Abstract
Polyketide natural products possess diverse architectures and biological functions and share a subset of biosynthetic steps with fatty acid synthesis. The final transformation catalyzed by both polyketide synthases (PKSs) and fatty acid synthases is most often carried out by a thioesterase (TE). The synthetic versatility of TE domains in fungal nonreducing, iterative PKSs (NR-PKSs) has been shown to extend to Claisen cyclase (CLC) chemistry by catalyzing C–C ring closure reactions as opposed to thioester hydrolysis or O–C/N–C macrocyclization observed in previously reported TE structures. Catalysis of C–C bond formation as a product release mechanism dramatically expands the synthetic potential of PKSs, but how this activity was acquired has remained a mystery. We report the biochemical and structural analyses of the TE/CLC domain in polyketide synthase A, the multidomain PKS central to the biosynthesis of aflatoxin B1, a potent environmental carcinogen. Mutagenesis experiments confirm the predicted identity of the catalytic triad and its role in catalyzing the final Claisen-type cyclization to the aflatoxin precursor, norsolorinic acid anthrone. The 1.7 Å crystal structure displays an α/β-hydrolase fold in the catalytic closed form with a distinct hydrophobic substrate-binding chamber. We propose that a key rotation of the substrate side chain coupled to a protein conformational change from the open to closed form spatially governs substrate positioning and C–C cyclization. The biochemical studies, the 1.7 Å crystal structure of the TE/CLC domain, and intermediate modeling afford the first mechanistic insights into this widely distributed C–C bond-forming class of TEs.
Keywords: biosynthesis, fungal metabolism, mutagenesis
Aflatoxin is a common contaminant of nuts and grains and is found particularly in the staple diets of many developing countries (1). The toxin is produced by fungal species (such as Aspergillus flavus, Aspergillus parasiticus, and Aspergillus nomius) that infect plants during growth, harvest, and storage. The toxin can withstand food processing and is considered by the Food and Drug Administration to be an unavoidable contaminant of foods. Chronic ingestion of dietary aflatoxin B1 (3), especially when combined with hepatitis B or C infections, leads to hepatocellular carcinoma (liver tumors), which is the third most common cause of cancer death globally (2). Aflatoxin biosynthesis presents problems of fundamental interest in natural products chemistry, and understanding its formation could afford means to control the occurrence of this environmental carcinogen (3).
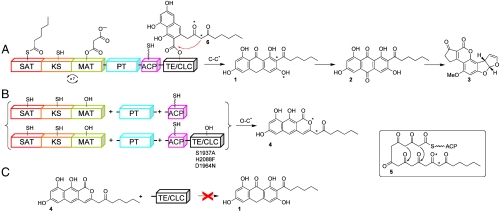
Aflatoxin biosynthesis is initiated by the multidomain enzyme, polyketide synthase A (PksA), which combines a hexanoyl starter unit and seven malonyl-CoA extender units to synthesize the precursor norsolorinic acid anthrone (noranthrone, 1) in the complex pathway to aflatoxin B1 (3, Fig. 1) (4–6). Identification of interdomainal linker regions using the Udwary–Merski Algorithm revealed six well-defined domains, but only four were readily identified by their homology to ketosynthase (KS), malonyl-CoA:ACP transacylase (MAT), acyl–carrier protein (ACP), and thioesterase (TE) domains (7). The N-terminal domain was shown to function as a starter unit:ACP transacylase (SAT), which selects the hexanoyl starter unit from a pair of specialized fungal fatty acid synthase (FAS) subunits (HexA/HexB) and transfers it onto the PksA ACP to prime polyketide chain elongation (8). In contrast to the chemically unreactive hydrocarbon chain generated in fatty acid biosynthesis, a poly-β-keto intermediate 5 is thought to be created by the nonreducing polyketide synthase (PKS) system. The second newly recognized domain, coined the product template (PT) domain (7), was shown to be critical in the biosynthetic process to control the correct cyclization of this highly reactive intermediate (9). In PksA, the KS, MAT, and ACP join with PT to extend the hexanoyl starter and carry out selective cyclization/aromatization chemistry (intermediate 6), which, either in the absence of TE, or with a nonfunctional TE gives rise to the pyrone 4, whereas in the presence of TE, a new C–C bond is formed to yield the Claisen product noranthrone 1 (Fig. 1). That is, the bicyclic intermediate 6 partitions either by C–C cyclization in the presence of TE/CLC to 1 or, in its absence, by spontaneous O–C cyclization to give pyrone 4. The Claisen cyclase (CLC) behavior for nonreducing fungal PKS TE domains was first observed in the work of Ebizuka, Fujii, and Watanabe where a normal Claisen product was diverted to its isomeric pyrone when the TE/CLC domain was either truncated (10) or inactivated by site-specific mutagenesis (11). How the PksA TE conducts Claisen cyclization (C–C bond formation) instead of hydrolysis or macrolactonization (O–C bond formation), which is typically observed in FAS and PKS proteins, is not understood. The lack of mechanistic knowledge about TE-catalyzed C–C bond formation has hindered attempts to rationally control the chain-terminating Claisen cyclization specificity in fungal PKSs.
Fig. 1.
PksA-catalyzed biosynthesis of norsolorinic acid anthrone (noranthrone, 1), the polyketide precursor of aflatoxin B1 (3). The domain architecture for PksA is shown, which includes SAT, β-ketoacyl synthase, MAT, PT, ACP, and TE/CLC domains. (A) The SAT domain receives a C6-fatty acid starter unit from an associated fungal FAS (HexA/HexB, not shown) and transfers it onto the ACP for chain elongation. The KS accepts the hexanoyl-ACP unit and subsequent malonate extender units are loaded onto the ACP from the MAT domain for chain extension to generate the linear poly-β-keto ACP-bound intermediate 5. The linear intermediate is then cyclized and aromatized in the PT domain. The resulting bicyclic intermediate is ultimately transferred from the ACP to the TE/CLC domain (intermediate 6) and undergoes Claisen-type C–C bond cyclization to release the product norsolorinic acid anthrone (noranthrone, 1), which spontaneously oxidizes in vitro to norsolorinic acid (2). (B) Three-part multidomain combination strategy. Product derailment (O–C cyclization, 4) primarily occurs without a competent TE/CLC domain (catalytic triad mutations S1937A, H2088F, and D1964N). (C) TE/CLC must accept a bicyclic ACP-bound intermediate to yield 6. The norpyrone product 4 is not a substrate for the TE/CLC domain.
Apart from the 3.1 Å x-ray structures of the primary metabolic yeast and filamentous fungal FASs (12, 13), there are very few high-resolution structures of fungal natural product biosynthetic enzymes, particularly of the multidomain fungal PKSs whose domain architectures are organized similarly to animal FASs (14). Here, we report the crystal structure of a dissected TE/CLC domain from the multidomain fungal PKS PksA solved to 1.7 Å resolution. TEs bear the canonical TE catalytic triad consisting of active site Ser-His-Asp (15) residues, which were tentatively identified in the PksA TE/CLC domain by primary sequence comparisons and confirmed here in mutagenesis studies and the crystal structure demonstrating their involvement in Claisen cyclization. The structure shows that fungal CLCs belong to the α/β-hydrolase family but harbor a deep, hydrophobic substrate-binding chamber that is distinct from previously reported TE structures from bacterial modular PKSs (16–18), nonribosomal peptide synthetases (NRPSs) (19, 20) and human FAS (21, 22). The structure-function studies presented herein provide a picture that is likely general for C–C bond formation in polyketide chain-terminating CLCs. Combined with the recently published x-ray crystal structure of the PksA PT domain (23), these studies provide a comprehensive view of the key polyketide cyclization reactions characteristic of NR-PKSs.
Results and Discussion
CLC Catalytic Activity.
The PksA ACP-TE/CLC didomain (holo-form) was previously shown to promote the hydrolysis of benzoyl N-acetylcysteamine thioester (benzoyl-SNAC; kcat0.84 s-1, Km 1.9 mM, and kcat/Km0.44 s-1 mM-1) in vitro (7). While the N-acetyl cysteamine (NAC) thioester mimics part of the ACP-phosphopantetheine (PPT) moiety that carries biosynthetic intermediates through the catalytic cycle, it is presumed that water acts as the nucleophile in the PksA TE/CLC domain to release the free acids in the absence of the native substrate. To examine the TE/CLC hydrolytic reaction, a mutant (S1746A, “apo-mutant”) was generated to remove the PPT attachment site from the ACP domain and to isolate the TE/CLC reaction component in the ACP-TE didomain. The apo-mutant showed a reduced ability to catalyze benzoyl-SNAC hydrolysis (kcat0.050 ± 0.004 s-1, Km1.89 ± 0.49 mM, and kcat/Km0.027 ± 0.007 s-1 mM-1) compared to the “wild-type” construct, suggesting that the ACP posttranslational modification contributes to TE hydrolysis activity. Subsequently, the TE/CLC catalytic triad residues were individually mutated (S1937A, H2088F, and D1964N) in the apo-mutant to assess their involvement in TE-catalyzed hydrolysis. These mutated constructs showed no detectable hydrolytic activity even at elevated protein concentrations to the limits of the assay. As a control, a conserved Asp not involved in the triad was mutated (D2070N) and gave only a slightly reduced rate of hydrolysis compared to the apo-mutant (SI Text). Therefore, the residues of the proposed catalytic triad (S1937-H2088-D1964) are essential for hydrolytic activity. CD analyses of the apo-mutant and each of the additional active and inactive mutants were highly similar, indicating no gross structural differences (Fig. S1).
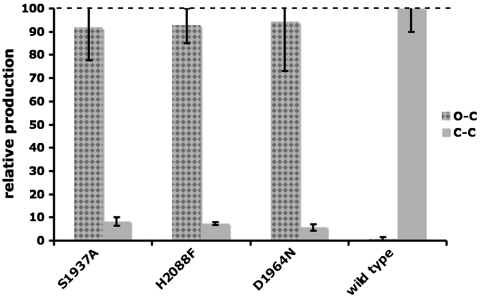
To determine if the same catalytic triad residues are involved in TE-catalyzed Claisen cyclization in the context of a fully reconstituted PksA, the holo-form didomain constructs (ACP-TE/CLC) containing the catalytic triad mutations (denoted with * below ) also were generated. PksA activity was reconstituted using a 3-part multidomain combination strategy (SAT-KS-MAT + PT + holo-ACP-TE/CLC∗). CD measurements again verified that the wild-type holo-ACP-TE/CLC and each of the catalytic triad mutants were similarly folded. Products of the enzymatic reactions were quantified by HPLC. The catalytic triad mutations in the PksA TE/CLC domain led predominantly to the production of pyrone 4 via O–C cyclization and product release (Fig. 1B). In comparison, the wild-type construct facilitates the correct C–C cyclization event to release noranthrone 1 (observed as its spontaneous oxidation product norsolorinic acid 2, Fig. 1A), confirming that S1937, H2088, and D1964 are critical in catalyzing the final Claisen (Dieckmann) cyclization (Fig. 2). As a control, the TE/CLC domain was incubated with increasing concentrations (1–500 μM) of the synthetic pyrone standard 4 to determine if the O–C cyclized pyrone could be converted to norsolorinic acid (Fig. 1C). The reactions were extracted and analyzed as above for direct comparison. Conversion to norsolorinic acid was not detected, implying that the TE/CLC domain must accept the partially cyclized, ACP-bound intermediate (the bicyclic 6) to drive the final cyclization reaction and to release noranthrone 1 (Fig. 1A).
Fig. 2.
TE/CLC activity and mutagenesis. The ratio (%) of pyrone 4 (Gray Checkered, O–C cyclization) vs. anthraquinone 2 (Gray Solid, C–C cyclization) is plotted for each individual experiment. The PksA ACP-TE/CLC* component was varied in 3-part (SAT-KS-MAT + PT + ACP-TE/CLC∗) multidomain combinations. The mutations (*) are listed on the x-axis.
Overall Structure: Conserved α/β Hydrolase Fold as a Monomer.
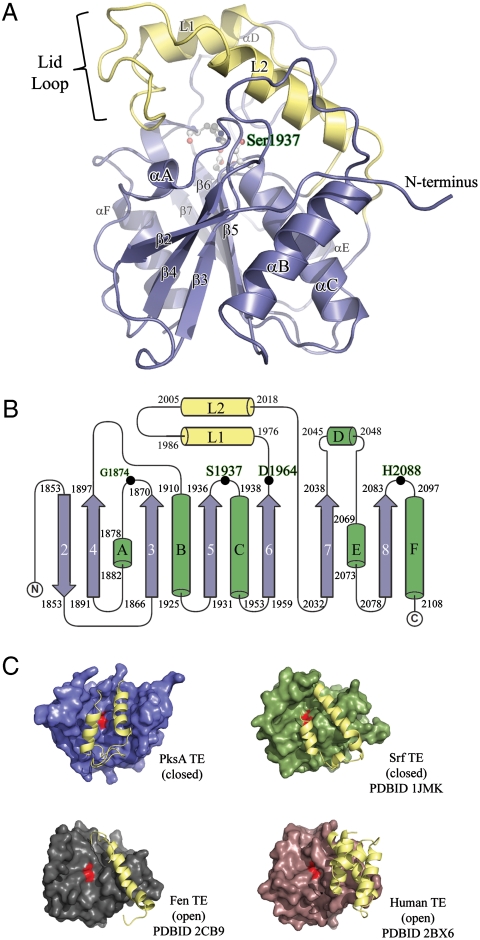
The PksA TE/CLC adopts an α/β-hydrolase fold that consists of a central β-sheet with the second strand (β2) antiparallel to the remaining strands (Fig. 3 A and B). Similar to other reported TE structures in NRPS, PKS, and mammalian FAS, the β1 strand typical of the α/β-fold is not present in PksA TE/CLC. There are two possible reasons for its absence: (i) this is a conserved structural feature of fungal PKS TE/CLC domains, or (ii) the PksA TE/CLC was proteolytically cleaved to lose β1. The latter explanation is less likely because the first eight residues of the N-terminus are present as a loop region that extends along the protein exterior around α1 with the semiconserved Arg1850 forming a salt bridge with conserved Asp1903 and stabilizing the protein structure (Fig. S2, Protein Sequence Alignment). In contrast, N-terminal helices have been reported to be important for TE dimer formation in DEBS and PICS TEs (17). The absence of these helices in PksA TE/CLC supports the observation that the PksA TE/CLC is a monomer, as observed in the crystal structure, as well as in solution by gel filtration and native PAGE mobility experiments. Sequence alignment further reveals that the lack of N-terminal β1 and N-terminal helices is a conserved structural feature among iterative fungal PKS TE/CLCs.
Fig. 3.
Overall structure of the PksA TE domain. (A) Ribbon diagram of the TE α/β-hydrolase fold with the core domain shown in blue and the unique lid in yellow. The location of the S1937-H2088-D1964 catalytic triad and the oxyanion site residue G1874 is shown in balls-and-sticks and labeled green. The position of the lid-loop, which presumably opens upon presentation of substrate by ACP, is closed. (B) Topology diagram of PksA TE showing the location of the catalytic triad residues along with G1874 (Black Dots), which is thought to act as the oxyanion hole for the reaction, and the lid region (Yellow) as an insertion between β6 and β7, a common feature of FAS, PKS and NRPS TE domains. The core sheets and helices are colored blue and green, respectively, and are numbered as the canonical α/β-hydrolase fold showing that PksA TE lacks the first N-terminal sheet necessary for dimerization. (C) Comparison of open and closed lid conformations found in TE crystal structures.
PksA TE/CLC also lacks the characteristic αD helix of the α/β-hydrolase family, instead having an inserted “lid” region consisting of helices αL1 and αL2. Such a lid is also seen in the DEBS (17), PICS (18), surfactin synthetase (SrfA-C) (19), fengycin synthetase (FenB) (20), and enterobactin (EntF) (24) TE structures, although there is considerable variability among these individual lid regions. A second small region of structural variability between these enzymes and PksA TE is found in the loop between β7 and the vestigial αE. This loop region in PksA TE/CLC has a short extra inserted helix (residues 2045–2048, Fig. 3B).
The Catalytic Triad and Hydrophobic Substrate-Binding Chamber.
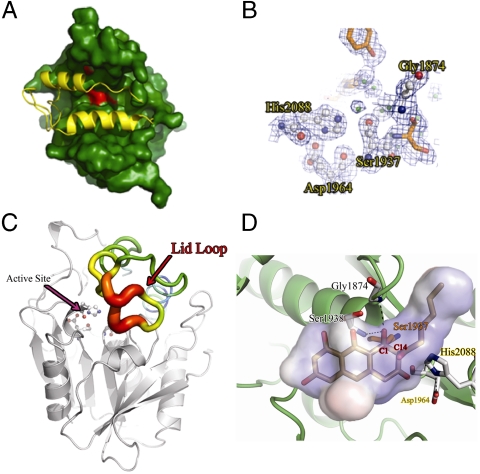
PksA TE/CLC residues Ser1937, His2088, and Asp1964 constitute a catalytic triad conserved in the α/β-hydrolase family. The nucleophilic Ser1937 is canonically located on a loop between β5 and αC within the motif GXSXG, with its side-chain oxygen within hydrogen bonding distance of Nε2 of His2088, which canonically resides on loop β8-αF. The carboxylate residue of the α/β-hydrolase catalytic triad is typically found on loop β7-αE. However, in the modular PKS and NRPS TEs it has moved to the C-terminal end of β6. Asp1964 in PksA is likewise found at this noncanonical position (Fig. 4B).
Fig. 4.
The TE active site chamber. (A) Surface representation of the PksA TE substrate chamber in green, and the active site in red. The lid in yellow completely closes off the entrance to the substrate-binding chamber. (B) Representative electron density from the 1.7 Å PksA TE structure, contoured to 1.5 Å, calculated from the 2Fo-Fc map and displayed around S1937, D1964, and H2088. (C) PksA TE displayed by B-factor putty (Red: high B-factor to Blue: low B-factor). The lid loop region (Red Arrow) has the highest B-factor suggesting this flexible region opens to allow access to the active site. (D) The docked model (Sticks) of the second tetrahedral intermediate of the TE/CLC reaction generated by CHARMm minimization. C1 is covalently attached to S1937 (Orange). Hydrogen bonds between the oxyanion and G1874 and S1938 are shown with black dashed lines. The internal chamber volume is shown as a transparent cloud. Active site residues (except for S1937) are shown as white sticks and intermediate carbon atoms in gray.
Two groups independently reported crystal structures of TE domains containing a covalently linked substrate analog mimicking the tetrahedral intermediate of the transesterification reaction. Giraldes et al. cocrystallized a series of phosphonate affinity labels with modular PKS domain PICS-TE (16), while Samel et al. obtained crystals with the general serine protease inhibitor phenylmethylsulfonyl fluoride covalently attached to NRPS domain FenTE (20). Both structures identified a probable oxyanion hole in which the backbone amide N-H that is one residue downstream of the catalytic serine forms a hydrogen bond with what would be the oxyanion in the actual transesterification. In PksA TE/CLC, this N-H corresponds to the amide of Ser1938. This residue is generally regarded as one component of the oxyanion hole for the entire α/β hydrolase superfamily (25). Furthermore, in the FenTE structure a second amide-to-oxyanion hydrogen bond could be observed from Ile1069 on loop β3-αA, which is also implicated for the majority of the enzymes in the family (in the PICS-TE structure, however, this residue is replaced by a crystallographic water molecule tethered to Thr1125 on the same loop) (16, 20). The corresponding residue in PksA TE/CLC is Gly1874 (Fig. 3B, Fig. 4B), which can plausibly complete the oxyanion hole in the TE/CLC.
Similar to the TE domains from FASs, modular PKSs, and NRPSs, the substrate-binding region of PksA TE/CLC is formed between the α/β hydrolase core and the lid region inserted between β6 and β7. In the modular DEBS and PICS-TE domains, the substrate-binding region is a long, open, predominantly hydrophilic channel that passes through the full length of the enzyme with the catalytic triad that carries out macrolactonization of a hydrophilic polyketide substrate located at the center (16–18). In contrast the NRPS TE structures show a channel that is closed off at both ends and instead opens from the side to reveal a bowl or crevice where the macrocyclization reaction occurs (19, 20). Crystal structures of the monomeric SrfTE fortuitously capture both “open” and “closed” forms in which a flexible lid covers a hydrophobic binding pocket for the surfactin substrate (19). In the open form, the crevice and the catalytic triad are exposed to solvent, while in the closed form, a region between strand β6 and helix αL1 moves to conceal the active site to all but small molecules.
The substrate-binding pocket of PksA TE/CLC resembles that of the NRPS TE structures more than the modular PKS TE structures in that no channel traverses the length of the protein. In fact, there does not appear to be any solvent access to the central chamber, because the lid region has sealed off the enzyme in the observed closed conformation where cyclization would occur (Fig. 4A). Presumably, docking of the ACP initiates a conformational change that exposes the chamber for substrate delivery. The reaction chamber is highly hydrophobic and is lined with the side chains of well-conserved residues Gly1875, Trp1936, Ala1941, Phe1942, Ala1965, Met1971, Phe2010, Val2013, Val2014, Met2017, His2055, Phe2056, and Phe2089, in addition to the catalytic residues. The high conservation of these chamber-forming residues implies that the hydrophobic and highly aromatic substrate chamber is a feature maintained in TE/CLC domains (Fig. S2).
Analysis of the B factors for each residue in the PksA TE/CLC structure reveals a region of high disorder in the loop between lid helices αL1 and αL2, suggesting that this could be the flexible entrance to the substrate chamber (Fig. 4 A and C). This flexible region is also conserved among the other nonreducing, iterative PKS TE domains, underscoring its structural and catalytic importance. Indeed, NMR titration experiments implicate this helix-turn-helix region with connecting loops to be important for acyl-thiolation substrate specificity in the assembly line TE1 involved in enterobactin biosynthesis (24, 26). It should be noted that while TEs adopt open and closed forms, SrfTE, for example, opens from the side of the lid (N-terminal to αL1), whereas we propose PksA TE opens from the tip of the lid (loop αL1-αL2, Fig. 4C).
Covalent Intermediate Minimization and Proposed Mechanism.
In the two PICS-TE structures with covalently bound phosphonate affinity labels, what would be one of the sulfur lone pairs from the phosphopantetheine moiety points directly toward the key nitrogen atom of the catalytic histidine (16). In the PksA TE we suggest that the histidine involved in the CLC reaction not only serves to deprotonate the serine for initial nucleophilic attack on the incoming thioester, but also acts to protonate the outgoing thiolate leaving group as the tetrahedral intermediate collapses. In simple TE-catalyzed hydrolytic reactions a water molecule, most likely deprotonated by the now neutral histidine, acts as the second nucleophile, and the histidinium reprotonates the seryl oxygen as the elimination step proceeds. In PksA TE/CLC, however, the nucleophile must be the substrate C-14 instead of a water molecule, suggesting also that the lid has closed to preclude water and the ACP-bound phosphopeantetheine arm has departed the active site. With these considerations in mind, an in silico model was generated for the proposed tetrahedral intermediate/transition state of the reaction of C-14 that attacks on the covalently bound intermediate. The model was minimized using CHARMm with constraints to place the C-1 oxygen atom within hydrogen bonding distance of the oxyanion hole comprised of Ser1938 and Gly1874 (Fig. 4D).
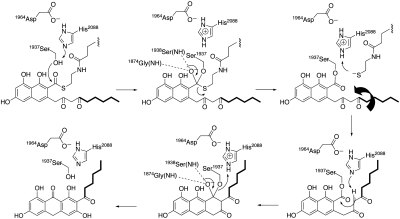
Based on the PksA TE/CLC crystal structure, associated modeling studies and mutational analyses, the following overall mechanism is proposed for PksA TE/CLC-catalyzed noranthrone (1) formation and termination of fungal aromatic polyketide biosynthesis (Fig. 5). Transfer of the penultimate bicyclic intermediate 6 from the PT domain to the TE/CLC domain and its final cyclization is facilitated by the direct tethering (27) of the ACP and TE/CLC domains. The PPT-ACP phosphodiester linkage then mediates docking of the substrate-bound ACP domain to the TE/CLC active site. Substrate and/or ACP binding likely triggers a conformational change in the lid region of the TE from closed to open, thus allowing the delivery of the bicyclic thioester substrate. The polyketide intermediate 6 then undergoes transesterification from the PPT arm of the ACP to the nucleophilic active site Ser1937, which is deprotonated by His2088 with the assistance of Asp1964. The backbone nitrogens of Gly1874 and Ser1938 act as the oxyanion hole to stabilize the transacylation reaction (Fig. 4D and Fig. 5). As the tetrahedral intermediate collapses, the leaving thiolate is in position to abstract the histidinium (His2088) proton as it exits the PPT-ACP channel. When the PPT group leaves the active site, C-14 of the substrate side chain can rotate to occupy the now empty space left by the departing PPT sulfur atom, and the lid can reclose once the substrate diketo side chain swings into this channel. The hexyl portion of the substrate is now enveloped by residues Gly1875, Trp1936, Gln1991, and Phe2010. The conformational change of the hexyl group locks C-14 close to catalytic His2088 and controls substrate positioning for enolate formation and subsequent C–C (Claisen/Dieckmann) cyclization through a second, virtually identical tetrahedral intermediate. Upon its collapse, the seryl oxygen is displaced and reprotonated by His2088, ending the catalytic cycle and restoring the resting state of PksA TE/CLC.
Fig. 5.
Proposed mechanism of TE/CLC-catalyzed chain-termination of fungal aromatic polyketide biosynthesis. The ACP of the ACP-bound substrate is displaced upon TE-catalyzed transesterification (Upper). The catalytic triad and oxyanion site residues are shown. Rotation of the substrate side chain can occur once the ACP leaves the pocket, and the TE can then close. TE conformational constraints as observed in the closed-form crystal structure guide Claisen-type cyclization to release noranthrone 1 (Lower).
Conclusions
We report the first crystal structure of a TE/CLC that catalyzes a C–C cyclization (Claisen cyclization) reaction, and we find that the TE/CLC imposes strict conformational control on the enzyme-bound intermediate to drive C–C cyclization and to prevent spontaneous O–C cyclization, which occurs with a defective or absent TE/CLC. The inherent hydrolytic activity of the TE/CLC domain supports an editing function in the absence of the correct substrate. Although primary sequence similarity is low compared to other TEs, the tertiary structure of this C–C bond-forming enzyme is similar to known enzymes that promote O–C or N–C forming reactions—hydrolysis, macrolactonization, and macrolactamization. The mutagenesis, structural studies and in silico models provide a picture of the catalytic cycle that is likely general for the CLC-type TEs that are widespread in filamentous fungi. The characteristic polyketide cyclization reactions exemplified by NR-PKSs are known to be carried out in the PT and TE domains (9). The mechanistic insights afforded by the structure of the PksA TE reported here can be combined with those of its companion PT domain (23) for a complete structural description of the fungal polyketide cyclization reactions. These new findings build a foundation for developing inhibitors of aflatoxin production that target polyketide assembly, ring closure and the unique aspects of fungal TE-catalyzed chain release as well as for engineering aromatic polyketide biosynthesis in the prolific secondary metabolite-producing fungi to produce alternative bioactive products.
Materials and Methods
Protein Production and Purification.
Recombinant PksA TE monodomain and PksA ACP-TE didomain constructs containing N-terminal His6-tags were produced and purified as previously described (7, 9). The proteins were dialyzed against 50 mM Tris-HCl pH 7.5, and the polyhistidine tag was cleaved with bovine thrombin (10 unit per 1 mg sample). The protein mixture was adjusted to 300 mM NaCl, and the polyhistidine tag was removed with Ni resin (Qiagen). The flow-through containing the cleaved protein was dialyzed against 50 mM Tris-HCl pH 7.5 and further purified by Q-Sepharose anion exchange chromatography using a 10–500 mM KCl gradient (Fig. S3). The purified protein was exchanged into 20 mM Tris-HCl pH 7.5 containing 5% glycerol and 2 mM DTT, concentrated to 5 mg/mL, and frozen in liquid nitrogen for crystallographic studies. Selenomethionine labeled PksA TE was produced in Escherichia coli methionine auxotroph B834(DE3) (Novagen) and was purified as above for phase determination.
Enzyme Assays.
The hydrolytic activity of the TE/CLC was determined as previously described (7). Briefly, the purified protein was exchanged into 50 mM potassium phosphate buffer (pH 7.5) using an EconoPac-10 desalting column (BioRad), and concentrations were determined using the Bradford assay with bovine albumin as a standard. Benzoyl-SNAC (250 mM) was dissolved in DMSO for substrate addition. The hydrolysis of a series of concentrations of benzoyl-SNAC was monitored in triplicate after the addition of enzyme (2 μM final), and 50 μl aliquots of the reaction were quenched in 50 μl aliquots of 8 M urea at 2, 5, 10, and 15 min. The resulting free thiols produced were reacted with 5,5′-dithiobis-2-nitrobenzoate (8 mg/mL in ethanol, 4 μl) for 15 min at 25 °C and quantified by UV-visible spectrophotometry at 412 nm. The catalytic triad mutants exhibited no detectable activity even at 10 mM substrate concentration.
Reconstitution of PksA activity utilizing a 3-part multidomain combination strategy and quantification of the resulting products were carried out as previously described (9). The SAT-KS-MAT, PT, and ACP-TE/CLC∗ components (10 μM each) were incubated with 0.5 mM hexanoyl-CoA and 2 mM malonyl-CoA in 100 mM potassium phosphate buffer (pH 7.4) for 14 h, and the extracted organic products were analyzed by HPLC. The holo-ACP-TE/CLC mutants (*) were monitored for their ability to carry out Claisen chemistry.
The PksA TE/CLC monodomain (10 μM) was treated with a series of substrate concentrations (1, 10, 50, 100, 250, and 500 μM pyrone) for 1 h in 100 mM potassium phosphate buffer pH 7.4 at 25 °C. The products were extracted and analyzed as above by HPLC to determine if pyrone 4 could be converted to noranthrone 1.
Mutagenesis.
The ACP-TE expression construct (pETA1) (7), generated by amplifying the pksA cDNA with oligonucleotides APACP5.1 (5′-GAATTCCCTTATAAACCAGCTAAT-3′) and APTE3.1 (5′-AAGCTTCTAAGCCATGACCCGGTC-3′) and inserting the amplified product into the EcoRI/HindIII sites in pET28a (Novagen) to create an N-terminal His6-tag fusion, was selected for mutagenesis studies. The catalytic triad mutations (S1937A, H2088F, and D1964N) were created in both the “holo-construct” that were activated by the promiscuous phosphopantetheinyltransferase, Svp (28), and the apo-construct lacking the phosphopantetheine attachment site (S1746A) by overlap extension PCR (29).
Crystallization of PksA TE.
Crystals of the selenomethionine derivative of PksA TE were grown as sitting drops at 25 °C by vapor diffusion. The well buffer contained 0.2 M ammonium acetate, 0.1 M sodium citrate pH 5.6, and 30% PEG 4K. Protein concentration was 5 mg/mL. Drops were generated by mixing 2 μL of purified protein with 2 μL of well solution. The twinned, orthorhombic crystals grew within 2 wk. Crystals of the PksA ACP-TE didomain construct grew in the same condition, although analysis of the PksA ACP-TE crystals by SDS-PAGE showed that the TE alone crystallized as a protein degradation product.
Data Collection.
The crystals were transferred to a well solution containing 85% of the above crystallization buffer plus 15% glycerol, and flash frozen in liquid nitrogen. Multiwavelength anomalous dispersion (MAD) data from the selenomethionine derivative was collected to 3.5 Å on a MarUSA MarMosaic -325 charge coupled detector at beamline 9-2 of the Stanford Synchrotron Radiation Laboratory. Native data were collected to 1.7 Å on an ADSQ Q210 charge coupled detector at beamline 5.0.2 of the Advanced Light Source. The space group for both native and derivative crystals was P212121 with isomorphous cell dimensions of a = 66.934 Å, b = 66.941 Å, c = 67.924 Å, α = β = γ = 90.000° (one monomer per asymmetric unit). Intensities were indexed and scaled with DENZO and SCALEPACK using the HKL2000 package. Scaled data from the Selenomethionine derivative was unmerged. A summary of the crystallographic statistics is shown in Table S1.
MAD Phasing, Model Building, and Refinement.
Initial phases were calculated by the MAD method. Using the program SOLVE (30), 12 initial Se sites were found, and initial electron density maps were generated using RESOLVE (30). The phases based on the Se sites from the MAD dataset were then used to calculate electron density maps that were improved using “solvent flipping” in CNS. The solvent boundary and protein regions in the initial electron density map were well defined, and > 80% of the protein backbone and side-chains were built into the initial map using QUANTA and COOT (31). This partial model was then used as a starting point for molecular replacement in CNS using the native data to 1.7 Å. One round of rigid-body refinement followed by simulated annealing, energy minimization, and B-factor refinement gave initial R and Rfree values of 30% and 35%, respectively, over the resolution range 50 - 1.7 Å. The remaining model was then rebuilt and problem areas were refined via iterative rounds of model building and refinement, although the R and Rfree values did not decrease below 27% and 30%. At this time the data were passed through the twinning server at University of California Los Angeles, which suggested that the native dataset is moderately twinned with a twinning factor of 0.42. Application of the twinning operator k, h, -l to correct for the crystal twinning lowered the R and Rfree to 22% and 25%, at which point waters were added using COOT. A final round of energy minimization and B-factor refinement gave final R and Rfree values of 18% and 20%.
CHARMm Minimization.
All steps were performed with the Discovery Studio (DS) 2.1 software suite. The transition state intermediate (Fig. 5D) was generated with the structure drawing functionality of DS 2.1, manually docked in the active site of PksA TE/CLC, and attached to Ser1937 by a sigma bond. The entire system was typed with the CHARMm forcefield (32) and minimized using an adopted basis NR algorithm with a generalized Born implicit solvent model with molecular volume. The oxyanion was constrained between 2.5 and 3.5 Å each of the amide nitrogens of Gly1874 and Ser1938. The algorithm exited after achieving an rms gradient of < 0.1 kcal/(mol Å). All other settings were default.
Supplementary Material
Acknowledgments.
This work was supported by the Pew Foundation (S.-C.T.). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a nationl user facility operated by Stanford University on behalf of the US Department of Enery Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Services. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy Contract DE-AC02-05CH11231. C.A.T. gratefully acknowledges support by the US National Institutes of Health Grant ES001670. J.M.C. is currently a fellow supported by the Damon Runyon Cancer Research Foundation Harvard Medical School Grant DRG-2002-09.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of PksA TE have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3D4H).
This article contains supporting information online at www.pnas.org/cgi/content/full/0913531107/DCSupplemental.
References
- 1.Williams JH, et al. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am J Clin Nutr. 2004;80:1106–1122. doi: 10.1093/ajcn/80.5.1106. [DOI] [PubMed] [Google Scholar]
- 2.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 3.Townsend C, Minto R. Comprehensive Natural Products. Amsterdam: Elsevier; 1999. Biosynthesis of Aflatoxin; pp. 443–471. [Google Scholar]
- 4.Minto R, Townsend C. Enzymology and molecular biology of aflatoxin biosynthesis. Chem Rev. 1997;97:2537–2555. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]
- 5.Yabe K, Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2004;64:745–755. doi: 10.1007/s00253-004-1566-x. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Udwary D, Merski M, Townsend C. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe A, et al. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett. 1999;40:91–94. [Google Scholar]
- 11.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 12.Jenni S, et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316:254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 13.Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 14.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 15.Carter P, Wells J. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 16.Giraldes JW, et al. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat Chem Biol. 2006;2:531–536. doi: 10.1038/nchembio822. [DOI] [PubMed] [Google Scholar]
- 17.Tsai S-C, et al. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: Versatility from a unique substrate channel. Proc Natl Acad Sci USA. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai S-C, Lu H, Cane D, Khosla C, Stroud R. Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry. 2002;41:12598–12606. doi: 10.1021/bi0260177. [DOI] [PubMed] [Google Scholar]
- 19.Bruner SD, et al. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure. 2002;10:301–310. doi: 10.1016/s0969-2126(02)00716-5. [DOI] [PubMed] [Google Scholar]
- 20.Samel SA, Wagner B, Marahiel MA, Essen LO. The thioesterase domain of the fengycin biosynthesis cluster: a structural base for the macrocyclization of a non-ribosomal lipopeptide. J Mol Biol. 2006;359:876–889. doi: 10.1016/j.jmb.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci USA. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pemble CW, IV, Johnson LC, Kridel SJ, Lowther WT. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nat Struct Mol Biol. 2007;14:704–709. doi: 10.1038/nsmb1265. [DOI] [PubMed] [Google Scholar]
- 23.Crawford JM, et al. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frueh DP, et al. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 26.Koglin A, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran L, Tosin M, Spencer JB, Leadlay PF, Weissman KJ. Covalent linkage mediates communication between ACP and TE domains in modular polyketide synthases. Chembiochem. 2008;9:905–915. doi: 10.1002/cbic.200700738. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez C, Du L, Edwards DJ, Toney MD, Shen B. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
- 29.Ho S, Hunt H, Horton R, Pullen J, Pease L. Site-directed mutagenesis by overlap extesion using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Brooks BR, et al. CHARMm: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.