Abstract
Histone deacetylase (HDAC) inhibitors are emergent cancer drugs. HR23B is a candidate cancer biomarker identified in a genome-wide loss-of-function screen which influences sensitivity to HDAC inhibitors. Because HDAC inhibitors have found clinical utility in cutaneous T-cell lymphoma (CTCL), we evaluated the role of HR23B in CTCL cells. Our results show that HR23B governs the sensitivity of CTCL cells to HDAC inhibitors. Furthermore, proteasome activity is deregulated in HDAC inhibitor-treated CTCL cells through a mechanism dependent upon HR23B, and HDAC inhibitors sensitize CTCL cells to the effects of proteasome inhibitors. The predictive power of HR23B for clinical response to HDAC inhibitors was investigated through an analysis of a unique collection of CTCL biopsies taken from a phase II clinical trial, where there was a frequent coincidence between HR23B expression and clinical response to HDAC inhibitor. Our study supports the personalized medicine approach for treating cancer and the increasing drive to translate laboratory-based findings into clinical utility.
Keywords: cancer, histone deacetylase inhibitor, biomarker, proteasome, clinical trial
It is widely recognized that one of the most significant hurdles to developing new and efficacious cancer drugs is the absence of proven strategies that enable responsive tumors to be identified and treated with the most effective drug. Predictive biomarkers that inform on the clinical response to cancer therapies will not only assist in planning focused and hypothesis-driven clinical development programs but, once approved, provide a companion biomarker that enables patients to be stratified and aligned with an appropriate therapeutic regime (1).
Aberrant epigenetic control is an early event in the onset of tumor progression, and there have been intense efforts to develop drugs that modulate epigenesis in cancer (2). One of the most promising approaches has been seen in the development of inhibitors of histone deacetylase (HDAC). HDAC is a family of enzymes that regulate the acetylation level of chromatin, together with a variety of nonhistone proteins (3). Proteins that are involved with tumor progression, including the cell cycle, apoptosis, angiogenesis, and cell invasion, are influenced by acetylation (4).
HDAC inhibitors are potent antiproliferative agents in cell-based studies, where they exhibit striking effects on tumor cell proliferation (5–7). Consequently, there has been great interest in developing HDAC inhibitors as new anticancer agents, and an extensive number of clinical trials are underway in which HDAC inhibitors either alone or in combination with other agents are being evaluated (8). To date, SAHA (also known as Vorinostat and Zolinza) and more recently romidepsin (also known as depsipeptide, FK228, and Istodax) are the only HDAC inhibitor-based therapies that have achieved regulatory approval, being marketed for the treatment of advanced and refractory cutaneous T-cell lymphoma (CTCL) (9–11) (http://istodax.com) . Other than CTCL, tumor types that have been established to undergo a favorable response to HDAC inhibitor-based therapies remain to be identified. In part, this reflects the widespread expression of HDAC subunits in both normal and malignant cells and the absence of information suggesting which tumors are likely to be sensitive.
In previous studies, we used a genome-wide loss-of-function screen to identify genes that govern the sensitivity of tumor cells to HDAC inhibitors (12). The screen identified HR23B which, in cell culture, sensitizes osteosarcoma cells to HDAC inhibitor-dependent cell death. At a mechanistic level, HR23B has an important role in shuttling ubiquitinated cargo proteins to the proteasome (13, 14). Indeed, proteasomes display aberrant activity in tumor cells treated with HDAC inhibitors, and HR23B is in part responsible for the effect on proteasome activity (12).
Because HR23B governs the sensitivity of tumor cells to HDAC inhibitors in vitro, we reasoned that it might find utility as a predictive biomarker in the clinical setting. To this end, we have explored the role of HR23B in CTCL cell lines, where we have found HR23B to be a sensitivity determinant for response to HDAC inhibitors. Importantly, proteasome activity is deregulated by HDAC inhibitors through a mechanism involving HR23B, and HDAC inhibitors sensitize CTCL cells to the effects of proteasome inhibitors. Remarkably, in CTCL biopsies taken from a phase II clinical trial there was a frequent coincidence between HR23B and clinical response to HDAC inhibitor therapy, supporting the idea that HR23B is a clinically useful biomarker. As such, HR23B represents a biomarker defined through a functional loss-of-function screen to find clinical utility as a predictor of therapeutic response. Our study supports the personalized medicine approach for treating cancer, and the increasing drive to translate laboratory-based findings into clinically useful tools.
Results
HR23B Regulates Tumor Cell Sensitivity to HDAC Inhibitors.
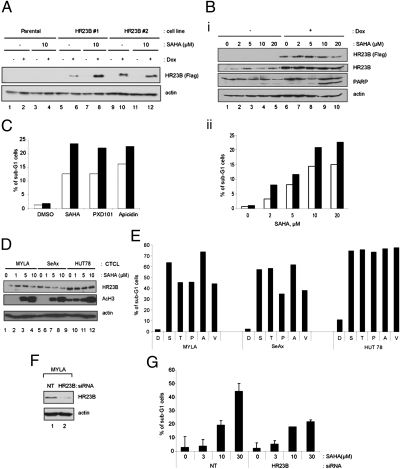
We have evaluated the role of HR23B in HDAC inhibitor-induced cell death by means of depleting HR23B with siRNA in U2OS osteo-sarcoma cells, which became less sensitive to apoptosis upon SAHA treatment (12). To establish whether HR23B has a dominant role in regulating tumor cell sensitivity to HDAC inhibitors, U2OS cell lines in which the expression of stable ectopic HR23B was under conditional control (induction upon doxycycline treatment) were prepared (Fig. 1 A and B). When the sensitivity of stable cells in which HR23B had been induced (+Dox) was compared with their uninduced counterparts (−Dox), a significant increase in sensitivity to HDAC inhibitors, which included SAHA, was apparent [on average a 40% increase in sub-G1 cells compared with the control (−Dox) treatment; Fig. 1 Bii and C]. Increased levels of poly (ADP ribose) polymerase cleavage upon SAHA treatment were also apparent in the induced compared with uninduced cells (Fig. 1B). These results therefore support a dominant role for HR23B in regulating the cellular outcome of HDAC inhibitor treatment.
Fig. 1.
HR23B in CTCL cells. (A) Immunoblot showing protein levels of FLAG-HR23B in uninduced (−) and induced (+) U2OS-TET cells (#1 and #2) with actin as a loading control. Dox, doxycycline. (B) U2OS-TET cells expressing ectopic FLAG-HR23B were treated with doxycycline for 16 h before an additional 48-h treatment with SAHA (2, 5, 10, and 20 μM) or DMSO control (0). (i) Immunoblot showing protein levels of FLAG-HR23B in uninduced (−) and induced (+) U2OS-TET cells (#1), HR23B, and PARP cleavage. Actin was used as a loading control. (ii) Graph showing the percentage sub-G1 cells in induced (black bar) cell lines treated with SAHA compared with DMSO control (clear bar). (C) The levels of sub-G1 apoptotic cells determined by FACS analysis after treating the U2OS cell line (#1) expressing ectopic FLAG-HR23B. Cells were treated with doxycycline for 16 h before an additional 48-h treatment with SAHA (10 μM), PXD101 (4 μM), apicidin (4 μM), or DMSO control. The graph shows the percentage of sub-G1 cells in induced (black bar) cell lines treated with HDAC inhibitors compared with DMSO control (clear bar). (D) Immunoblot showing protein levels of HR23B in the CTCL cell lines MYLA, SeAx, and HUT78 treated with the indicated dose of SAHA (μM). Acetylated H3 and actin (loading control) are shown for comparison. (E) Level of sub-G1 apoptotic cells determined by FACS after treating MYLA, SeAx, or HUT78 with the HDAC inhibitors SAHA (S), TSA (T), PXD101 (P), and apicidin (A), all at 5 μM, and valproic acid (V) at 5 mM. (F) MYLA cells were treated with HR23B or nontargeting control (NT) siRNA for 48 h, followed by immunoblotting. Actin served as the loading control. (G) The levels of sub-G1 MYLA cells determined by FACS after treatment with HR23B siRNA or nontargeting (NT) control siRNA for 48 h followed by treatment with DMSO control (0) or increasing concentrations of SAHA, 3, 10, or 30 (μM), as indicated for 72 h (n = 4; error bars, SEM).
HR23B in CTCL Cell Lines.
CTCL is sensitive to the effects of HDAC inhibitor-based therapy (9, 11, 15, 16). As a consequence, we evaluated the role of HR23B as a predictive biomarker for HDAC inhibitors in CTCL, and began by studying HR23B in CTCL cell lines, including MYLA, SeAx, and HUT78. The level of HR23B was generally similar in treated and untreated cells, although in SeAx there was a modest decline upon increasing SAHA dose (Fig. 1D). As previously reported (7, 15), the CTCL cell lines were sensitive to apoptosis induced by SAHA and a variety of other HDAC inhibitors (TSA, PXD101, apicidin, and valproic acid), although HUT78 was moderately more sensitive than SeAx and MYLA cells (Fig. 1E).
To assess whether HR23B is a sensitivity determinant for HDAC inhibitors in CTCL cells, we examined the effect of de-pleting HR23B in MYLA cells. The introduction of siRNA against HR23B RNA reduced the level of endogenous HR23B compared with the siRNA control treatment (Fig. 1F), and HR23B depletion coincided with reduced sensitivity to apoptosis of MYLA cells upon SAHA treatment (Fig. 1G). Thus, HR23B is functionally important in determining the effect of HDAC inhibitors in CTCL cell lines.
Proteasome Activity in CTCL Cells Treated with HDAC Inhibitors.
HR23B shuttles ubiquitinated cargo proteins to the proteasome for subsequent degradation (13, 14). In U2OS cells treated with HDAC inhibitors, HR23B contributes to the inhibition of proteasome activity (12). We were interested to establish whether there were mechanistic parallels in CTCL, and therefore evaluated proteasome activity in CTCL cells upon HDAC inhibitor treatment.
When proteasomes were purified from HDAC inhibitor-treated MYLA cells and protease activity was measured in vitro using a fluorogenic substrate peptide (12), a significant decrease in catalytic activity was apparent compared with the control treatment (Fig. 2A). The inhibition of proteasome activity was both concentration-of-drug- and time-of-treatment-dependent (Fig. 2A). To assess the role of HR23B in regulating proteasome activity in treated cells, HR23B was depleted in HDAC inhibitor-treated MYLA cells (Fig. 2B) and then proteasome activity was measured in vitro (Fig. 2C). Under conditions of reduced levels of HR23B, the decrease in proteasome activity was no longer apparent (Fig. 2C), suggesting that proteasome activity in HDAC inhibitor-treated cells is in part influenced by HR23B.
Fig. 2.
Proteasome activity in HDAC inhibitor-treated CTCL cells. (A) MYLA cells were treated with vehicle (blue), SAHA 10 μM (red), SAHA 30 μM (green), or proteasome inhibitor (yellow) and harvested at 16 h. Purified proteasomes were then incubated with fluorogenic substrate. Cleaved substrate as arbitrary fluorescence units (AFU) representative of active proteasome was measured at 2-min intervals for 60 min (n = 3; error bars, SEM). (B) Immunoblot showing the expression of HR23B, with actin as a loading control, for the experiment described in C. (C) MYLA cells were treated with HR23B siRNA for 48 h followed by a 48-h treatment with vehicle (blue), SAHA 10 μM (red), SAHA 30 μM (green), or proteasome inhibitor (yellow) and harvested at 24 h. Purified proteasomes were then incubated with fluorogenic substrate. Cleaved substrate representative of active proteasome was measured at 2-min intervals for 60 min (n = 3; error bars, SEM). (D) The levels of sub-G1 cells determined by FACS after treating MYLA cells with increasing concentrations of SAHA (clear) and bortezomib (gray) [0.2, 0.5, 1, and 2× IC50 (1× IC50 = 5 μM SAHA and 50 nM bortezomib)] or a combination (black) for 48 h (n = 3; error bars, SEM). (E and F) Effect of SAHA (S) and bortezomib (B) alone and in combination, in the uninduced (clear) or induced (black) FLAG-HR23B (HR23B #1) stable cell line (described in Fig. 1; HR23B #1), showing the percentage of sub-G1 cells from FACS analysis (E) and relative change in sub-G1 percentage (F); + and − indicate treatment and not with doxycycline, respectively. (G) The level of ectopic HR23B in HR23B #1 cells (− or + doxycycline) treated with 0.5× (upper) or 2× (lower) IC50 of SAHA (S) or bortezomib (B) as indicated.
Because the proteasome is becoming an increasingly well validated cancer target (17), we considered that HR23B might in addition provide useful information on the cellular effects of proteasome inhibitors, particularly in combination with HDAC inhibitors. To pursue this idea, we measured the sensitivity of MYLA cells to bortezomib-induced apoptosis. MYLA cells were sensitive to bortezomib (Fig. 2D), and the combined treatment of SAHA with bortezomib caused increased levels of apoptosis (Fig. 2D). To assess the impact of HR23B, we studied the properties of bortezomib and SAHA in the stable HR23B cell lines (Figs. 1A and 2G). The activity of bortezomib was enhanced upon expressing ectopic HR23B (Fig. 2 E and F; usually between 40 and 50%). Similarly, ectopic HR23B increased the level of apoptosis in the combined SAHA/bortezomib treatment (at 0.5× IC50 usually about 25%; Fig. 2 E and F). However, at high IC50 (2.0×), the effect of the ectopic HR23B on the fraction of apoptotic cells was less marked (Fig. 2 E and F), which might reflect the greater inhibition of proteasome activity under increased drug concentration (0.5 compared with 2.0× IC50).
HR23B Levels in CTCL Biopsies from a Phase II Clinical Trial with SAHA.
To determine whether the expression level of HR23B is a clinically useful biomarker for the response of CTCL to HDAC inhibitor therapy, an analysis was performed on a collection of CTCL biopsies taken from a phase II clinical trial of SAHA (9). Paired skin-lesion biopsies from before and after SAHA treatment were assessed in 20 out of the 33 trial patients, using immunohistochemistry on paraffin-embedded formalin-fixed tissue. A total of 21 samples were available for analysis of pretreatment expression (Table 1) because one patient (patient 8) had two distinct courses of treatment with SAHA and was therefore counted twice. A further 44 posttreatment biopsies taken from patients undergoing active SAHA therapy were also analyzed for HR23B expression, giving a total of 65 biopsies from the phase II clinical trial.
Table 1.
Summary of phase II clinical trial patients, total staining score, and clinical outcome
| Patient number | Total staining score | Clinical outcome |
| 1 | 7 | PR |
| 2 | 8 | PD |
| 3 | 7 | PD |
| 4 | 6 | PD |
| 5 | 8 | PR |
| 6 | 7 | PR |
| 7 | 6 | PR |
| 8a | 9 | PR |
| 8b | 5 | PD |
| 9 | 5 | SD |
| 10 | 10 | SD |
| 11 | 8 | PD |
| 12 | 5 | PD |
| 13 | 8 | SD |
| 14 | 7 | PD |
| 15 | 7 | PR |
| 16 | 6 | SD |
| 17 | 8 | PR |
| 18 | 5 | SD |
| 19 | 7 | PR |
| 20 | 5 | PD |
Total staining scores for expression of HR23B in pretreatment biopsies from patients with advanced CTCL from the phase II trial, with the clinical outcome after treatment with SAHA indicated. PR, partial response; SD, stable disease; PD, progressive disease. The clinical and pathological criteria assessed to define PR, SD, or PD were as described in ref. 9. To determine the TSS, HR23B expression was assessed in the malignant lymphocytes in each biopsy.
The biopsies were scored blind by two independent pathologists and a semiquantitative analysis based upon intensity and frequency of HR23B staining and the type of dermal and epidermal infiltrate was performed on each biopsy (Table S1). A total staining score (TSS) was determined out of 13, where a score of greater than or equal to 6 was considered to be strong HR23B staining. The threshold for differentiating between positive and negative immunostaining was set at a TSS of 6. This optimal cutoff was determined by the receiver operating characteristic curve distribution analysis (18, 19). The TSS was then compared with clinical outcome, namely partial response (PR), stable disease (SD), or progressive disease (PD) (Table 1). Using pretreatment expression of HR23B in the 20 trial patients, the proportion of patients with strong staining who had a clinical response was 68.8% (Table 2). Therefore, the positive predictive value (PPV) of HR23B in this sample set was determined to be 68.8%. When the pre- and posttreatment biopsies were combined (a total of 65 biopsies), a similar PPV was evident at 71.7% (Table 3).
Table 2.
Summary of TSS for pretreatment expression of HR23B in phase II trial CTCL biopsies
| TSS | PR/SD | PD | Total |
| ≥6 | 11 | 5 | 16 |
| <6 | 2 | 3 | 5 |
| Total | 13 | 8 | 21 |
Distribution of patients categorized as having high (≥6) or low (<6) TSS for HR23B according to response to treatment (PR/SD) or no response to treatment (PD) in patients from the phase II trial. Values relating to the pretreatment expression of HR23B are shown. Statistical significance was assessed using Fisher's exact test. Positive predictive value: 68.8%; one-tailed P = 0.262126; two-tailed P = 0.325372.
Table 3.
Summary of TSS for pretreatment and posttreatment expression of HR23B in phase II trial CTCL biopsies
| TSS | PR/SD | PD | Total |
| ≥6 | 38 | 15 | 53 |
| <6 | 5 | 7 | 12 |
| Total | 43 | 22 | 65 |
Distribution of patients categorized as having high (≥6) or low (<6) TSS for HR23B according to response to treatment (PR/SD) or no response to treatment (PD) in patients from the phase II trial treated with SAHA. Values for pretreatment and posttreatment expression of HR23B are shown. Statistical significance was assessed using Fisher's exact test. Positive predictive value: 71.7%; one-tailed P = 0.0522323; two-tailed P = 0.0874581.
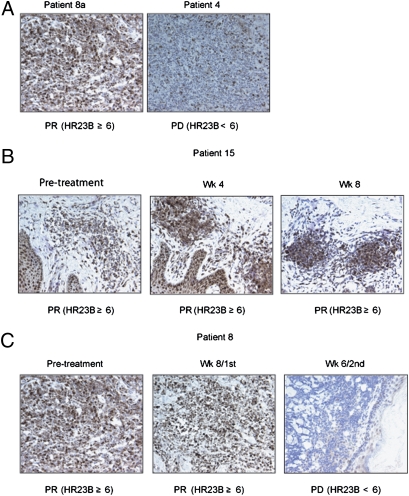
Pretreatment expression of HR23B in patient 8a provided an example of a high level of expression where a partial response to SAHA was observed and, by contrast, expression in patient 4 was low and this patient's disease progressed after SAHA treatment (Fig. 3A). Biopsies from patient 15 demonstrated that expression of HR23B remained high throughout the time that the patient was undergoing a partial response to SAHA (Fig. 3B). Very interestingly, the expression of HR23B coincided with both a response and lack of response in the same patient who was treated with two separate courses of SAHA (Fig. 3C). Thus, patient 8 had high baseline expression of HR23B and derived clinical benefit from SAHA after the first course of treatment, undergoing a partial response. However, 6 weeks into the second course of treatment, the patient's disease became unresponsive and progressive disease was apparent. This lack of response coincided with an almost complete absence of HR23B expression in the tumor mass (Fig. 3C). Together, these results strongly suggest that HR23B is an informative biomarker to identify CTCL patients who will undergo a favorable clinical response to SAHA.
Fig. 3.
HR23B expression in SAHA-treated CTCL biopsies. (A) Representative images of the CTCL biopsies for patient 8a, who underwent a partial response (PR) following treatment with SAHA for 27 weeks before relapsing, and patient 4, who exhibited progressive disease (PD) following 2 weeks on SAHA. Note that HR23B immunostaining is noticeably stronger in the former compared with the latter. The TSS for HR23B is shown in brackets. (B) Representative images of HR23B immunostaining in CTCL biopsies performed on pretreatment and week-4 and week-8 biopsies from patient 15, who presented with Sezary syndrome and underwent a PR throughout SAHA treatment. The TSS for HR23B is indicated. (C) Representative images of HR23B immunostaining in CTCL biopsies performed on pretreatment and posttreatment week-8 biopsies from patient 8, who underwent a PR following SAHA treatment. Following 27 weeks of SAHA treatment, the patient underwent PD and was subsequently placed on a different dose of SAHA. The patient exhibited PD after 6 weeks of treatment. HR23B immunostaining performed on posttreatment week-6 biopsy from this second dose of SAHA is illustrated. The TSS for HR23B is indicated.
HR23B as a Pan-Biomarker for HDAC Inhibitors.
We were interested to evaluate HR23B in CTCL patients treated with HDAC inhibitors other than SAHA. We therefore studied pretreatment biopsies taken from CTCL patients treated with PXD101 (1 patient), depsipeptide/FK228 (2 patients), and LBH589 (4 patients), both pre- and posttreatment. A total of 21 biopsies were tested and ranked according to the HR23B TSS as described earlier (Table 4). One of the patients provided an example of pretreatment HR23B staining and following treatment with PXD101 after 7 cycles (Fig. 4A). The pretreatment HR23B TSS was greater than 6, when the patient underwent a partial response. This contrasted with later during the treatment regime (cycle 7), when the TSS was less than 6, which coincided with disease progression. Another patient provided an example of HR23B biopsy staining and TSS score after treatment with depsipeptide. HR23B remained high during the time when the patient underwent a partial response to depsipeptide (pretreatment and cycle 2), and was low at drug cycle 3 when it coincided with disease progression (Fig. 4B).
Table 4.
Summary of TSS in CTCL biopsies taken from patients treated with PXD101, depsipeptide, or LBH589
| TSS | PR/SD | PD | Total |
| ≥6 | 9 | 4 | 13 |
| <6 | 4 | 4 | 8 |
| Total | 13 | 8 | 21 |
Distribution of patients categorized as having high (≥6) or low (<6) TSS for HR23B according to response to treatment (PR/SD) or no response (PD) in patients treated with PXD101, depsipeptide, or LBH589. Positive predictive value: 64.7%.
Fig. 4.
HR23B expression in PXD101, LBH589, and depsipeptide-treated CTCL biopsies. (A) Representative images are shown of the CTCL biopsies from a patient treated with PXD101 who underwent a partial response following treatment but relapsed when the disease progressed after treatment with seven cycles of PXD101. Note that HR23B immunostaining is noticeably stronger at pretreatment compared with when the patient's disease was progressing (after seven cycles of treatment). The TSS for HR23B is shown in parentheses. (B) Representative images of HR23B immunostaining performed on pretreatment and after cycle-2 and cycle-3 biopsies from a patient who underwent a partial response following depsipeptide treatment for two cycles but then the disease progressed after the third cycle of treatment. Note that HR23B immunostaining is noticeably stronger at pretreatment and up to cycle 2 compared with when the patient's disease was progressing after cycle 3. The TSS for HR23B is shown in parentheses.
A total of 21 biopsies were characterized according to HR23B TSS (Table 4). Comparing the TSS value with clinical outcome gave a PPV of 64.7%. This value rests on a smaller number of bi-opsies analyzed compared with the SAHA trial biopsies (Table 3), and as a consequence was not open to statistical analysis. Nevertheless, the PPV continues to support the favorable coincidence between HR23B levels and clinical outcome. Thus, HR23B could be considered as a biomarker for HDAC inhibitor-based therapies in addition to SAHA.
Discussion
HR23B and CTCL.
Although HDAC inhibitors represent a group of emerging and promising anticancer agents, knowledge of their clinical utility remains rudimentary. This reflects in part the paucity of information available on the tumor types that are likely to undergo a favorable clinical response. It was against this background that we sought to identify predictive biomarkers that inform on the tumor response to HDAC inhibitor-based therapies. HR23B represents a candidate biomarker that was identified in a genome-wide loss-of-function screen for HDAC inhibitor-induced apoptosis (12).
Because the validation of HR23B has relied upon studies in cell culture, it was important for us to identify ways in which its clini-cal utility as a predictive biomarker could be assessed. Because SAHA has gained regulatory approval (as Zolinza) for late-stage refractory CTCL (9, 11), we considered that evaluating HR23B in CTCL cell lines, followed by an analysis of its expression and properties in the clinical setting, might provide useful information about its longer-term clinical potential. Our results suggest that in CTCL cell lines, HR23B influences sensitivity to HDAC inhibitor-induced apoptosis. Specifically, manipulating HR23B levels in CTCL cells resulted in altered sensitivity to the effects of HDAC inhibitors which, our results suggest, is partly mediated through the proteasome.
Most significantly, an analysis of HR23B levels in a unique collection of CTCL biopsies taken from a phase II trial of SAHA suggested that HR23B shows considerable promise as an informative biomarker. Specifically, relating the levels of HR23B in 65 CTCL biopsies to clinical response gave a PPV of 71.7%. Although we recognize that the number of biopsies surveyed is limited, we are nevertheless very encouraged that this high PPV supports the informative nature of HR23B levels as a predictive biomarker in CTCL. A scenario in which a CTCL patient could be predicted 70% of the time to undergo clinical benefit from HDAC inhibitor treatment is significantly better than the existing frequency of response in unselected patients (9, 11). A similar trend was apparent in biopsies taken from CTCL patients treated with other HDAC inhibitors, including PXD101 and depsipeptide/FK228, implying that HR23B is a generally useful biomarker for predicting clinical responses to HDAC inhibitors.
The Proteasome in HDAC Inhibitor-Treated Cells.
Whereas HR23B is a potential biomarker for HDAC inhibitor-based therapy, it also has provided us with mechanistic insights into how HDAC inhibitors kill tumor cells (12). HR23B shuttles cargo proteins for degradation to the proteasome, and proteasomal activity is under aberrant control in CTCL cells treated with HDAC inhibitors (12). In this respect, the outcome of the combined treatment of HDAC inhibitor and proteasome inhibitor resulted in increased inhibitory effects on proteasome activity in a fashion that reflected levels of HR23B. From the clinical perspective, HDAC inhibitors have been studied in combination with an increasing array of anticancer agents (6, 8, 16), and clinical studies in which HDAC inhibitors were combined with proteasome inhibitors yielded results that suggest additive effects (7, 16, 20, 21). For example, the combined treatment of SAHA/vorinostat with bortezomib produced encouraging clinical effects in patients with relapsed and refractory multiple myeloma (20). Based on the results described here, we surmise that the improved clinical profile reflects the additive inhibition of the proteasome, resulting in greater levels of associated tumor cell death. Indeed, under some circumstances, this outcome could influence autophagy (22). Moreover, we suggest too that tumors expressing high levels of HR23B might be expected to undergo a particularly favorable response to such a combination treatment. Future studies in which HR23B levels are assessed in a combined-therapy clinical trial will allow this important possibility to be assessed.
In conclusion, our results provide strong foundation for HR23B as a predictive biomarker in CTCL. As such, HR23B represents a striking example of “bench to clinic” translation, and in turn supports the drive to progress laboratory research findings into clinical utility. Our studies have focused on HR23B in CTCL but, given the high positive predictive value of the biomarker, it will be interesting to evaluate HR23B as a predictive biomarker in other types of tumors as a basis for selecting patients who respond favorably to HDAC inhibitor-based therapy.
Materials and Methods
Cell Lines.
The CTCL cell lines MYLA, HUT78, and SeAx were maintained in RPMI1640 supplemented with 20% fetal calf serum and 1% pen/strep.
Electroporation.
siRNA was introduced into CTCL cell lines using electroporation. HR23B siRNA (100 nM) or nontargeting (NT) control siRNA (100 nM) was added to 500 μL of hypo-osmolar buffer and 1 × 106 cells and the mixture was incubated for 10 min at room temperature. An Eppendorf multiporator was then used to pulse cells at 1,100 V for 100 μs and cells were incubated for a further 10 min at room temperature. Cell-culture media at 37 °C was then added and the mixture was added to cell-culture dishes. Cells were allowed to grow in media for 48 h before a 1-mL aliquot was removed to allow determination of protein depletion via immunoblotting. Cells were then treated with appropriate drug concentrations for a further 72 h as indicated. siRNA treatment of other cell types was as previously described (12).
Stable Cell Line Production.
The stable inducible cell lines ectopically expressing FLAG-HR23B were developed using the TET-ON system with U2OS-TET cells obtained from Clontech. Human HR23B was PCR-amplified from pET-HR23B (kind gift from F. Hanaoka, RIKEN, Saitama, Japan) using the primers forward 5′-GTGGTGGGATCCCCATGGATGGACTACAAAGACGATGACGACAAGCATATGCAGGTCACCCTGAAGACC-3′ and reverse 5′-CTTATCGATGGCGGCCGCAGTTCAGTCTTCGTCAAAGTTCTGCTG-3′ and subcloned into the pTRE vector using BamHI and NotI restriction enzyme sites. pTRE and pTRE-FLAG-HR23B were then transfected into U2OS-TET cells. Cells were then selected for the pTRE construct using media containing 100 μg/mL hygromycin for 10 days. Remaining cells were selected and pooled to provide growing cultures of pTRE-containing and pTRE-FLAG-HR23B-containing cells. Expression of FLAG-HR23B was induced by addition of 1 μg/mL doxycyline to culture media for 16 h followed by the appropriate drug treatment.
Pathology Examination.
CTCL diagnosis was verified by examination by two independent pathologists. A total of 86 paraffin-embedded formalin-fixed specimens from 27 patients were sectioned (5 mm) onto glass slides, cleared of paraffin in Citroclear, and rehydrated through graded alcohol baths. After a rinse in water, sections were “pressure-cooked” for 2 min in 1 mM EDTA at pH 8 for heat-induced epitope retrieval. Slides were incubated in 0.03% hydrogen peroxide for 5 min to inactivate endogenous peroxidases, incubated with 0.1% Triton X-100 for 10 min to improve antibody penetration, and treated with an anti-HR23B followed by rabbit anti-mouse secondary antibody and then substrate (DAB; DAKO). Slides were placed in hematoxylin (Sigma) for 1–2 s for nuclear counterstaining and then mounted with coverslips using AquaTex (Merck). Sections were examined under a Zeiss Axioskop light microscope. Images were captured with a MicroPublisher 5.0 RTV camera (JH Technologies), and imaging was performed with Adobe Photoshop 8.0. Use of human tissues in this study was approved by the Institutional Review Board of the MD Anderson Cancer Center, and written informed consent was obtained from each patient before study enrollment.
Assay for Proteasome Activity from HDAC Inhibitor-Treated Cells.
MYLA cells were harvested after 16-h treatment with SAHA (10 and 30 μM), bortezomib (100 nM), or vehicle control. Following a wash in PBS, cells were resuspended in 130 mL Hepes buffer (5 mM Hepes, 1 mM EDTA [pH 7.5]) and sonicated for 1 min. Samples were centrifuged for 10 min (13,200 rpm at 4 °C) and the supernatant was removed, stored on ice, and used immediately. Proteasome activity per sample was measured by adding 20 μL cell lysate to each well in triplicate. Assay buffer (95 μL) was added as a blank. Five microliters of the fluorogenic substrate Suc-LLVY-AMC was added to all samples and incubated for 10 min at room temperature. The plate was read every 2 min for 1 h on a plate reader (360-nm excitation, 460-nm emission, gain = 58).
Supplementary Material
Acknowledgments
We thank Rosemary Williams for help in preparation of the manuscript. Work in our laboratory is supported by Cancer Research UK, Medical Research Council, Leukemia Research Council, the Association for International Cancer Research, and the European Union. O.K. was supported by the NIHR Biomedical Research Centre, Oxford, United Kingdom.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913912107/DCSupplemental.
References
- 1.Stimson L, La Thangue NB. Biomarkers for predicting clinical responses to HDAC inhibitors. Cancer Lett. 2009;280:177–183. doi: 10.1016/j.canlet.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Inche AG, La Thangue NB. Chromatin control and cancer-drug discovery: Realizing the promise. Drug Discov Today. 2006;11:97–109. doi: 10.1016/S1359-6446(05)03691-3. [DOI] [PubMed] [Google Scholar]
- 3.Marks P, et al. Histone deacetylases and cancer: Causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 4.Marks PA, Richon VM, Breslow R, Rifkind RA. Histone deacetylase inhibitors as new cancer drugs. Curr Opin Oncol. 2001;13:477–483. doi: 10.1097/00001622-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 6.Richon VM, Zhou X, Rifkind RA, Marks PA. Histone deacetylase inhibitors: Development of suberoylanilide hydroxamic acid (SAHA) for the treatment of cancers. Blood Cells Mol Dis. 2001;27:260–264. doi: 10.1006/bcmd.2000.0376. [DOI] [PubMed] [Google Scholar]
- 7.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: Current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Khan O, La Thangue NB. Drug insight: Histone deacetylase inhibitor-based therapies for cutaneous T-cell lymphomas. Nat Clin Pract Oncol. 2008;5:714–726. doi: 10.1038/ncponc1238. [DOI] [PubMed] [Google Scholar]
- 9.Duvic M, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duvic M, Vu J. Vorinostat: A new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 11.Olsen EA, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 12.Fotheringham S, et al. Genome-wide loss-of-function screen reveals an im-portant role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell. 2009;15:57–66. doi: 10.1016/j.ccr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Richon V, Ni X, Talpur R, Duvic M. Selective induction of apoptosis by histone deacetylase inhibitor SAHA in cutaneous T-cell lymphoma cells: Relevance to mechanism of therapeutic action. J Invest Dermatol. 2005;125:1045–1052. doi: 10.1111/j.0022-202X.2005.23925.x. [DOI] [PubMed] [Google Scholar]
- 16.Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–1302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- 17.Zavrski I, et al. Proteasome as an emerging therapeutic target in cancer. Curr Pharm Des. 2007;13:471–485. doi: 10.2174/138161207780162908. [DOI] [PubMed] [Google Scholar]
- 18.Metz CE. Basic principles of ROC analysis. Semin Nucl Med. 1978;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- 19.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 20.Badros A, et al. Phase I study of vorinostat in combination with bortezomib for relapsed and refractory multiple myeloma. Clin Cancer Res. 2009;15:5250–5257. doi: 10.1158/1078-0432.CCR-08-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller CP, et al. Caspase-8 dependent histone acetylation by a novel proteasome inhibitor, NPI-0052: A mechanism for synergy in leukemia cells. Blood. 2009;113:4289–4299. doi: 10.1182/blood-2008-08-174797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. January 14, 2010 doi: 10.1038/emboj.2009.405. 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.