Abstract
Little is known about the mechanisms by which Treponema pallidum (Tp), the causative agent of syphilis, copes with oxidative stress as it establishes persistent infection within its obligate human host. The Tp genomic sequence indicates that the bacterium’s antioxidant defenses do not include glutathione and are limited to just a few proteins, with only one, TP0509, offering direct defense against peroxides. Although this Tp peroxiredoxin (Prx) closely resembles AhpC-like Prxs, Tp lacks AhpF, the typical reductant for such enzymes. Functionally, TpAhpC resembles largely eukaryotic, nonAhpC typical 2-Cys Prx proteins in using thioredoxin (Trx, TP0919) as an efficient electron donor and exhibiting broad specificity toward hydroperoxide substrates. Unlike many of the eukaryotic Prxs, however, TpAhpC is relatively resistant to inactivation during turnover with hydroperoxide substrates. As is often observed in typical 2-Cys Prxs, TpAhpC undergoes redox-sensitive oligomer formation. Quantitative immunoblotting revealed that TpTrx and TpAhpC are present at very high levels (over 100 and 300 μM, respectively) in treponemes infecting rabbit testes; their redox potentials, at -242 ± 1 and -192 ± 2 mV, respectively, are consistent with the role of TpTrx as the cellular reductant of TpAhpC. Transcriptional analysis of select antioxidant genes confirmed the presence of high mRNA levels for ahpC and trx which diminish greatly when spirochetes replicate under in vitro growth conditions. Thus, T. pallidum has evolved an extraordinarily robust, broad-spectrum AhpC as its sole mechanism for peroxide defense to combat this significant threat to treponemal growth and survival during infection.
Keywords: alkyl hydroperoxide reductase, peroxidases, syphilis, syphilis spirochete, oxidative stress
Despite the availability of effective antimicrobial therapy for more than six decades, syphilis, a sexually transmitted infection caused by the spirochetal pathogen Treponema pallidum, remains a public health problem of global dimensions (1). Following inoculation, usually in the genital area, T. pallidum disseminates to diverse organs where it can establish persistent, even lifelong, infection (2, 3). Investigation of the mechanisms the syphilis spirochete employs to establish and maintain infection traditionally have centered on the characterization of surface-exposed molecules which function at the host-pathogen interface (2–4). T. pallidum, an extracellular organism with an exceptionally limited biosynthetic capacity (5, 6), also must acquire essential nutrients from every microenvironment in which it takes up residence, while fending off measures by the host to disrupt bacterial homeostasis (7–9).
T. pallidum is classified as a microaerophile because of its limited oxygen tolerance in vitro (10, 11); however, like all bacterial pathogens, the bacterium must cope with endogenous and exogenous sources of oxidative stress as it disseminates throughout the human body. In other bacteria, such as Escherichia coli, incomplete reduction of oxygen by the aerobic respiratory chain is one endogenous source of reactive oxygen species (ROS), some of which can react with iron (Fenton chemistry) and produce protein- and DNA-damaging molecules (12). In lacking the ability to perform oxidative phosphorylation and maintaining minuscule intracellular concentrations of iron (Fe) (5, 6), T. pallidum inherently limits the production of such oxidants. On the other hand, upon colonization of genital mucosal sites prior to dissemination, spirochetes likely encounter ROS produced both by commensal flora to ward-off noncommensal intruders (13, 14) as well as by macrophages and neutrophils to combat the microbes (8, 15, 16). Despite these obstacles, T. pallidum readily disseminates hematogenously and survives in well oxygenated tissues and chronically inflamed sites (2, 3), suggesting that it possesses robust defenses against oxidative stress (11, 17) a more thorough examination of which would enhance our understanding of syphilis pathogenesis.
T. pallidum encodes some antioxidant proteins common to other pathogenic bacteria but is surprisingly deficient in other systems which support oxidant defense and redox homeostasis in many organisms (Table 1; SI Results) (6, 11, 17). Two of the principal ROS bacteria must protect themselves against are superoxide (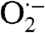 ) and peroxides (12). While T. pallidum lacks an ortholog for superoxide dismutase (SOD), the classical enzyme that detoxifies
) and peroxides (12). While T. pallidum lacks an ortholog for superoxide dismutase (SOD), the classical enzyme that detoxifies 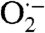 (12, 18), it does encode a superoxide reductase (SOR, TP0823) and its putative reductant, rubredoxin (TP0991) (19, 20). T. pallidum lacks catalase and contains no homolog for glutathione peroxidase; instead, the spirochete encodes only one known peroxide scavenging enzyme, an alkyl hydroperoxide reductase C (AhpC; TP0509), which belongs to a widespread family of cysteine-based peroxiredoxins with activity against hydrogen peroxide (H2O2), organic peroxides, and peroxynitrite (21). Among peroxiredoxins, the AhpC-type enzymes have been best characterized in Salmonella typhimurium, Streptococcus mutans, and E. coli (21–23). These closely related bacterial enzymes (> 50% amino acid sequence identity) are recycled by a dedicated flavoprotein disulfide reductase, alkyl hydroperoxide reductase F (AhpF) (21). While AhpF acts as a highly efficient electron donor to AhpC in these organisms, thioredoxin (Trx) can substitute as a less efficient reductant and is apparently the natural, physiological reductant of most nonAhpC peroxiredoxins (21). Remarkably, T. pallidum lacks AhpF altogether (6), suggesting that the AhpC from this organism relies on a single Trx (TP0919) present in the cytoplasm (11, 21). Without catalase or other peroxide-metabolizing enzymes, including other peroxiredoxin homologs, T. pallidum is likely to be highly dependent on AhpC and its reducing partner for protection against H2O2 and other hydroperoxides. Moreover, given the absence of other typical cellular thiol reductants such as glutaredoxins and glutathione in T. pallidum (Table 1 and SI Results), one might expect the single Trx protein to be of exceptional importance in maintaining the proper thiol-disulfide homeostasis in this organism (6).
(12, 18), it does encode a superoxide reductase (SOR, TP0823) and its putative reductant, rubredoxin (TP0991) (19, 20). T. pallidum lacks catalase and contains no homolog for glutathione peroxidase; instead, the spirochete encodes only one known peroxide scavenging enzyme, an alkyl hydroperoxide reductase C (AhpC; TP0509), which belongs to a widespread family of cysteine-based peroxiredoxins with activity against hydrogen peroxide (H2O2), organic peroxides, and peroxynitrite (21). Among peroxiredoxins, the AhpC-type enzymes have been best characterized in Salmonella typhimurium, Streptococcus mutans, and E. coli (21–23). These closely related bacterial enzymes (> 50% amino acid sequence identity) are recycled by a dedicated flavoprotein disulfide reductase, alkyl hydroperoxide reductase F (AhpF) (21). While AhpF acts as a highly efficient electron donor to AhpC in these organisms, thioredoxin (Trx) can substitute as a less efficient reductant and is apparently the natural, physiological reductant of most nonAhpC peroxiredoxins (21). Remarkably, T. pallidum lacks AhpF altogether (6), suggesting that the AhpC from this organism relies on a single Trx (TP0919) present in the cytoplasm (11, 21). Without catalase or other peroxide-metabolizing enzymes, including other peroxiredoxin homologs, T. pallidum is likely to be highly dependent on AhpC and its reducing partner for protection against H2O2 and other hydroperoxides. Moreover, given the absence of other typical cellular thiol reductants such as glutaredoxins and glutathione in T. pallidum (Table 1 and SI Results), one might expect the single Trx protein to be of exceptional importance in maintaining the proper thiol-disulfide homeostasis in this organism (6).
Table 1.
Redox-relevant proteins encoded within the genomes of Treponema and Borrelia spirochetes
| Protein family | Treponema pallidum | Treponema denticola | Borrelia burgdorferi |
| Peroxiredoxin | TP0509 | TDE0011 | No |
| (AhpC group) | (Prx6 group) | ||
| AhpF | No | No | No |
| TrxR | TP0814 | TDE0743 | BB0515 |
| NADH oxidase | TP0921 | TDE0096 | No |
| CoA disulfide reductase | No | TDE0153 (with rhodanese) | BB0728 |
| Thioredoxin | TP0919 | TDE0744 | BB0061 |
| TP0100 | TDE0238 (with Se) | ||
| Catalase | No | No | No |
| BtuE (Gpx homologue) | No | TDE1729 (with Se) | No |
| Ohr | No | No | No |
| Dps/NapA | TP1038 | No | BB0690 |
| Superoxide dismutase | No | No | BB0153 |
| Superoxide reductase | TP0823 | TDE1754 | No |
| Rubredoxin | TP0991 | TDE1052 | No |
| Glutaredoxin | No | No | No |
| Glutathione reductase | No | No | No |
| γ-Glutamate-cysteine ligase | No | No | No |
| OxyR | No | No | No |
| SoxR | No | No | No |
We report herein that the Trx and AhpC proteins of T. pallidum together constitute a very active and robust peroxidase system with broad specificity toward primary and tertiary hydroperoxide substrates. Moreover, these two enzymes are highly abundant in treponemes from inflamed rabbit testes but markedly down-regulated in spirochetes grown in an in vitro epithelial cell cocultivation system replete with exogenous antioxidants. Our findings suggest that peroxides are a major form of oxidative stress encountered by T. pallidum during infection and also provide an explanation for the paradoxical observation that T. pallidum has limited oxygen tolerance in vitro but flourishes in blood and highly oxygenated tissues within the human host (2, 3).
Results and Discussion
Bioinformatic Analysis of Peroxiredoxins and Associated Redox Proteins Within the T. pallidum Genome.
Prx proteins are highly abundant and widely distributed in biology; few organisms lack identifiable Prx(s) in their genomes, and many organisms express multiple Prx family members (Table S1). In spirochetes, however, the situation is quite unusual; species of Borrelia, Spirochaeta, and Brachyspira lack Prx orthologs, while T. pallidum and Treponema denticola have single Prx proteins encoded in their genomes which belong to separate Prx classes and are only 27.0% identical in amino acid sequence (Table S1). The T. pallidum Prx (TP0509) under investigation herein is best classified as an AhpC-like protein (within the broader category of typical 2-Cys Prxs) (Fig. 1); in addition to the peroxide-reactive (peroxidatic) cysteine at the active site, AhpC-like proteins, as well as those within the broader typical 2-Cys Prx group, also conserve a second (resolving) cysteine which participates in catalysis by forming a disulfide bond with the oxidized, sulfenic acid form of the peroxidatic cysteine. The T. denticola Prx (TDE0011), on the other hand, falls into the Prx6-like group, proteins which typically conserve only the peroxidatic cysteine (24, 25).
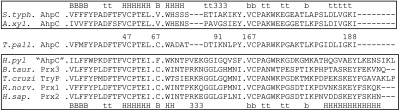
Fig. 1.
Alignment of fragments within representative typical 2-Cys peroxiredoxins (Prxs). Shown in the rectangle above the T. pallidum AhpC (TpAhpC) sequence are select sequence regions of S. typhimurium AhpC (1yep, 51.1% identity with TpAhpC) and Amphibacillus xylanus AhpC (1we0, 46.6% identity) and their corresponding secondary structure elements (B = beta strand, H = α helix, 3 = 3/10 helix, t = hydrogen-bonded turn, and b = beta bridge). In the rectangle below the T. pallidum sequence are representative typical 2-Cys Prx proteins of known structure from a wide variety of organisms that group separately from AhpC-like proteins in multiple sequence alignments, including proteins from Helicobacter pylori (1zof, 33.0% identity with TpAhpC), Bos taurus (1zye, 33.5%), Trypanosoma cruzi (1uul, 38.4%), Rattus norvegicus (1qq2, 37.0%) and Homo sapiens (1qmv, 38.4%). Shown are sequence segments (separated by a dot), from (A) regions around the peroxidatic cysteine (Cys 47 in TpAhpC), (B) the additional (third) cysteine (Cys67) present in TpAhpC but not in the two other AhpC proteins, (C) the Prx1-linked GG(i/l/v)G motif, and (D) the C terminus including the resolving cysteine (Cys167 in TpAhpC).
The typical 2-Cys Prx group, which includes the T. pallidum Prx, can be subdivided into (i) the widespread group represented by human Prx1-like proteins, and (ii) the predominantly bacterial AhpC proteins (Fig. 1 and SI Results). Based on sequence alignments and conservation of key residues, the T. pallidum Prx groups with the AhpC proteins (TpAhpC) (Fig. 1). Earlier analyses suggested that all bacterial genomes with readily identified ahpC genes (≥50% amino acid sequence identity compared to S. typhimurium AhpC) invariably encode an ahpF orthologue immediately downstream of ahpC (26) (the name “AhpC” has been used for proteins linked with AhpD or reduced by other Trx-like proteins, but, using our recent bioinformatic analyses, we are not defining these as “canonical” AhpC proteins; SI Results). In the case of TpAhpC, which exhibits 51.1% amino acid sequence identity with S. typhimurium AhpC (StAhpC), no ahpF or other putative reductase gene is found near the ahpC. As shown in Fig. 1, TpAhpC is 46.6% identical with another structurally characterized representative of the AhpC class, Amphibacillus xylanus AhpC, which also is expressed from an ahpC gene proximal to an ahpF orthologue (27).
The most likely alternative source of electrons for TpAhpC is thioredoxin (Trx); the single T. pallidum Trx (TpTrx; TP0919) is similar to both E. coli Trx proteins and an Arabidopsis thaliana Trxh1 (amino acid identities of 29.4%, 25.9%, and 26.2% with E. coli Trx1, E. coli Trx2 and A. thaliana Trxh1, respectively; Fig. S1A). The T. pallidum genome contains one additional Trx-related protein, TP0100, which is similar to Bacillus subtilis ResA (28) and about twice the size of canonical Trx proteins (17.5% identity between the TP0919 and TP0100). TP0100 is periplasmic, however (29), and could not serve as an electron donor to cytosolic AhpC. The thioredoxin reductase (TrxR) protein required to reduce Trx, TP0814, is 37.4% identical in amino acid sequence with the well-characterized E. coli TrxR, with the highest degree of conservation around the functionally important regions (Fig. S1B) (30).
Based on these bioinformatic analyses, the two proteins studied herein, TpAhpC and TpTrx, along with the TrxR protein, appear to be the only protein-mediated peroxide defense and redox-buffering system present in T. pallidum (Table 1). Given that the biosynthetic machinery for glutathione (e.g., γ-glutamate cysteine ligase in Table 1) is lacking in Treponema and Borrelia species (6, 31, 32), the possibility exists that these organisms, like some others, use the low molecular weight thiol coenzyme A (CoASH) as their primary “redox buffer” (33). However, the lack of a specialized CoA disulfide reductase (CoADR) in T. pallidum, unlike in Borrelia burgdorferi, suggests that CoASH is not a major thiol in T. pallidum (Table 1 and SI Results) (34, 35). Thus, the presence or nature of any low molecular weight thiol in T. pallidum remains to be resolved.
TpAhpC Peroxidase Activity Is Supported By Reductive Turnover with TpTrx and E. coli Trx1, but Not with S. typhimurium AhpF, and Is Higher in Non-His-Tagged Protein.
Despite the sequence similarity between TpAhpC and AhpF-dependent AhpC in many bacteria, the absence of an AhpF ortholog in T. pallidum suggests TpAhpC might have diverged from other AhpC proteins to the point that it can no longer use AhpF as an electron donor. Indeed, TpAhpC showed substantial peroxidase activity in an NADPH-dependent reductase system containing TpTrx, but none with NADH and S. typhimurium AhpF as the electron-donating system (SI Experimental Procedures describes assays used).
Our initial characterization of the enzymatic activity of TpAhpC employed His-tagged protein and included determination of Vmax ,app and Km,app values using varying concentrations of four hydroperoxide substrates and a single concentration (10 μM) of either E. coli Trx1 (EcTrx1) or TpTrx. In comparative assays using both Trx proteins, the Km for H2O2 was identical and the maximal rate (Vmax) was actually about twofold higher with the EcTrx1 enzyme (Table S2). Given these results, EcTrx1 was used as the reductant for most experiments in place of TpTrx primarily due to the large amounts of pure protein available to us. Using this assay system (including 10 μM EcTrx1), His-tagged TpAhpC was shown to exhibit a Vmax ,app of about 1 μM s-1 (μM enzyme)-1 with all four hydroperoxide substrates tested [H2O2, ethyl hydroperoxide, cumene hydroperoxide (CHP), and t-butyl hydroperoxide (tBHP)], and Km values that ranged from 3.5 to 10 μM for the first three substrates and about 100 μM for tBHP (Table S3). These results yielded Vmax ,app/Km,app values of around 1 or 2 × 105 M-1 s-1 for reactivity of TpAhpC toward all four hydroperoxide substrates in the presence of 10 μM E. coli Trx. Activity measured in the presence of increasing amounts of Trx (as high as 80 μM) increased proportionately with Trx concentration, suggesting a nonsaturable interaction of Trx with His-tagged TpAhpC.
In order to determine whether or not the His tag was impairing activity in the recombinant protein, a non-His-tagged version of the protein was expressed and purified. Retesting of H2O2, CHP, and tBHP with 10 μM EcTrx1 and varying concentrations of peroxides demonstrated an increase (by 3 to 13-fold) in the catalytic efficiency (Vmax ,app/Km,app) of the enzyme with each substrate (Table S3). The nontagged protein exhibited both a higher Vmax ,app (by 2- to 4-fold) and a lower Km,app (by 1.2- to 6-fold) toward all three substrates relative to the tagged protein (Table S3). In contrast to results obtained with the His-tagged enzyme, the EcTrx1 reductant also interacted with the non-His-tagged TpAhpC in a saturable manner, allowing for the true kinetic parameters for the bisubstrate enzyme to be determined. Further biochemical characterization, therefore, employed the non-His-tagged version of the protein (below).
TpAhpC Is a Broad Specificity Peroxiredoxin.
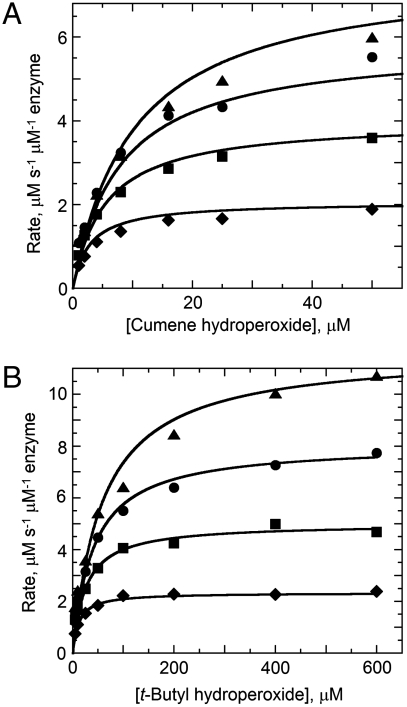
For detailed analysis of the substrate specificity of TpAhpC, a sensitive, fluorescence-based assay was used in which prereduced EcTrx1 (10 to 100 μM), TpAhpC (500 nM), and various concentrations of hydroperoxide substrates were mixed rapidly and monitored for fluorescence decreases reflecting oxidation of EcTrx1; computed rates of reaction were fit globally to the rate equation for a ping pong reaction (SI Experimental Procedures) to yield true, bisubstrate turnover numbers (kcat) and Km values for both the reducing and oxidizing substrates (datasets and fits using CHP and tBHP substrates are shown in Fig. 2). A summary of the overall results for all three of the hydroperoxide substrates studied in detail is provided in Table 2. Several features are notable; first, the catalytic efficiency with hydroperoxide substrates (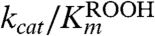 ) varies over a relatively narrow range of sevenfold for substrates as distinct as H2O2, CHP and, tBHP, with a value of 1.4 × 106 M-1 s-1 for the best substrate (H2O2). In fact, the most complex and bulky hydroperoxide, CHP, is very similar to H2O2 as a substrate for TpAhpC. Interestingly, the smaller of the tertiary hydroperoxide substrates, tBHP, has about a sevenfold higher Km than CHP but also about a twofold increase in kcat. Not surprisingly, kcat/Km of TpAhpC with Trx reflecting the reductive half-reaction does not change at all with varying hydroperoxide substrates.
) varies over a relatively narrow range of sevenfold for substrates as distinct as H2O2, CHP and, tBHP, with a value of 1.4 × 106 M-1 s-1 for the best substrate (H2O2). In fact, the most complex and bulky hydroperoxide, CHP, is very similar to H2O2 as a substrate for TpAhpC. Interestingly, the smaller of the tertiary hydroperoxide substrates, tBHP, has about a sevenfold higher Km than CHP but also about a twofold increase in kcat. Not surprisingly, kcat/Km of TpAhpC with Trx reflecting the reductive half-reaction does not change at all with varying hydroperoxide substrates.
Fig. 2.
Activities of Treponema pallidum AhpC with tertiary hydroperoxide substrates. Peroxidase activity was measured by mixing E. coli thioredoxin (Trx, prereduced by dithiothreitol) and AhpC (0.5 μM) in 50 mM potassium phosphate pH 7.0, 0.5 mM EDTA and 100 mM ammonium sulfate with various concentrations of peroxide substrate in a stopped-flow spectrophotometer at 25 °C. The decrease in Trx fluorescence due to oxidation was monitored using excitation at 280 nm and emission > 320 nm. Fixed concentrations of Trx assayed over a range of cumene hydroperoxide (A) and t-butyl hydroperoxide (B) concentrations were 10 μM (diamonds), 25 μM (squares), 50 μM (circles), and 100 μM (triangles). Results using H2O2 as substrate closely matched those with cumene hydroperoxide (A).
Table 2.
Kinetic parameters for reaction of TpAhpC with various peroxide substrates
| Peroxide substrate | kcat, s-1 |
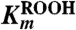 , μM , μM |
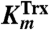 , μM , μM |
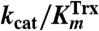 , μM-1 s-1 , μM-1 s-1
|
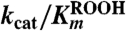 , μM-1 s-1 , μM-1 s-1
|
| Hydrogen peroxide | 17 ± 2 | 12 ± 2 | 62 ± 17 | 0.27 | 1.4 |
| Cumene hydroperoxide | 9.8 ± 0.6 | 9.6 ± 1.1 | 38.8 ± 4.9 | 0.25 | 1.0 |
| t-Butyl hydroperoxide | 20.9 ± 1.4 | 104 ± 9 | 79.4 ± 9.7 | 0.26 | 0.20 |
The relatively high reactivity of TpAhpC with multiple substrates, at ∼106 M-1 s-1, supports the view that it must act as a potent antioxidant defense across a range of peroxide substrates, unlike AhpC in E. coli or S. typhimurium, where the need for broad specificity is less critical given the presence of other Prxs including the organic hydroperoxide defense enzyme Tpx (36). In fact, StAhpC, which exhibits a somewhat higher reactivity than TpAhpC with H2O2 (kcat/Km of 3.7 × 107 M-1 s-1), is about two orders of magnitude less reactive with tBHP and CHP substrates (2.3 × 105 and 4.9 × 105 M-1 s-1, respectively) (37). One limitation for the TpAhpC enzyme may be imposed by the need for Trx to interact with many other substrate proteins, unlike the dedicated reductant of StAhpC, AhpF. Another implication of the lack of an AhpF homologue in T. pallidum is that the electrons for both peroxide defense and thiol-disulfide redox homeostasis must come from NADPH and presumably funnel through the single TrxR and Trx proteins. In E. coli and S. typhimurium, peroxide defense through AhpC is supported primarily by NADH because of the selectivity of AhpF for this cofactor, while disulfide stress is generally alleviated by NADPH through thioredoxin reductase and glutathione reductase pathways.
Another interesting comparison emerging from these biochemical studies of TpAhpC is the distinction between CHP and t-BHP as substrates for this protein (Fig. 2). The difference between these two tertiary hydroperoxides is the presence of a phenyl group in CHP in place of a methyl on tBHP; whereas one might expect steric hindrance issues with the bulkier substituent of CHP, the data reflect an opposite trend, with the phenyl group contributing in a positive way to the enzyme-substrate interaction (Km values of 14 and 104 μM for CHP and tBHP, respectively). This suggests the presence of a rather large, hydrophobic pocket for substrate binding in TpAhpC which might potentially also accommodate macromolecular substrates like lipid and protein hydroperoxides in vivo; comparisons of a structural model of TpAhpC with the known structure of StAhpC suggest enhanced hydrophobicity in the environment around the putative substrate binding region (Fig. S2).
TpAhpC Is Resistant Toward Substrate-Mediated Inactivation.
Because some Prxs, particularly those from eukaryotes, are inactivated during turnover with high levels of substrate (38), the sensitivity of TpAhpC for this type of inactivation was tested with its most potent substrate, H2O2. Using a full assay system containing NADPH, E. coli TrxR, EcTrx1, TpAhpC, and as much as 10 and 30 mM H2O2, a curvature in the loss of 340 nm absorbance over time was observed after multiple turnovers, indicating a very low degree of sensitivity toward inactivation by substrate, slightly less than that observed with the StAhpC protein (Fig. S3). Surprisingly, His-tagged TpAhpC under the same conditions exhibited no evidence of inactivation during turnover even at 30 mM H2O2 (Fig. S3). Thus, TpAhpC is a very robust enzyme, similar to but even less sensitive than StAhpC toward inactivation by peroxide substrates.
The Midpoint Reduction Potentials for TpTrx and TpAhpC Are -242 and -192 mV, Consistent with Their Redox Functions.
Knowing the midpoint reduction (redox) potential of redox-active proteins enables one to place them within the hierarchy of known cellular electron donors or acceptors. Measurements of this parameter for TpTrx and TpAhpC were carried out by equilibration of the proteins with E. coli glutaredoxin-1 (Grx1) in reduced and oxidized forms, then assessment of the redox status of both proteins following acidification and HPLC analysis (37, 39). Given the known redox potential of Grx1 of -233 mV at pH 7 (39), the redox potential for TpAhpC was computed to be -192 ± 2 mV (n = 3, Fig. S4). Like the value previously obtained for StAhpC (-178 mV) (37), this is a relatively high redox potential compared with those of many other redox disulfide-containing proteins. The only other redox potentials reported for Prx proteins, to our knowledge, have been for proteins from chloroplast or mitochondria and range from -288 to -325 mV at pH 7 (40, 41), values that are apparently more suited to their specialized function(s) in these organelles. The redox potential for TpTrx, at -242 ± 1 mV (n = 4), is higher than that of EcTrx [at -270 mV (42)], perhaps in part due to the alteration of the “WCGPC” motif at the active site (WCGSC for TpTrx; Fig. S1). This is similar to the value of -249 mV obtained for a specialized Trx-like protein, tryparedoxin, from another human pathogen, Trypanosoma brucei (43). With redox potentials separated by 50 mV, transfer of electrons from TpTrx to TpAhpC will be thermodynamically favorable even under conditions where the reduced form of TpTrx becomes scarce.
Reduced TpAhpC Forms Stable Octamers or Decamers in Solution Whereas Oxidation Generates a Heterogeneous Mixture of Oligomeric Species.
Members of the typical 2-Cys group of Prxs have been shown to form decameric, as well as octameric and dodecameric, complexes in solution through a redox-sensitive interface (called the “A interface”), while maintaining in all states a dimeric protomer form through the “B interface” (which brings the edges of two beta sheets together) (44). When analyzed by sedimentation velocity analytical ultracentrifugation (45), TpAhpC was found to exist in a stable octameric or perhaps decameric form (MW ∼ 162 kDa) when fully reduced (Fig. S5). Based on mutational analysis of StAhpC (46), decamers are more active than dimers as peroxide scavengers; thus, stabilization of a higher order species for reduced TpAhpC is in line with its apparently critical role in peroxide defense within this organism. At concentrations of 300 μM and higher, a small amount of an additional higher order species was detected in the reduced sample as well (sedimentation coefficient ∼14 S versus 8 S for the predominant species; Fig. S5). Following oxidation of TpAhpC, a heterogenous mixture of species was observed which did not fit well to single or two species models and suggested the presence of a range of species (s = 6 to 16 S) with overall sedimentation coefficients increasing with concentration (Fig. S5). This suggests that TpAhpC has a greater diversity of oligomeric forms than StAhpC, a protein which is decameric when reduced and exhibits a tendency to dissociate into dimers upon oxidation (45).
AhpC and Trx Are Highly Expressed Proteins in T. pallidum.
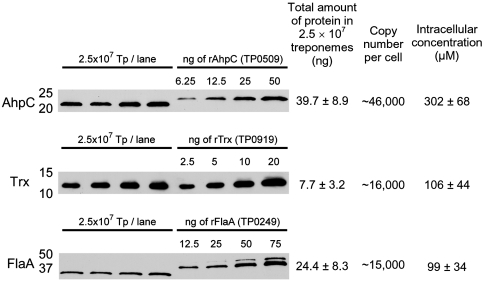
Antisera directed against purified, recombinant TpAhpC and TpTrx was used to probe T. pallidum lysates and quantitatively evaluate expression of the native proteins in motile treponemes freshly harvested from rabbit testes based on recombinant protein standards (Fig. 3). Strong bands detected with apparent molecular masses of 21,000 Da and 12,000 Da were consistent with those predicted for TpAhpC (20,709 Da) and TpTrx (11,391 Da). Copy numbers calculated per T. pallidum cell for TpAhpC and TpTrx were 46,200 ± 10,400 and 16,300 ± 6,800, respectively; the abundant flagellar protein, FlaA, yielded a value of 15,100 ± 5,100 copies per spirochete. Using an estimate of the volume of a single T. pallidum cell, the intracellular concentrations of AhpC, Trx, and FlaA in T. pallidum were approximately 302, 106, and 99 μM, respectively (Fig. 3). Thus, TpAhpC and TpTrx are very abundant enzymes in the cytoplasm, with Trx concentrations reaching levels above the measured Km of TpAhpC for EcTrx1 and levels of TpAhpC as much as threefold over that. Such high concentrations of the peroxidase ensure rapid detoxification of any H2O2 or macromolecular hydroperoxides generated endogenously, while maintenance of such a large pool of reduced and active TpAhpC would allow for immediate defense against a sudden influx of high levels of peroxides from the environment.
Fig. 3.
Immunoblot analysis to evaluate the amounts of peroxiredoxin and thioredoxin proteins expressed in T. pallidum. Estimated expression levels of AhpC, Trx, and FlaA in T. pallidum harvested from rabbit testes (left side of gel) based on purified recombinant protein standards (right side) are given as the total amount of native protein (ng) in 2.5 × 107 treponemes (averages ± standard deviations calculated from four samples out of a single harvest). The lower mobility of recombinant AhpC and FlaA reflects the additional His tag on these proteins. Intracellular protein concentrations were calculated based on the estimated volume of T. pallidum (2.54 × 10-16 L) (SI Experimental Procedures). Concentrations shown for TpAhpC and TpTrx correspond to 6.3 and 1.2 mg/mL in the cell, respectively. Numbers to the left of the immunoblots represent molecular weight standards in kilodaltons.
Transcriptional Analysis of Select Antioxidant Genes Underscores the Importance of AhpC and Trx and Reveals That the Corresponding Genes Are Regulated by Changing Growth Conditions.
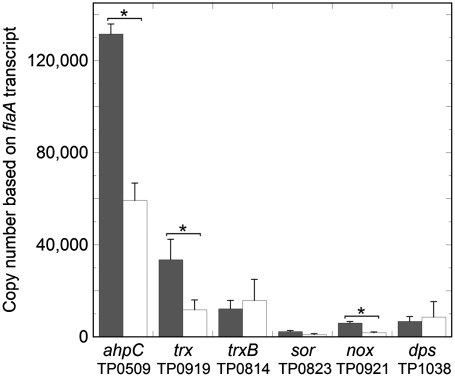
To put into perspective the contribution of the Trx-AhpC detoxification system to T. pallidum’s oxidative stress response, we used quantitative real-time reverse transcription-PCR (qRT-PCR) to survey antioxidant gene expression in the bacterium. The panel of genes chosen for these experiments (Fig. 4) was based on a previous in silico analysis of oxidative stress defense pathways encoded in the spirochete’s genome (Table 1) (11, 17). As described above, trxB (TP0814) encodes a TrxR homologue that is predicted to provide reducing equivalents to Trx and indirectly to AhpC. sor (TP0823) encodes superoxide reductase (19, 20), while the nox (TP0991) gene product, NADH oxidase, is predicted to detoxify free oxygen by four-electron reduction to form water through the oxidation of two molecules of NADH. Lastly, the dps gene product (TP1038) is a dodecameric, ferritin-like protein which binds and sequesters intracellular Fe and may protect DNA from oxidative damage (47). As shown in Fig. 4, flaA-normalized values of antioxidant gene expression in treponemes isolated from inflamed rabbit testis (peak orchitis) covered a broad range of transcript levels. The abundance of transcripts for ahpC and trx was particularly striking and consistent with the high levels of the corresponding proteins detected by quantitative immunoblotting (Fig. 3).
Fig. 4.
Quantitative real-time reverse transcription-PCR (qRT-PCR) of antioxidant genes. Shown are the flaA-normalized transcript copy numbers (assuming 10,000 copies of flaA mRNA) for T. pallidum genes involved in oxidative stress protection following harvest from rabbit testes (black bars) or subsequent cocultivation with rabbit epithelial cells (white bars). Bars represent the means ± SEM for four independent experiments, each analyzed in duplicate. The asterisk indicates a significant difference (p < 0.05 using the Student t-test) between rabbit-harvested and cocultivated transcript copy numbers.
T. pallidum undergoes limited replication when cultivated with rabbit Sf1Ep cells in the presence of a low partial pressure of oxygen (4%) and antioxidants (e.g., SOD and catalase) (48). We used this in vitro cocultivation system to study the expression of T. pallidum genes under conditions in which exogenous sources of oxidative stress are presumably markedly reduced as compared to those encountered by the spirochete within the inflamed rabbit testis. As shown in Fig. 4, transcript levels of three genes (ahpC, trx, and nox) showed highly significant reductions during in vitro cultivation, while the reduction in a fourth (sor) just missed statistical significance. It is noteworthy that the ahpC transcript copy number remained well above that of the constitutively expressed flaA gene.
Demonstration of decreased mRNA levels for ahpC and trx genes from treponemes isolated from cocultures with epithelial cells compared with those from the inflamed rabbit testes suggest that the high concentrations of these antioxidant proteins reported in Fig. 3 are regulated and in part a consequence of their inflammatory environment. While this may seem to be of little surprise given the known OxyR and PerR regulons in a variety of bacteria (12), T. pallidum lacks any of the known bacterial regulators of antioxidant gene expression, leaving unresolved the mechanism underlying the apparent response observed in the present studies.
Conclusions
The broad substrate specificity of TpAhpC and the antioxidant gene/protein expression data clearly indicate that the Prx-Trx peroxide defense system is a major source of protection against oxidative stress in T. pallidum. In support of this conclusion, the spirochete’s streamlined metabolism, which lacks the ability to perform oxidative phosphorylation and maintains low intracellular concentrations of Fe (5, 6), would inherently limit the production of superoxide, and by extension, the likelihood that biomolecules (e.g., proteins and DNA) will be damaged by it (11). The functionality of TpAhpC toward a wide range of peroxide substrates (Table 2) suggests that the enzyme is present to combat both H2O2 and oxidatively damaged cellular constituents like protein and lipid hydroperoxides [the suggestion that activity with CHP can correlate with activity toward lipid hydroperoxides is supported by work with other peroxiredoxins (36, 49, 50)]. Indeed, in the iron-deficient Lyme disease spirochete, B. burgdorferi, membrane lipids (e.g., linoleic and linolenic acids) derived from the host have been shown to be the major target of ROS (51). In E. coli, AhpC scavenges basal metabolic levels of endogenous H2O2, whereas catalase provides protection from high levels of exogenous H2O2 (23). In T. pallidum, the decrease in expression of antioxidant genes in vitro, when compared to the in vivo transcript levels, implies that a substantial amount of peroxide-related stress is exogenous with the in vitro expression values depicting the spirochete’s response to ROS of endogenous origin (Fig. 4). Thus, unlike E. coli, but consistent with its genomic parsimony, the syphilis spirochete encodes a single robust AhpC-Trx system which appears to be evolutionarily tailored to protect the bacterium from both endogenous and exogenous sources of peroxide-related stress. The dramatic up-regulation of ahpC and trx in vivo suggests a substantial amount of peroxide-related stress coming from exogenous sources within the testes and also provides a potential explanation to the longstanding paradox created by the spirochete’s oxygen sensitivity in vitro and its presumed ability to thrive in oxygenated tissues within its obligate human host (10, 52). Ironically, from this insight a new paradox emerges: how does T. pallidum, lacking all known redox regulatory proteins (6), sense and respond to oxidative stress?
Experimental Procedures
Details of materials and methods used are given in SI Experimental Procedures. Methods described include bioinformatics, propagation and in vitro cocultivation of T. pallidum, cloning, expression and purification of proteins, enzyme assays, redox potential determination, analytical ultracentrifugation, quantitative immunoblotting, and qRT-PCR.
Supplementary Material
Acknowledgments.
The authors thank Samantha Alley for contributing experimental data. This work was supported by National Institutes of Health Grant R01 GM050389 (to L.B.P) and Grant R01 AI026756 (to J.D.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910057107/DCSupplemental.
References
- 1.Peeling RW, Mabey DC. Syphilis. Nat Rev Microbiol. 2004;2:448–449. doi: 10.1038/nrmicro914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaFond RE, Lukehart SA. Biological basis for syphilis. Clin Microbiol Rev. 2006;19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radolf JD, Hazlett KRO, Lukehart SA. Pathogenesis of syphilis. In: Radolf JD, Lukehart SA, editors. Pathogenic treponemes: Cellular and molecular biology. Norfolk, United Kingdom: Caister Academic Press; 2006. pp. 197–236. [Google Scholar]
- 4.Cameron CE, Radolf JD, Lukehart SA. The T. pallidum outer membrane and outer membrane proteins. In: Radolf JD, Lukehart SA, editors. Pathogenic treponema: Molecular and cellular biology. Norwich, United Kingdom: Caister Academic Press; 2006. pp. 237–266. [Google Scholar]
- 5.Cox DL, Radolf JD. Metabolism of the Treponema. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponema: Molecular and cellular biology. Norwich, United Kingdom: Caister Academic Press; 2006. pp. 61–100. [Google Scholar]
- 6.Fraser CM, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 8.Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaible UE, Kaufmann SH. A nutritive view on the host-pathogen interplay. Trends Microbiol. 2005;13:373–380. doi: 10.1016/j.tim.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Cox DL, et al. Effects of molecular oxygen, oxidation-reduction potential, and antioxidants upon in vitro replication of Treponema pallidum subsp. pallidum. Appl Environ Microbiol. 1990;56:3063–3072. doi: 10.1128/aem.56.10.3063-3072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazlett KRO, et al. Contribution of neelaredoxin to oxygen tolerance by Treponema pallidum. Methods Enzymol. 2002;353:140–156. doi: 10.1016/s0076-6879(02)53044-5. [DOI] [PubMed] [Google Scholar]
- 12.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschenbach DA, et al. Prevalence of hydrogen peroxide-producing Lactobacillus species in normal women and women with bacterial vaginosis. J Clin Microbiol. 1989;27:251–256. doi: 10.1128/jcm.27.2.251-256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan CS, Kleinberg I. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch Oral Biol. 1995;40:753–763. doi: 10.1016/0003-9969(95)00029-o. [DOI] [PubMed] [Google Scholar]
- 15.Lukehart SA. Scientific monogamy: Thirty years dancing with the same bug: 2007 Thomas Parran Award Lecture. Sex Transm Dis. 2008;35:2–7. doi: 10.1097/OLQ.0b013e318162c4f2. [DOI] [PubMed] [Google Scholar]
- 16.Radolf JD, Lukehart SA. Immunology of syphilis. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponemes: Cellular and molecular biology. Norfolk, United Kingdom: Caister Academic Press; 2006. pp. 285–322. [Google Scholar]
- 17.Gherardini FC, Boylan JA, Brett PJ. Metal utilization and oxidative stress. In: Radolf JD, Lukehart SA, editors. Pathogenic Treponema: Molecular and cellular biology. Norwich, United Kingdom: Caister Academic Press; 2006. pp. 101–126. [Google Scholar]
- 18.Fridovich I. Superoxide radical and superoxide dismutases. Ann Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 19.Jovanovic T, et al. Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J Biol Chem. 2000;275:28439–28448. doi: 10.1074/jbc.M003314200. [DOI] [PubMed] [Google Scholar]
- 20.Lombard M, Touati D, Fontecave M, Niviere V. Superoxide reductase as a unique defense system against superoxide stress in the microaerophile Treponema pallidum. J Biol Chem. 2000;275:27021–27026. doi: 10.1074/jbc.M004201200. [DOI] [PubMed] [Google Scholar]
- 21.Poole LB. Bacterial defenses against oxidants: Mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Poole LB, Higuchi M, Shimada M, Calzi ML, Kamio Y. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radical Biol Med. 2000;28:108–120. doi: 10.1016/s0891-5849(99)00218-x. [DOI] [PubMed] [Google Scholar]
- 23.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann B, Hecht H-J, Flohé L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 25.Karplus PA, Hall A. Structural survey of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin systems. New York: Springer; 2007. pp. 41–60. [Google Scholar]
- 26.Poole LB. Bacterial peroxiredoxins. In: Torres M, Fukuto JM, Forman HJ, editors. Signal transduction by reactive oxygen and nitrogen species: Pathways and chemical principles. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2003. pp. 80–101. [Google Scholar]
- 27.Kitano K, Kita A, Hakoshima T, Niimura Y, Miki K. Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus. Proteins. 2005;59:644–647. doi: 10.1002/prot.20412. [DOI] [PubMed] [Google Scholar]
- 28.Lewin A, Crow A, Oubrie A, Le Brun NE. Molecular basis for specificity of the extracytoplasmic thioredoxin ResA. J Biol Chem. 2006;281:35467–35477. doi: 10.1074/jbc.M607047200. [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko DV, et al. Identification of homologs for thioredoxin, peptidyl prolyl cis-trans isomerase, and glycerophosphodiester phosphodiesterase in outer membrane fractions from Treponema pallidum, the syphilis spirochete. Infect Immun. 1997;65:4179–4189. doi: 10.1128/iai.65.10.4179-4189.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waksman G, Krishna TS, Williams CH, Jr, Kuriyan J. Crystal structure of Escherichia coli thioredoxin reductase refined at 2 Å resolution. Implications for a large conformational change during catalysis. J Mol Biol. 1994;236:800–816. [PubMed] [Google Scholar]
- 31.Fraser CM, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 32.Seshadri R, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 34.Boylan JA, et al. Borrelia burgdorferi bb0728 encodes a coenzyme A disulphide reductase whose function suggests a role in intracellular redox and the oxidative stress response. Mol Microbiol. 2006;59:475–486. doi: 10.1111/j.1365-2958.2005.04963.x. [DOI] [PubMed] [Google Scholar]
- 35.Mallett TC, et al. Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 Å resolution. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker LM, Poole LB. Catalytic mechanism of thiol peroxidase from Escherichia coli. Sulfenic acid formation and overoxidation of essential CYS61. J Biol Chem. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsonage D, Karplus PA, Poole LB. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci USA. 2008;105:8209–8214. doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang KS, et al. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 39.Åslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 40.Cox AG, Peskin AV, Paton LN, Winterbourn CC, Hampton MB. Redox potential and peroxide reactivity of human peroxiredoxin 3. Biochemistry. 2009;48:6495–6501. doi: 10.1021/bi900558g. [DOI] [PubMed] [Google Scholar]
- 41.Dietz KJ, et al. The function of peroxiredoxins in plant organelle redox metabolism. J Exp Bot. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- 42.Krause G, Lundstrom J, Barea JL, Pueyo de la Cuesta C, Holmgren A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J Biol Chem. 1991;266:9494–9500. [PubMed] [Google Scholar]
- 43.Reckenfelderbaumer N, Krauth-Siegel RL. Catalytic properties, thiol pK value, and redox potential of Trypanosoma brucei tryparedoxin. J Biol Chem. 2002;277:17548–17555. doi: 10.1074/jbc.M112115200. [DOI] [PubMed] [Google Scholar]
- 44.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins—structures, mechanisms, and functions. Febs J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: Redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 46.Parsonage D, et al. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–10592. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thumiger A, et al. Crystal structure of antigen TpF1 from Treponema pallidum. Proteins. 2006;62:827–830. doi: 10.1002/prot.20828. [DOI] [PubMed] [Google Scholar]
- 48.Cox DL. Culture of Treponema pallidum. Methods Enzymol. 1994;236:390–405. doi: 10.1016/0076-6879(94)36029-4. [DOI] [PubMed] [Google Scholar]
- 49.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: Genetic and kinetic characterization. J Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Budde H, et al. Kinetics and redox-sensitive oligomerisation reveal negative subunit cooperativity in tryparedoxin peroxidase of Trypanosoma brucei brucei. Biol Chem. 2003;384:619–633. doi: 10.1515/BC.2003.069. [DOI] [PubMed] [Google Scholar]
- 51.Boylan JA, Lawrence KA, Downey JS, Gherardini FC. Borrelia burgdorferi membranes are the primary targets of reactive oxygen species. Mol Microbiol. 2008;68:786–799. doi: 10.1111/j.1365-2958.2008.06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norris SJ, Edmondson DG. Factors affecting the multiplication and subculture of Treponema pallidum subsp. pallidum in a tissue culture system. Infect Immun. 1986;53:534–539. doi: 10.1128/iai.53.3.534-539.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.