Abstract
Melatonin is a pineal hormone produced at night; however, many strains of laboratory mice are deficient in melatonin. Strangely enough, the gene encoding HIOMT enzyme (also known as ASMT) that catalyzes the last step of melatonin synthesis is still unidentified in the house mouse (Mus musculus) despite the completion of the genome sequence. Here we report the identification of the mouse Hiomt gene, which was mapped to the pseudoautosomal region (PAR) of sex chromosomes. The gene was highly polymorphic, and nonsynonymous SNPs were found in melatonin-deficient strains. In C57BL/6 strain, there are two mutations, both of which markedly reduce protein expression. Mutability of the Hiomt likely due to a high recombination rate in the PAR could be the genomic basis for the high prevalence of melatonin deficiency. To understand the physiologic basis, we examined a wild-derived strain, MSM/Ms, which produced melatonin more under a short-day condition than a long-day condition, accompanied by increased Hiomt expression. We generated F2 intercrosses between MSM/Ms and C57BL/6 strains and N2 backcrosses to investigate the role of melatonin productivity on the physiology of mice. Although there was no apparent effect of melatonin productivity on the circadian behaviors, testis development was significantly promoted in melatonin-deficient mice. Exogenous melatonin also had the antigonadal action in mice of a melatonin-deficient strain. These findings suggest a favorable impact of melatonin deficiency due to Hiomt mutations on domestic mice in breeding colonies.
Keywords: Hiomt, Asmt, pseudoautosomal, photoperiodism, circadian
Melatonin is a hormone produced primarily in the pineal gland (1). In most animals, melatonin is synthesized and secreted predominantly at night, and it has been implicated in the regulation of the circadian rhythm and seasonal reproduction (2, 3). Melatonin is synthesized by two enzymatic steps from serotonin. First, serotonin is converted to N-acetylserotonin (NAS) by arylalkylamine N-acetyltransferase (AANAT; EC 2.1.3.87). NAS is subsequently methylated by hydroxyindole O-methyltransfease [HIOMT; EC 2.1.1.4; also known as N-acetylserotonin O-methyltransferase (ASMT)] to form melatonin. The daily rhythm of melatonin synthesis is thought to be due to a marked nocturnal increase of AANAT activity (4). HIOMT activity is, in contrast, almost constant throughout day and night. Although the mechanisms of expression and posttranslational regulation of AANAT have been well studied after the cloning of Aanat cDNA (5), HIOMT has been poorly characterized despite the relative stability of the enzymatic activity. Curiously, the mouse Hiomt gene has not been identified yet, even after a high-quality genome assembly of mouse was finished by the Mouse Genome Sequencing Consortium (6). The assembly of the mouse genome does not contain a sequence, which is homologous to Hiomt of other species. A second curious fact about the melatonin synthesis in mice is that many strains of laboratory mice produce no or less melatonin (7–9). Enzymatic activities of AANAT or HIOMT (or both) are compromised in mice of the melatonin-deficient inbred strains, such as BALB/c and C57BL/6J (B6J) (Table S1). In the history of domestication of mice, the genetic defects must have been introduced and fixed. Only one defect has so far been found, a point mutation in the Aanat gene, which causes an aberrant splicing and a frameshift in B6J strain (10).
In this study, we identified the mouse Hiomt gene, which is located in the pseudoautosomal region (PAR), a short region of homology between the X and Y chromosomes. PAR mediates proper sex chromosome pairing and segregation, and meiotic recombination between the sex chromosomes occurs in the PAR, which is therefore termed pseudoautosomal (11, 12). Because the PAR of mouse is highly variable due to a high rate of recombination (13), the mouse Hiomt gene may have undergone rapid divergence. We present evidence that melatonin deficiency has a favorable impact on testis development of mice, suggesting some selection for defective alleles of the Hiomt in breeding colonies of fancy/laboratory mice.
Results
Identification of Mouse Hiomt cDNA.
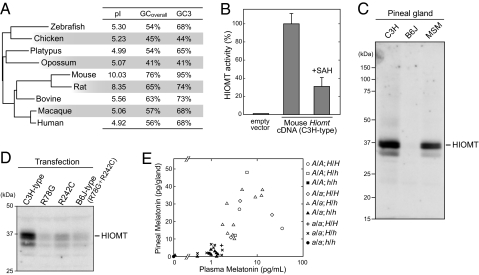
TBLASTN search, comparing rat HIOMT protein sequence against the mouse genome database, detected two statistically significant hits (SI Materials and Methods). Both contigs were obtained by Celera's whole-genome shotgun project. Several PCR primers were designed based on the mouse sequences and rat Hiomt, RT-PCR and rapid amplification of cDNA ends (RACE) techniques were used to clone the mouse Hiomt cDNA (Fig. S1A). We have succeeded in obtaining the full-length coding sequence (CDS) of the mouse Hiomt CDS from pineal RNA of C3H/HeJ (C3H), one of the melatonin-proficient strains of classical laboratory mice (Table S1). Although the deduced amino acid sequence of mouse HIOMT (Fig. S1B) is highly divergent from HIOMT of other species (69% and 52% identical to rat and human HIOMT proteins, respectively), the phylogenetic tree constructed on the basis of the amino acid sequences of HIOMT is almost consistent with the expected vertebrate phylogeny (Fig. 1A). This analysis reveals the rapid evolution of the Hiomt gene in the rodent species; notably, the theoretical isoelectric point of rodent HIOMT is quite different from (higher than) that of the other species (Fig. 1A). The other characteristic aspect of mouse Hiomt is a high GC content, and, in particular, the GC content at the third codon position (GC3) is extremely high (95%) (Fig. 1A).
Fig. 1.
Mouse Hiomt has a highly diverged sequence but encodes a functional enzyme. (A) Comparison of characteristics of Hiomt sequences and a phylogenetic tree of vertebrate HIOMT proteins. The tree was constructed by using the neighbor-joining method, and the lengths of the horizontal lines represent evolutionary distances. The C3H-type sequence is used as mouse HIOMT. The theoretical isoelectric point (pI), the overall GC content of Hiomt CDS (GCoverall), and the GC content at the third codon position (GC3) are shown. (B) HIOMT enzymatic activity. HIOMT activity was assayed with 5.0 μg of soluble protein of HEK293 cells transfected with mouse Hiomt-expressing vector or empty vector. Results are expressed as means ± SEM of assays performed in triplicate. The value (100%) obtained from mouse HIOMT-expressing cells is 5.8 nmol/h/mg protein. Addition of SAH in the reaction at a concentration equimolar to SAM (0.1 mM) inhibited the activity by ≈70%. (C) Western blot analysis of mouse pineal HIOMT. Mouse pineal homogenates (1 gland/lane) were subjected to SDS/PAGE and the gel was electroblotted onto a PVDF membrane, which was immunostained by the antiserum against mouse HIOMT. (D) Western blot analysis of mouse HIOMT variants overexpressed in HEK293 cells. (E) Melatonin productivity of F2 intercrosses (C3H × B6J). Pineal gland and plasma were collected at ZT 22–23, and the melatonin contents were assayed. The Aanat and Hiomt alleles derived from C3H, a melatonin-proficient strain, are shown in capital letters (A and H); and the alleles from B6J, a melatonin-deficient strain, are shown in lowercase letters (a and h).
The mouse Hiomt cDNA that we cloned was expressed in HEK293 cells, and the HIOMT enzymatic activity was assayed. Although the mouse HIOMT has weak sequence homology to those of other species, robust HIOMT activity was detected in the soluble fraction of the transfected cells (Fig. 1B). The activity was inhibited by S-adenosyl-homocysteine, a common inhibitor of methyl transfer reactions involving S-adenosyl-methionine (SAM).
We examined the B6J strain, which reportedly had no HIOMT activity (7). RT-PCR amplified the Hiomt from B6J pineal RNA, but HIOMT protein was not detected in the pineal gland by immunoblot analysis using anti-mouse HIOMT antiserum (Fig. 1C). There are seven SNPs in the Hiomt CDS between C3H and B6J. Two were nonsynonymous SNPs, resulting in amino acid substitutions (R78G and R242C) (Fig. S1B). We overexpressed mouse HIOMT variants, C3H type, R78G, R242C, and B6J type (R78G+R242C), in HEK293 cells. Immunoblot analysis showed that each of the mutations markedly reduced the expression level of HIOMT protein (Fig. 1D).
In addition to C3H and B6J strains, we examined MSM/Ms inbred strain (MSM), which was recently established from wild mice, Mus musculus molossinus, and retains many traits of wild mice (14). MSM mice produced melatonin during nighttime with a peak just before the light came on (Fig. S2A). HIOMT protein in the pineal gland was detected (Fig. 1C) and the two mutations, R78G and R242C, were absent in the Hiomt cDNA sequence of MSM (Fig. S1B). We investigated the daily changes of Aanat and Hiomt expression levels by quantitative RT-PCR analysis of the pineal glands of MSM mice maintained in a 12-h light/12-h dark cycle (LD12:12). The Aanat transcript showed the robust daily fluctuation in abundance with a peak at midnight [zeitgeber time (ZT) 20] and a trough at early evening (Fig. S2C). In sharp contrast, subtle change was observed in the Hiomt mRNA level, in agreement with reports in other species (4). Tissue distribution study showed that the mouse Hiomt was predominantly expressed in the pineal gland and very slightly in the retina (Fig. S3).
The previous studies showed that two loci responsible for AANAT and HIOMT activities are autosomally located and segregate independently in mice (7, 15, 16). To confirm the genetics of melatonin production using the gene sequence information of Aanat and Hiomt, we analyzed the melatonin productivity trait of (B6J × C3H) F2 intercross mice. The genotypes of the Aanat and Hiomt loci were determined by PCR amplification and direct sequencing (Fig. S4). Both the plasma and the pineal gland were collected from each animal at ZT 22–23, and melatonin levels were assayed. Mice homozygous for the B6J-derived Aanat allele or homozygous for the B6J-derived Hiomt allele produced very little melatonin (Fig. 1E). The result indicated that the gene we cloned is the only gene encoding authentic HIOMT enzyme. Melatonin productivity of mice can be determined for by the combination of Aanat and Hiomt genotypes.
Mouse Hiomt Is a Pseudoautosomal Gene.
The mouse Hiomt gene, from the start codon to the stop codon, was amplified by long PCR from the B6J and C3H genomic DNAs. The exon/intron structure of the mouse Hiomt gene, which comprises eight exons and seven introns, corresponds exactly to those of the rat and human genes. The gene size of the mouse Hiomt (B6J, 5.4 kb; C3H, 5.8 kb) differs greatly from the human HIOMT (27.8 kb) but is similar to the rat Hiomt gene (supposedly ∼5–6 kb). In contrast to the CDS characterized by a high GC content (Fig. 1A), the intron sequences of the mouse Hiomt gene exhibits 40–60% GC content (Fig. S5A). The intron sequences, however, extensively consist of repeat sequences, which are distinctive of each intron (Fig. S5B).
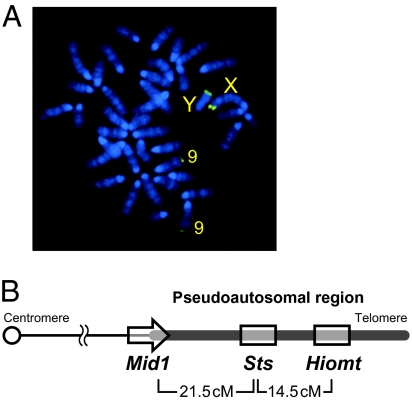
To obtain further information on the mouse Hiomt gene, we screened a bacterial artificial chromosome (BAC) library of genomic DNA of MSM mice (17). We screened the library (equivalent to ≈60,000 clones) by PCR and isolated two positive clones. One of them (MSMg01-318O12) was used in further studies. Both ends of the BAC clone insert were sequenced and analyzed using BLAST search. One end sequence showed high homology to a number of mouse genomic contigs, almost all of which were mapped to most proximal (subtelomeric) regions of various chromosomes. The other end sequence showed homology to only one genomic contig (NW_001035897). The contig contains a part of the Mid1 gene (also known as Fxy), which spans the pseudoautosomal boundary on X chromosome (18). The result of the BAC end sequencing suggested that the BAC clone contained a part of the mouse PAR. This is reasonably accounted for by chromosomal synteny between mouse and human; that is, the human HIOMT/ASMT gene is located in the PAR1 that is homologous to mouse PAR.
Using the BAC clone, we performed FISH on metaphase chromosome preparations from B6J mice. We obtained strong hybridization signals of apparently identical intensity on most proximal segments of the X and Y chromosomes corresponding to PAR (Fig. 2A). A weak signal was detected on the most proximal segment of chromosome 9. Mouse PAR and the subtelomeric region of chromosome 9 reportedly share a homologous sequence, which is highly repetitive (19, 20).
Fig. 2.
Chromosomal localization of mouse Hiomt gene. (A) FISH using the BAC DNA probe containing Hiomt gene. The strong signals were observed at the distal region of the sex chromosomes with equal signal intensity. Weak signals were found at the distal region of chromosome 9. (B) Schematic map of the mouse PAR. Because the directions of Sts and Hiomt genes are unknown, these genes are shown without arrowheads.
Sts (steroid sulfatase; U37545) is the only gene that has been known to be located in the mouse PAR (21, 22), whereas it has not mapped on the reference genome sequence (Build 37). We confirmed that the BAC clone contained the Sts gene. Next, we examined the linkage between Hiomt, Sts, and Mid1 (exon 10, located in the PAR). Male (B6J × MSM) F1 mice and female B6J were mated to produce a total of 103 progenies, and the three PAR loci were genotyped. The nucleotide sequences used for the genotyping are shown in Fig. S4. The recombination fractions during male meiosis were 0.126 (Hiomt–Sts), 0.175 (Sts–Mid1), and 0.262 (Mid1–Hiomt). The result indicated that Hiomt maps distal to Sts, and the genetic distances estimated using Haldane's map function were 21.5 cM (Mid1–Sts) and 14.5 cM (Sts–Hiomt) (Fig. 2B).
Interstrain Variation of Mouse Hiomt.
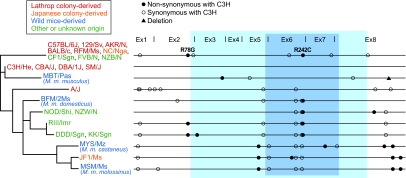
Ebihara et al. (7–9) reported the genetic defects of HIOMT enzyme in many mouse strains as well as B6J (Table S1). To investigate genetic diversity of Hiomt in the divergent lineages, we sequenced the entire CDS of Hiomt gene in 24 inbred strains: nine laboratory strains derived from Lathrop's stock (23); two strains derived from Japanese fancy mice; four strains derived from wild mice of different Mus musculus subspecies (domesticus, musculus, castaneus, and molossinus); and seven strains of other or unknown origin. We defined the nucleotide sequence of C3H as a reference and sorted out the SNPs and deletions identified. On average, 7.2 SNPs/deletion were found in Hiomt CDS of each strain, and 2.3 of these caused amino acid changes (Fig. 3 and Table S2). The B6J-type allele was shared not only in many strains from Lathrop's stock but also in several strains of other origins. This cannot exclude the possibility of genetic contamination of mouse strains. Genealogy and mutation analysis of the Hiomt gene in different strains showed that (i) R78G and R242C were found in various strains, whereas synonymous mutations of the B6J-type allele were scarcely present in the other strains having R78G and/or R242C mutations; (ii) M. m. castaneus, M. m. molossinus (a natural hybrid between musculus and castaneus), and JF1 were grouped into a cluster, supporting the validity of the lineage; (iii) the C3H-type allele, shared only by its related strains (CBA/J, DBA/1J, and SM/J), was likely to be originated from M. m. musculus; and (iv) the origin of B6J-type allele was unclear.
Fig. 3.
Sequence variation in Hiomt CDS of different mouse strains. Twenty-four strains were analyzed and are classified into four origins (essentially according to ref. 23). Bars on right schematically show the CDSs from the initiator codon to the stop codon. Nonsynonymous and synonymous base changes are indicated by closed and open circles, respectively, and a deletion is denoted by a closed triangle. Detailed information on the SNPs, deletions, and amino acid substitutions is presented in Table S2. The functional domain conserved among SAM-dependent methyltransferases and the amino acid sequence highly conserved among vertebrate HIOMT proteins are indicated by blue and cyan backgrounds. A phylogenetic tree was constructed by using the maximum likelihood method. The deepest root was determined by a separate calculation using the sequence of rat Hiomt.
Melatonin Productivity and Circadian Behaviors.
Melatonin receptors are highly expressed in the hypothalamic suprachiasmatic nucleus (SCN), where the self-sustained circadian oscillator located, and exogenous melatonin influences the circadian rhythm of mice (24, 25). However, the role of endogenous melatonin on regulation of the circadian oscillator has been unclear. We characterized wheel-running behavior of (B6J × C3H) F2 intercross mice to evaluate the effect of endogenous melatonin on the circadian rhythm and its related behaviors. The male F2 intercrosses were genotyped and classified into two groups, melatonin-proficient and melatonin-deficient mice (each n = 15), according to the result mentioned above (Fig. 1E). Mice were maintained in LD12:12, and all of the mice tested displayed higher activity at night (Fig. S6). Next, mice were transferred to an inverted LD12:12, and all of the mice were re-entrained within 1 week. Both melatonin-proficient and melatonin-deficient mice were entrained also to a skeleton photoperiod (L1:D10:L1:D12), which is ecologically more relevant. After a 2-week LD12:12, mice were placed in constant darkness. There was no significant difference in free-running period of circadian locomotor activity (melatonin-proficient, 23.74 ± 0.20 h (mean ± SD); melatonin-deficient, 23.77 ± 0.17 h). Mice were exposed to a 1-h light pulse during the early subjective night. Acute suppression of locomotor during the light pulse (light masking effect) and light-induced phase shift were equally observed in melatonin-proficient and melatonin-deficient mice.
Melatonin Levels Under Long- and Short-Day Photoperiods.
Next, we focused on a link between photoperiodism, a physiologic response to the length of day or night, and melatonin and Hiomt, because melatonin plays an essential role in conveying temporal information for reproductive function in seasonal mammals (2, 3, 26). In a wild field, mice (Mus musculus species) generally showed seasonal breeding with high reproductive activity in summer; however, the seasonality was considered to be the result of annual fluctuations in environmental conditions such as food and refuge availability (refs. 27–29 and references therein).
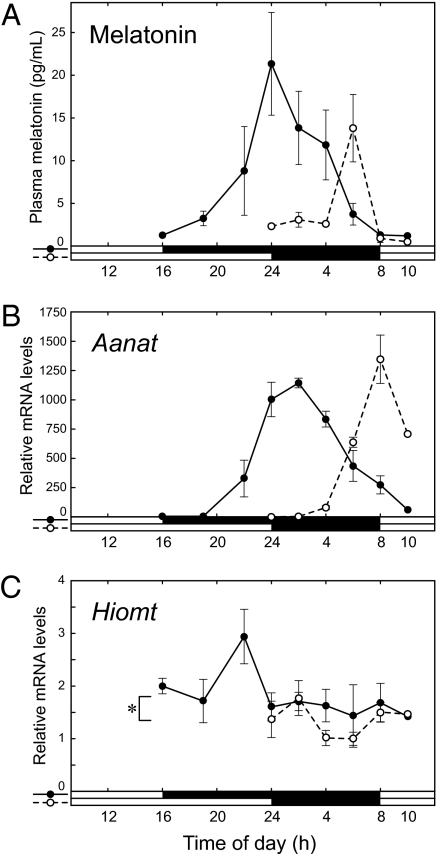
We used MSM mice to investigate the photoperiodic change in the melatonin secretion profile. MSM mice were maintained in LD12:12 and divided into two groups: a short-day photoperiod (SP) group transferred in LD8:16, and a long-day photoperiod (LP) group in LD16:8. Four weeks later, we examined nocturnal melatonin levels in the plasma and mRNA expression levels of Aanat and Hiomt in the pineal glands. The amount of plasma melatonin during nighttime under SP was >3 times higher than that under LP (Fig. 4A) and ≈1.5 times higher than that in LD12:12 (Fig. S2A). Strangely, Aanat mRNA level under SP peaked at dawn, lagging behind the peak of melatonin production (Fig. 4B). Whereas the peak value of Aanat mRNA under SP was similar to that under LP, Hiomt mRNA level under SP was significantly increased compared with that under LP (P = 0.015, general linear model statistics) (Fig. 4C). The increased Hiomt expression may contribute to the elevated melatonin production under SP (Fig. 4A).
Fig. 4.
Photoperiodic change in melatonin synthesis and secretion. (A) Nocturnal melatonin secretion profiles of MSM mice maintained in short-day and long-day photoperiods. Plasma melatonin levels of mice in LD8:16 (closed circles and solid line) and in LD16:8 (open circles and dashed line) were assayed using ELISA (mean ± SD, n = 3; except for 16:00 and 10:00, n = 1). (B, C) Nocturnal Aanat (B) and Hiomt (C) mRNA profiles in short-day and long-day photoperiods. Pineal glands were collected from the mice shown in A. Aanat, Hiomt, and Actb mRNAs were assayed by RT-PCR. Aanat and Hiomt mRNA levels normalized to Actb mRNA were shown as values (mean ± SD, n = 3; except for 16:00 and 10:00, n = 1) relative to the lowest values (Aanat, 24:00 of mice in LD16:8; Hiomt, 6:00 of mice in LD8:16). Nocturnal Hiomt mRNA level was significantly increased under short-day photoperiod (*P < 0.02). In this statistical analysis, which was performed by using R, we used a general linear model with two factors, photoperiod and ZT, following to conversion of local time (time of day) into ZT. For example, 4 AM in a short LD8:16 photoperiod is ZT21, and the same local time in a long LD16:8 photoperiod is ZT18.
Melatonin and Gonadal Development in Mice.
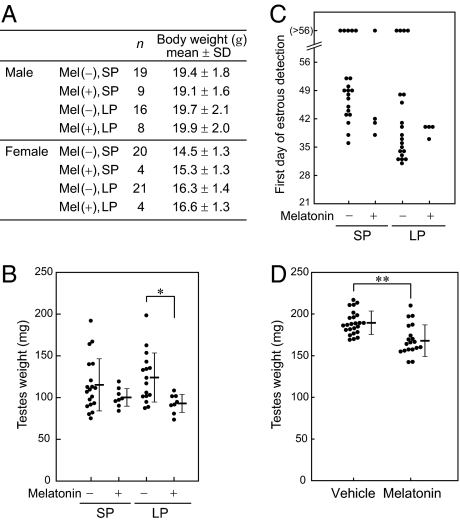
We examined the effect of melatonin productivity trait and light condition (LP or SP) on mouse gonadal development. Male (B6J × MSM) F1 mice and female B6J were mated to produce melatonin-proficient and melatonin-deficient N2 offspring. The dams were transferred to SP during gestation and maintained in the condition until weaning. The weaned mice were housed individually, and half the mice were transferred to LP. These mice were weighed at 7 weeks of age (Fig. 5A). There was no difference in body weight in males with regard to the melatonin productivity or the light conditions. In females, however, body weights of mice under LP were significantly greater than those under SP [F(1,45) = 9.03, P < 0.01, ANOVA], although the melatonin productivity did not influence the body weight.
Fig. 5.
Effect of melatonin on mouse gonadal development. (A–C) The offspring of (B6J × MSM) × B6J were maintained in LD8:16 (SP) or LD16:8 (LP) after weaning. Melatonin productivity is estimated from the genotypes of Aanat and Hiomt. (A) Effect on body weight at 7 weeks of age. (B) Effect on male gonadal development. Testes were weighed at 8 weeks of age. Values from individual animals (circles) as well as mean ± SD values (horizontal bars) are shown. Testicular weights of melatonin-deficient males were significantly greater than those of melatonin-deficient mice (P < 0.01, ANOVA), and it was particularly prominent under LP (*P < 0.002, posthoc t test). (C) Effect on female gonadal development. The first day of predominant appearance of cornified epithelial cells in a vaginal smear is plotted. (D) The effect of exogenous melatonin on testis development of melatonin-deficient ICR mice. Testes were weighed at 5 weeks of age. Values from individual animals (circles) as well as mean ± SD values (horizontal bars) are shown. Testicular weights of vehicle-treated males were significantly greater than those of melatonin-treated males (**P < 0.001, t test).
Testicular weight was measured at 8 weeks of age (Fig. 5B). The melatonin productivity trait had an impact on the testis development, and testicular weights of melatonin-proficient males were significantly lower than those of melatonin-deficient mice [F(1,47) = 8.01, P < 0.01, ANOVA]. Posthoc analysis showed a significant difference between melatonin-proficient and melatonin-deficient males under LP (P = 0.0018, t test). For females, the first vaginal opening day was logged and thereafter vaginal smear was collected every morning to check the estrous phase until 8 weeks of age (Fig. 5C); however, no difference was found between melatonin-proficient and melatonin-deficient females.
The effect of exogenous melatonin on gonadal development was examined in melatonin-deficient ICR males reared in LD12:12. ICR mice, although outbred, carry the B6J-type Hiomt allele in a homozygous form and are deficient in melatonin as far as we tested (n = 10). Melatonin was given to dams and offspring via drinking water or daily i.p. injection as described in Materials and Methods. Testicular weight at 5 weeks of age was significantly lower in melatonin-treated mice compared with controls treated with vehicle (P = 0.00057; t test) (Fig. 5D). Taken together, both endogenous melatonin and exogenous melatonin had an antigonadal impact on male mice.
Discussion
Mouse PAR is an enigmatic region of the genome. The current genome assembly of the mouse does not contain the sequence of PAR (6). Three properties of nucleotide sequence in the mouse PAR have been proposed based on the 3′ portion of the Mid1 gene, which is located within the PAR (30, 31): high GC3, shortened introns, and high-density minisatellites. These are also true of Sts and Hiomt (Table S3). The three properties most likely have a correlation with the high rate of recombination within the PAR, which is the sole region required for homologous chromosome pairing between sex chromosomes. Because the PAR is considerably smaller than other chromosomes, meiotic recombination frequently occurs. In this study, we analyzed the genetic distance between Hiomt and the proximal end of PAR (Mid1) was 37 cM (Fig. 2B). The physical size of mouse PAR was reported to be ≈0.7 Mb (32). Taken together, the recombination rate in the mouse PAR is speculated to be >50 cM/Mb, which is ∼100 times higher than that of the genome-wide average (0.56 cM/Mb) (33). Recombination is mutagenic (34) and is also GC biased, promoting mutations from A and T to G and C (31, 35). The mutability of the PAR was clearly shown by sequencing of mouse Hiomt of various strains (Fig. 3 and Table S2). Between B6J and MSM, 10 synonymous and five nonsynonymous SNPs were found in the Hiomt CDS (1,164 bp), and the SNP frequency is markedly higher than the genome-wide average (2.5 coding SNPs per 1,000 bp between B6J and MSM) (NIG Mouse Genome Database at http://molossinus.lab.nig.ac.jp/msmdb/). Localization of the Hiomt gene in the PAR could be the genomic basis for the prevalence of melatonin deficiency in mice.
We argue that the impact of melatonin on the gonadal development, not on the circadian rhythm, could be a key to understanding the physiologic basis for the fixation of melatonin deficiency in mouse breeding colonies. Melatonin has an antigonadal effect in long-day breeders, and contrarily, treatment of short-day breeders with melatonin has a progonadal effect (1–3). The present study demonstrated the inhibitory action of endogenous and exogenous melatonin on mouse testis development (Fig. 5B, D). Several experiments of melatonin injection have been conducted in mice, although the results are not consistent. As far as we know, however, the effect of melatonin treatment on the developing gonads of melatonin-deficient mice has not been examined. Ono et al. recently reported that a long-day breeder-like photoperiodic signal pathway was retained in laboratory mice (36). The report and our results suggest that mice are potentially a member of long-day breeder family. Day length (short or long photoperiod), however, did not affect testis development of mice, whereas melatonin was secreted more under short photoperiod (ref. 37 and this study). That means photoperiodic control of reproduction is unlikely in mice, which is also shown by ecological studies in the wild (27–29). Again, it should be stressed that genetically melatonin-deficient mice showed accelerated testis growth irrespective of day length (Fig. 5B). Artificial selection of melatonin-deficient mice based on rapid development of testis would be reasonable, whether it is an intentional or unconscious selection. We found that the B6J-type allele of Hiomt is shared in several inbred strains unrelated to B6J (Fig. 3). The expansive prevalence of the B6J-type allele is likely to support the selection pressure rather than genetic drift.
Some researchers claim that melatonin has an antioxidant effect (reviewed in ref. 38). Because laboratory mice were used principally for cancer research in the early years, oncogenic traits due to melatonin deficiency, if any, might also contribute to the artificial selection, although cancer risk caused by melatonin deficiency has not been elucidated. In addition, artificial selection based on other traits could not be ruled out. Melatonin receptors are expressed not only in the pituitary pars tuberalis and SCN, which are crucial sites for photoperiodism and circadian rhythm, but also slightly in several hypothalamic nuclei, hippocampus, cerebral cortex, amygdala, and so on (25). Identification of the mouse Hiomt gene will provide “melatonin knockout” or “melatonin knockin” mice (e.g., melatonin-deficient MSM mice or melatonin-proficient B6J mice), which would tell us the uncharacterized functions involving melatonin such as neural development (39) and mood (40).
Materials and Methods
Mouse Strains.
MSM/Ms mice were kindly supplied from T. Shiroishi (National Institute of Genetics, Mishima, Japan) and maintained in the laboratory animal facility in RIKEN Brain Science Institute (BSI). 129+Ter/SvJcl, BALB/cAJcl, C3H/HeJJcl, and C57BL/6JJcl mice were purchased from CLEA Japan; CBA/JNCrlj, DBA/1JNCrlj, and NC/NgaTndCrlj mice were from Charles River Laboratories Japan; and AKR/NSlc, NZW/NSlc, and Slc:ICR mice were from Japan SLC. The Wako Animal Experiment Committee, RIKEN, approved all animal experiment protocols.
Genomic DNA samples of the following had been extracted from the tail tips or kidneys: BFM/2Ms (from T. Shiroishi; RIKEN BioResource Center [BRC] strain no, RBRC00659), CF1/Sgn (RBRC01674), DDD/Sgn (RBRC01670), FVB/NJcl (CLEA Japan), JF1/Ms (from T. Shiroishi; RBRC00639), KK/Sgn (RBRC01677), MBT/Pas (from X. Montagutelli, Institut Pasteur; RBRC01164), MYS/Mz (from C. Koshimoto, University of Miyazaki; RBRC01196) (41), NOD/ShiJic (CLEA Japan), NZB/NSlc (Japan SLC), RIII/ImrKiKua (RBRC02519), RFM/MsStm (RBRC00216), and SM/JHam (RBRC01221). All of the strains, except for FVB/NJcl, NOD/ShiJic, and NZB/NSlc, were provided by RIKEN BRC, which participates in the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
Cloning of Mouse Hiomt cDNA.
A detailed procedure is provided in SI Materials and Methods, and the progress of the cloning and the locations of the primers used are shown in Fig. S1. The database accession numbers of mouse Hiomt cDNA sequences of B6J, C3H, and MSM are AB512670, AB512671, and AB512672, respectively.
Cloning of Mouse Hiomt Gene.
The mouse Hiomt gene was amplified by PCR from genomic DNA in three overlapping fragments and subjected to direct sequencing. Detailed procedures are provided in SI Materials and Methods. The database accession numbers of mouse Hiomt sequences of B6J and C3H are AB512673 and AB512674, respectively.
Genotyping of Mice.
Genomic DNA was extracted from mouse tail biopsy samples by proteinase K–SDS digestion. Genotyping of the Hiomt, Aanat, Mid1, and Sts alleles was performed by PCR and direct sequencing of the PCR products. The primers used are shown in Fig. S4.
Transient Expression of Mouse HIOMT, HIOMT Enzyme Assay, and Generation of Mouse HIOMT Antiserum.
HIOMT activity was determined by a method based on Bernard et al. (42). Detailed procedures are provided in SI Materials and Methods.
Melatonin Assay.
Mice were deeply anesthetized with diethyl ether in a very dim red light (at night), and blood was collected from the inferior vena cava followed by sampling of the pineal gland in dim red light (at night). A detailed description of the ELISA for melatonin is given in SI Materials and Methods.
Quantitative RT-PCR.
Details of quantitative RT-PCR are provided in SI Materials and Methods.
Recording of Circadian Behaviors.
Wheel-running activity was measured as previously described (40), and a detailed procedure is provided in SI Materials and Methods.
Administration of Melatonin.
Melatonin in drinking water (10 mg/L) was given to dams from the third trimester of pregnancy until weaning (day 21 postpartum). The weaned offspring were given melatonin in drinking water (10 mg/L) for 1 week and thereafter by daily i.p. injection (50 μg) at ZT11–12. The concentration of vehicle (ethanol) was 0.005% (drinking water) and 0.1% (ip injection).
Supplementary Material
Acknowledgments
We thank Dr. Toshihiko Shiroishi (National Institute of Genetics) for providing BFM/2Ms, JF1/Ms, and MSM strains; Drs. Kazuhiro Yamakawa and Tetsushi Yamagata (RIKEN BSI) for advice on breeding and rearing MSM mice; Dr. Eiki Takahashi (RIKEN BSI) for discussions concerning mouse breeding and development; Dr. Thomas Bourgeron and Cecile Pagan (Institut Pasteur) for advice on HIOMT enzyme assay; and Taeko Miyauchi, Mizuho Ishiwata, Fukiko Isono, Mizue Kametani, and Misako Yuzuriha for technical assistance. We are grateful to the Support Unit for Bio-material Analysis (Research Resources Center, RIKEN BSI) for help with DNA sequencing. This work was supported by grants to the Laboratory for Molecular Dynamics of Mental Disorders, RIKEN BSI; and Grants-in-Aid from MEXT.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (DDBJ/EMBL/GenBank accession nos. AB512670–AB512674).
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914399107/DCSupplemental.
References
- 1.Arendt J. Melatonin and the Mammalian Pineal Gland. London: Chapman and Hill; 1995. [Google Scholar]
- 2.Reppert SM, editor. Development of Circadian Rhythmicity and Photoperiodism in Mammals. Ithaca, NY: Perinatology Press; 1989. [Google Scholar]
- 3.Pévet P. Melatonin: From seasonal to circadian signal. J Neuroendocrinol. 2003;15:422–426. doi: 10.1046/j.1365-2826.2003.01017.x. [DOI] [PubMed] [Google Scholar]
- 4.Ganguly S, Coon SL, Klein DC. Control of melatonin synthesis in the mammalian pineal gland: The critical role of serotonin acetylation. Cell Tissue Res. 2002;309:127–137. doi: 10.1007/s00441-002-0579-y. [DOI] [PubMed] [Google Scholar]
- 5.Coon SL, et al. Pineal serotonin N-acetyltransferase: Expression cloning and molecular analysis. Science. 1995;270:1681–1683. doi: 10.1126/science.270.5242.1681. [DOI] [PubMed] [Google Scholar]
- 6.Church DM, et al. Mouse Genome Sequencing Consortium Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebihara S, Marks T, Hudson DJ, Menaker M. Genetic control of melatonin synthesis in the pineal gland of the mouse. Science. 1986;231:491–493. doi: 10.1126/science.3941912. [DOI] [PubMed] [Google Scholar]
- 8.Ebihara S, Hudson DJ, Marks T, Menaker M. Pineal indole metabolism in the mouse. Brain Res. 1987;416:136–140. doi: 10.1016/0006-8993(87)91505-8. [DOI] [PubMed] [Google Scholar]
- 9.Goto M, Oshima I, Tomita T, Ebihara S. Melatonin content of the pineal gland in different mouse strains. J Pineal Res. 1989;7:195–204. doi: 10.1111/j.1600-079x.1989.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 10.Roseboom PH, et al. Natural melatonin ‘knockdown’ in C57BL/6J mice: Rare mechanism truncates serotonin N-acetyltransferase. Brain Res Mol Brain Res. 1998;63:189–197. doi: 10.1016/s0169-328x(98)00273-3. [DOI] [PubMed] [Google Scholar]
- 11.Ellis N, Goodfellow PN. The mammalian pseudoautosomal region. Trends Genet. 1989;5:406–410. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- 12.Helena Mangs A, Morris BJ. The human pseudoautosomal region (PAR): Origin, function and future. Curr Genomics. 2007;8:129–136. doi: 10.2174/138920207780368141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soriano P, et al. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc Natl Acad Sci USA. 1987;84:7218–7220. doi: 10.1073/pnas.84.20.7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriwaki K, et al. Unique inbred strain MSM/Ms established from the Japanese wild mouse. Exp Anim. 2009;58:123–134. doi: 10.1538/expanim.58.123. [DOI] [PubMed] [Google Scholar]
- 15.Goto M, Oshima I, Hasegawa M, Ebihara S. The locus controlling pineal serotonin N-acetyltransferase activity (Nat-2) is located on mouse chromosome 11. Brain Res Mol Brain Res. 1994;21:349–354. doi: 10.1016/0169-328x(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 16.Kennaway DJ, Voultsios A, Varcoe TJ, Moyer RW. Melatonin and activity rhythm responses to light pulses in mice with the Clock mutation. Am J Physiol Regul Integr Comp Physiol. 2003;284:R1231–R1240. doi: 10.1152/ajpregu.00697.2002. [DOI] [PubMed] [Google Scholar]
- 17.Abe K, et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer S, Perry J, Kipling D, Ashworth A. A gene spans the pseudoautosomal boundary in mice. Proc Natl Acad Sci USA. 1997;94:12030–12035. doi: 10.1073/pnas.94.22.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbers K, Francke U, Soriano P, Jaenisch R, Müller U. Structure and chromosomal mapping of a highly polymorphic repetitive DNA sequence from the pseudoautosomal region of the mouse sex chromosomes. Cytogenet Cell Genet. 1990;53:129–133. doi: 10.1159/000132912. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, et al. Methylation imprinting was observed of mouse mo-2 macrosatellite on the pseudoautosomal region but not on chromosome 9. Chromosoma. 1994;103:450–458. doi: 10.1007/BF00337383. [DOI] [PubMed] [Google Scholar]
- 21.Keitges E, Rivest M, Siniscalco M, Gartler SM. X-linkage of steroid sulphatase in the mouse is evidence for a functional Y-linked allele. Nature. 1985;315:226–227. doi: 10.1038/315226a0. [DOI] [PubMed] [Google Scholar]
- 22.Salido EC, et al. Cloning and expression of the mouse pseudoautosomal steroid sulphatase gene (Sts) Nat Genet. 1996;13:83–86. doi: 10.1038/ng0596-83. [DOI] [PubMed] [Google Scholar]
- 23.Beck JA, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 24.Benloucif S, Masana MI, Dubocovich ML. Responsiveness to melatonin and its receptor expression in the aging circadian clock of mice. Am J Physiol. 1997;273:R1855–R1860. doi: 10.1152/ajpregu.1997.273.6.R1855. [DOI] [PubMed] [Google Scholar]
- 25.von Gall C, Stehle JH, Weaver DR. Mammalian melatonin receptors: Molecular biology and signal transduction. Cell Tissue Res. 2002;309:151–162. doi: 10.1007/s00441-002-0581-4. [DOI] [PubMed] [Google Scholar]
- 26.Stehle JH, von Gall C, Korf H-W. Melatonin: A clock-output, a clock-input. J Neuroendocrinol. 2003;15:383–389. doi: 10.1046/j.1365-2826.2003.01001.x. [DOI] [PubMed] [Google Scholar]
- 27.Bomford M. Food and reproduction of wild house mice. III. Experiments on the breeding performance of caged house mice fed rice-based diets. Aust Wildl Res. 1987;14:207–218. [Google Scholar]
- 28.Matthewson DC, van Aarde RJ, Skinner JD. Population biology of house mice (Mus musculus L.) on sub-Antarctic Marion Island. S Afr J Zool. 1994;29:99–106. [Google Scholar]
- 29.King CM, Innes JG, Flux M, Kimberley MO. Population biology of small mammals in Pureora Forest Park. 2. The feral house mouse (Mus musculus) N Z J Ecol. 1996;20:253–269. [Google Scholar]
- 30.Perry J, Ashworth A. Evolutionary rate of a gene affected by chromosomal position. Curr Biol. 1999;9:987–989. doi: 10.1016/s0960-9822(99)80430-8. [DOI] [PubMed] [Google Scholar]
- 31.Montoya-Burgos JI, Boursot P, Galtier N. Recombination explains isochores in mammalian genomes. Trends Genet. 2003;19:128–130. doi: 10.1016/S0168-9525(03)00021-0. [DOI] [PubMed] [Google Scholar]
- 32.Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachman MW. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 2001;17:481–485. doi: 10.1016/s0168-9525(01)02409-x. [DOI] [PubMed] [Google Scholar]
- 35.Marais G. Biased gene conversion: Implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. [DOI] [PubMed] [Google Scholar]
- 36.Ono H, et al. Involvement of thyrotropin in photoperiodic signal transduction in mice. Proc Natl Acad Sci USA. 2008;105:18238–18242. doi: 10.1073/pnas.0808952105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yellon SM, Tran LT. Photoperiod, reproduction, and immunity in select strains of inbred mice. J Biol Rhythms. 2002;17:65–75. doi: 10.1177/074873002129002348. [DOI] [PubMed] [Google Scholar]
- 38.Reppert SM, Weaver DR. Melatonin madness. Cell. 1995;83:1059–1062. doi: 10.1016/0092-8674(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 39.Melke J, et al. Abnormal melatonin synthesis in autism spectrum disorders. Mol Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasahara T, et al. Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol Psychiatry. 2006;11:577–593. doi: 10.1038/sj.mp.4001824. , 523. [DOI] [PubMed] [Google Scholar]
- 41.Tsuchiya K, et al. MYS/Mz, a novel inbred strain developed from Mus musculus castaneus (Translated from Japanese) Kyusyu J Exp Anim. 2000;16:23–25. [Google Scholar]
- 42.Bernard M, Donohue SJ, Klein DC. Human hydroxyindole-O-methyltransferase in pineal gland, retina and Y79 retinoblastoma cells. Brain Res. 1995;696:37–48. doi: 10.1016/0006-8993(95)00651-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.