Ruden T. Antibody discovery: Phage display. Curr Opin Biotechnol. 2002;13:598–602. doi: 10.1016/s0958-1669(02)00380-4. [DOI] [PubMed] [Google Scholar]
- 11.Bowley DR, Jones TM, Burton DR, Lerner RA. Libraries against libraries for combinatorial selection of replicating antigen–antibody pairs. Proc Natl Acad Sci USA. 2009;106:1380–1385. doi: 10.1073/pnas.0812291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 13.Yusuf-Makagiansar H, Anderson ME, Yakovleva TV, Murray JS, Siahaan TJ. Inhibition of LFA-1/ICAM-1 and VLA-4/VCAM-1 as a therapeutic approach to inflammation and autoimmune diseases. Med Res Rev. 2002;22:146–167. doi: 10.1002/med.10001. [DOI] [PubMed] [Google Scholar]
- 14.Shimaoka M, Springer TA. Therapeutic antagonists and the conformational regulation of the beta2 integrins. Curr Top Med Chem. 2004;4:1485–1495. doi: 10.2174/1568026043387575. [DOI] [PubMed] [Google Scholar]
- 15.Yonekawa K, Harlan JM. Targeting leukocyte integrins in human diseases. J Leukocyte Biol. 2005;77:129–140. doi: 10.1189/jlb.0804460. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz M, et al. Conformation-specific blockade of the integrin GPIIb/IIIa: A novel antiplatelet strategy that selectively targets activated platelets. Circ Res. 2006;99:25–33. doi: 10.1161/01.RES.0000232317.84122.0c. [DOI] [PubMed] [Google Scholar]
- 17.Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 18.Li R, Rieu P, Griffith DL, Scott D, Arnaout MA. Two functional states of the CD11b A-domain: Correlations with key features of two Mn2+-complexed crystal structures. J Cell Biol. 1998;143:1523–1534. doi: 10.1083/jcb.143.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimaoka M, et al. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Kang S, Chen X, Shoemaker CB, Jin MM. Yeast surface two-hybrid for quantitative in vivo detection of protein–protein interactions via the secretory pathway. J Biol Chem. 2009;284:16369–16376. doi: 10.1074/jbc.M109.001743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 22.Jin M, et al. Directed evolution to probe protein allostery and integrin I domains of 200,000-fold higher affinity. Proc Natl Acad Sci USA. 2006;103:5758–5763. doi: 10.1073/pnas.0601164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieke MC, Cho BK, Boder ET, Kranz DM, Wittrup KD. Isolation of anti-T cell receptor scFv mutants by yeast surface display. Protein Eng. 1997;10:1303–1310. doi: 10.1093/protein/10.11.1303. [DOI] [PubMed] [Google Scholar]
- 24.Rao BM, Driver I, Lauffenburger DA, Wittrup KD. Interleukin 2 (IL-2) variants engineered for increased IL-2 receptor alpha-subunit affinity exhibit increased potency arising from a cell surface ligand reservoir effect. Mol Pharmacol. 2004;66:864–869. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- 25.Kim YS, Bhandari R, Cochran JR, Kuriyan J, Wittrup KD. Directed evolution of the epidermal growth factor receptor extracellular domain for expression in yeast. Proteins. 2006;62:1026–1035. doi: 10.1002/prot.20618. [DOI] [PubMed] [Google Scholar]
- 26.Lee HW, et al. Inducing rigid local structure around the zinc-binding region by hydrophobic interactions enhances the homotrimerization and apoptotic activity of zinc-free TRAIL. Biochem Biophys Res Commun. 2007;362:766–772. doi: 10.1016/j.bbrc.2007.08.075. [DOI] [PubMed] [Google Scholar]
- 27.Xiong JP, Li R, Essafi M, Stehle T, Arnaout MA. An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain. J Biol Chem. 2000;275:38762–38767. doi: 10.1074/jbc.C000563200. [DOI] [PubMed] [Google Scholar]
- 28.McCleverty CJ, Liddington RC. Engineered allosteric mutants of the integrin alphaMbeta2 I domain: structural and functional studies. Biochem J. 2003;372:121–127. doi: 10.1042/BJ20021273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dransfield I, Cabanas C, Craig A, Hogg N. Divalent cation regulation of the function of the leukocyte integrin LFA-1. J Cell Biol. 1992;116:219–226. doi: 10.1083/jcb.116.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol R. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith J, Kontermann RE, Embleton J, Kumar S. Antibody phage display technologies with special reference to angiogenesis. Faseb J. 2005;19:331–341. doi: 10.1096/fj.04-2863rev. [DOI] [PubMed] [Google Scholar]
- 32.Shimaoka M, et al. Reversibly locking a protein fold in an active conformation with a disulfide bond: Integrin alphaL I domains with high affinity and antagonist activity in vivo. Proc Natl Acad Sci USA. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, et al. An unusual allosteric mobility of the C-terminal helix of a high-affinity alphaL integrin I domain variant bound to ICAM-5. Mol Cell. 2008;31:432–437. doi: 10.1016/j.molcel.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 35.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 36.Diamond MS, Garcia-Aguilar J, Bickford JK, Corbi AL, Springer TA. The I domain is a major recognition site on the leukocyte integrin Mac-1 (CD11b/CD18) for four distinct adhesion ligands. J Cell Biol. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harlan JM, Vedder NB, Winn RK, Rice CL. Mechanisms and consequences of leukocyte–endothelial interaction. West J Med. 1991;155:365–369. [PMC free article] [PubMed] [Google Scholar]
- 38.Curley GP, Blum H, Humphries MJ. Integrin antagonists. Cell Mol Life Sci. 1999;56:427–441. doi: 10.1007/s000180050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens P, et al. KIM127, an antibody that promotes adhesion, maps to a region of CD18 that includes cysteine-rich repeats. Cell Adhes Commun. 1995;3:375–384. doi: 10.3109/15419069509081292. [DOI] [PubMed] [Google Scholar]
- 40.Petruzzelli L, Maduzia L, Springer TA. Activation of lymphocyte function-associated molecule-1 (CD11a/CD18) and Mac-1 (CD11b/CD18) mimicked by an antibody directed against CD18. J Immunol. 1995;155:854–866. [PubMed] [Google Scholar]
- 41.de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18:989–994. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- 42.Elphick GF, et al. Recombinant human activated protein C inhibits integrin-mediated neutrophil migration. Blood. 2009;113:4078–4085. doi: 10.1182/blood-2008-09-180968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.