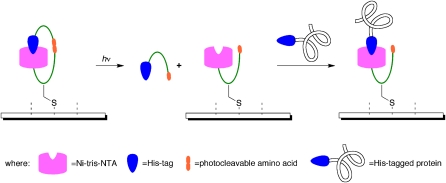
The ability to control molecular behavior in the four dimensions of time and space is almost uniquely afforded by light. Consequently, there has been a long history of the use of light to trigger molecular interactions in the study of biological phenomena or the creation of active biomolecules (1). One application of this principle has been in the fabrication of protein microarrays, a subset of the field of protein biochips (2), broadly defined as proteins immobilized on flat surfaces. The report by Grunwald et al. (3) provides a significant advance in the spatially defined immobilization of proteins on surfaces. It accomplishes this task using a novel paradigm for photochemical activation: Binding sites on the surface recognize a ligand on the proteins to be immobilized but also carry that ligand themselves. It competes with and inhibits protein binding, so the protein can only be bound once the competing ligand has been released via irradiation (Fig. 1). This autoinhibition strategy offers advantages over past methods for iterative, spatially directed protein immobilization, both in the size of the protein features that can be created and the specificity (that is, signal/background) with which this can be done.
Fig. 1.
Cartoon of the light-directed immobilization of a His-tagged protein on a microarray using the autoinhibition method. An undecapeptide construct (green) is created with a nickel-tris-(nitrilotriacetic acid) multivalent head group (nickel-tris-NTA) at the N terminus and a His-tag at the C terminus. Included within the peptide sequence is the photocleavable amino acid 3-amino-3-(2-nitrophenyl)-propionic acid and a single Cys for attachment of the construct to a microarray (or other molecules, in further applications). Initially, the construct is in the autoinhibited state. Irradiation (280–400 nm) cleaves the backbone of the construct and treatment with a competing ligand (imidazole) disrupts the intramolecular self-inactivation. Removal of the autoinhibitory His-tag opens up the nickel-tris-NTA head group for binding of His-tagged proteins.
Any protein microarray technology needs to exploit a broad-based method to link proteins to the surface to enhance the accessibility of the technology. Well-known methods used to immobilize proteins on surfaces such as Biacore chips, like N-hydroxysuccinimide active esters, are not relevant for microarrays when they are not site-selective. Typically, an affinity tag is added to the protein that is recognized by a complementary binding partner on the surface. Protein microarrays have been fabricated using a variety of ligand pairs, including azide-alkyne click chemistry, double-stranded DNA hybridization, and avidin–biotin interactions. An appealing feature of the Grunwald et al. (3) study is the use of the widely available His-tag system, where an immobilized metal ion recognizes an oligo-His affinity tag introduced into a protein using genetic engineering. The Ni-NTA (nitrilotriacetic acid)·oligo-His complex can be reversed by the addition of imidazole or EDTA. In this work, the binding strength of the complex is enhanced by a multivalent head group (4) that uses three nickel ions to bind the oligo-His affinity-tagged protein.
In the postgenomic era, DNA microarrays have been key contributors to systematic investigations of nucleic acid-based phenomena, such as gene expression, mutations, and single-nucleotide polymorphisms. They have whetted scientists’ appetites to interrogate protein-based phenomena such as posttranslational modifications or protein–protein interactions in the same massively parallel fashion, which has been achieved in only a few cases. The immobilization of 5,800 His-tagged yeast proteins to screen for interactions with other proteins could be performed on an Ni-NTA microarray surface using spotting technology, for example (5). Fabricating microarrays that are sufficiently miniaturized to address larger proteomes poses a far greater challenge, and protein microarrays have not met the challenge despite concerted efforts (6). It has been possible to create microscopic or nanoscopic protein domains using photolithographic microfabrication, soft lithography, or nano-technology methods. Past work has patterned NTA for protein immobilization on chips using microcontact printing (in domains of 10 μm) (4, 7) and nanoimprint lithography (in domains of 500 nm) (8). However, it has been a far more difficult task to iterate such processes to selectively immobilize a large number of different proteins (i.e., step and repeat). An important distinction between protein arrays and DNA microarray technology is that, for the latter, in situ synthesis permits multiple bonding events to occur in parallel, whereas for the former, it is essential that each protein be independently and sequentially immobilized. Methods of site-selective protein immobilization that rely on physical delivery either generate impractically large domains or cannot be used in step-and-repeat sequences. Ideally, protein microarrays could be prepared via iterative light-based assembly (9), because light offers the advantage that the domains of individual proteins that can be fabricated are in the size range of 10–50 μm (related to the near-UV light wavelength), much smaller than the physical delivery techniques (spotting, jetting) that can be iterated. Until the methodology of Grunwald et al. (3), such a capability had not been demonstrated.
Key to any protein microarray fabrication method is control of surface regions activated for protein immobilization while inhibiting reactions on the vast majority of the surface that should not be activated. The greater the contrast between these two regions, the more effective will be the site discrimination in protein immobilization and the greater the number of unique protein regions that can be fabricated. The distinguishing feature of the Grunwald approach to this problem is that the inhibition is not based on a specific chemical modification of the ligand-binding unit (i.e., with a photoremovable group), the strategy of past methods for molecular photolithography. Rather, it is based on the provision of a competing His-tag. A designed molecular construct for surface attachment and photochemical activation (Fig. 1) includes an internal affinity tag that ordinarily occupies the tris-NTA of its cognate molecule, like a snake biting its tail. Light activation severs the chain linking the binding site to the inhibitor; the thus freed His-tag is washed away, opening up the tris-NTA for binding to a protein bearing a His-tag. This concept is well precedented in biology, with a number of receptors, ion channels, and enzymes offering their own protein domains that bind to their active sites and compete with ligand binding (10).
Although conceptually unique, this method is most notable for its outstanding performance, particularly regarding the selectivity with which irradiated zones are exposed. The ratio of protein immobilized in intended versus unintended areas is as high as 60:1, and there is very clear distinction of regions where proteins are immobilized in successive steps. This level of selectivity has not been observed in past work, suggesting that methodologies like this should be useful in step-and-repeat photopatterning. That could lead to the development of protein microarrays of significantly greater complexity than have been available. The size of the protein features created in this work reached to and below 20 μm, commensurate with a miniaturized biosensor that could use small sample sizes and examine a multitude of analytes simultaneously.
The size of the protein features created in this work reached to and below 20 μm.
It is currently unclear why this photopatterning method is much more successful than traditional approaches based on photoremovable protecting groups. Notably, this autoinhibition approach ought to be applicable to many affinity tag/receptor pairs, and offers the advantage that it does not depend on the ability to discover a protecting group appropriate for a particular pair. The unique autoinhibition feature of this method can be exploited in other applications of light-based formation of macromolecular complexes, many of which are envisioned by the authors.
The parallel immobilization of multiple proteins on chips (11) has been developed into a robust multianalyte analysis technology with point-of-use capabilities (12). Until now, microfluidic methods have been used to generate such devices, with limitations in the number of proteins (antibodies) that could be immobilized. Using technologies like those described in this publication, the potential to create high-complexity protein chips that could truly impact biological research is significantly enhanced.
Footnotes
The author declares no conflict of interest.
See companion article on page 6146.
References
- 1.Goeldner M, Givens R, editors. Dynamic Studies in Biology: Phototriggers, Photoswitches and Caged Biomolecules. Weinheim: Wiley-VCH; 2005. [Google Scholar]
- 2.Jonkheijm P, Weinrich D, Schröder H, Niemeyer CM, Waldmann H. Chemical strategies for generating protein biochips. Angew Chem Int Ed. 2008;47:9618–9647. doi: 10.1002/anie.200801711. [DOI] [PubMed] [Google Scholar]
- 3.Grunwald C, et al. In situ assembly of macromolecular complexes triggered by light. Proc Natl Acad Sci USA. 2010;107:6149–6154. doi: 10.1073/pnas.0912617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valiokas R, et al. Differential protein assembly on micropatterned surfaces with tailored molecular and surface multivalency. ChemBioChem. 2006;7:1325–1329. doi: 10.1002/cbic.200600176. [DOI] [PubMed] [Google Scholar]
- 5.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 6.Weinrich D, Jonkheijm P, Niemeyer CM, Waldmann H. Applications of protein biochips in biomedical and biotechnological research. Angew Chem Int Ed. 2009;48:7744–7751. doi: 10.1002/anie.200901480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groll J, Ameringer T, Spatz JP, Möller M. Ultrathin coatings from isocyanate terminated star PEG prepolymers: Patterning of proteins on the layers. Langmuir. 2005;21:3076–3083. doi: 10.1021/la047438n. [DOI] [PubMed] [Google Scholar]
- 8.Maury P, et al. Creating nanopatterns of His-tagged proteins on surfaces by nanoimprint lithography using specific NiNTA-histidine interactions. Small. 2007;3:1584–1592. doi: 10.1002/smll.200700046. [DOI] [PubMed] [Google Scholar]
- 9.Blawas AS, Oliver TF, Pirrung MC, Reichert WM. Step-and-repeat photopatterning of protein features using caged-biotin−BSA. Langmuir. 1998;14:4243–4250. [Google Scholar]
- 10.Goldberg J, Nairn AC, Kuriyan J. Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell. 1996;84:875–887. doi: 10.1016/s0092-8674(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 11.Rowe-Taitt CA, et al. Simultaneous detection of six biohazardous agents using a planar waveguide array biosensor. Biosens Bioelectron. 2000;15:579–589. doi: 10.1016/s0956-5663(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 12.Taitt CR, Shriver-Lake LC, Ngundi MM, Ligler FS. Array biosensor for toxin detection: Continued advances. Sensors. 2008;8:8361–8377. doi: 10.3390/s8128361. [DOI] [PMC free article] [PubMed] [Google Scholar]