Abstract
Upon mating, females of many animal species undergo dramatic changes in their behavior. In Drosophila melanogaster, postmating behaviors are triggered by sex peptide (SP), which is produced in the male seminal fluid and transferred to female during copulation. SP modulates female behaviors via sex peptide receptor (SPR) located in a small subset of internal sensory neurons that innervate the female uterus and project to the CNS. Although required for postmating responses only in these female sensory neurons, SPR is expressed broadly in the CNS of both sexes. Moreover, SPR is also encoded in the genomes of insects that lack obvious SP orthologs. These observations suggest that SPR may have additional ligands and functions. Here, we identify myoinhibitory peptides (MIPs) as a second family of SPR ligands that is conserved across a wide range of invertebrate species. MIPs are potent agonists for Drosophila, Aedes, and Aplysia SPRs in vitro, yet are unable to trigger postmating responses in vivo. In contrast to SP, MIPs are not produced in male reproductive organs, and are not required for postmating behaviors in Drosophila females. We conclude that MIPs are evolutionarily conserved ligands for SPR, which are likely to mediate functions other than the regulation of female reproductive behaviors.
Keywords: Drosophila, female post-mating behavior, G protein–coupled receptor, neuropeptide, neuromodulation
Peptide signaling through G protein–coupled receptors is a widely used mechanism for reversibly modulating the behavioral output of innate neural circuits (for review see ref. 1). A prominent example of peptidergic modulation of behavior is the regulation of female reproductive behavior in Drosophila melanogaster by the male's sex peptide (SP). SP is a small peptide present in the male seminal fluid. Upon mating, SP is transferred to the female, where it triggers dramatic changes in reproductive and other behaviors (2, 3). These behavioral modifications typically last for about a week, the period for which the female is able to store and use sperm from the initial mating (for review see ref. 4). Within females, SP is thought to activate a specific G protein-coupled receptor (SPR) (5) in a small set of internal sensory neurons of the female reproductive tract (6, 7). Signaling by SP and SPR is essential for the modulation of female behavior as these changes do not occur if the male lacks SP (8, 9) or the female lacks SPR (5).
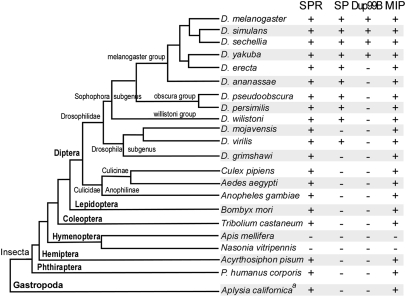
The modulation of female reproductive behavior in response to SP (and its closely related homolog DUP99B) (10, 11) is currently the only known role of SPR. Nonetheless, several lines of evidence have hinted that SPR may have other ligands and possibly also other functions. First, SPR is broadly expressed throughout the central nervous system (5), yet it is required only in reproductive tract sensory neurons for the postmating behavioral switch (6, 7). Second, SPR is also expressed in the central nervous system of males (5), where it is not likely to be exposed to SP. Third, orthologs of SPR are clearly detectable in most (but not all) sequenced genomes from the lophotrochozoa and ecdysozoa (5), yet genes encoding SP-like peptides can only be discerned in the genomes of a few closely related Drosophila species (Fig. 1).
Fig. 1.
SPR and MIP genes occur in most sequenced insect genomes, whereas SP and Dup99B orthologs are detected only in small groups of Drosophila genus. The presence of corresponding orthologs was revealed by BLAST analysis on the corresponding genome assemblies and is indicated by + (presence) or − (absence). The phylogram showing relationships among insect species was adopted and modified from http://flybase.org/blast/. aThe presence of SPR and MIP genes in Aplysia californica were inferred from the identification of corresponding ESTs from a CNS-specific cDNA library (39).
Based on these observations, we speculated that SPR may have additional ligands that are only distantly related to SP, if at all, and that these ligands may function in processes other than the regulation of female reproductive behavior. If so, then this also raises the question of the ancestral function of SPR. Does SPR have an ancient role in the regulation of female reproductive behavior, with SP becoming the principle ligand for this function only recently in Drosophila evolution? Or does SPR have a different ancestral function, with a role in postmating behavior arising only recently in Drosophila evolution, concomitant with the emergence of its novel SP ligand? To begin to address these questions, we set out to identify additional ligands for SPR. As we expected these other ligands to be conserved across insect species, we focused on identifying SPR ligands present in the yellow fever mosquito Aedes aegypti, as well as in D. melanogaster.
We report here that the evolutionary conserved family of myoinhibitory peptides (MIPs), also known as allastostatin-B (AST-B) or prothoracicostatic peptides (PTSPs), are potent ligands for SPR in both Aedes and Drosophila, as well as the sea slug Aplysia. In contrast to SP, MIPs do not appear to mediate postmating behavioral changes in Drosophila and are not present in seminal fluid in either insect species. We infer that MIP signaling via SPR is a conserved pathway that functions in some process other than the regulation of female reproductive behavior.
Results and Discussion
SP and Dup99B Arose Recently in Drosophila Evolution.
To investigate the phylogenetic distribution of SP across the insects, we performed BLAST analysis on all publically available insect genome assemblies. We located SP orthologs in 10 out of 12 sequenced Drosophila species, but not in any of the non-Drosophilidae insects (Fig. 1 and Table S1). SP-like peptides have also been described in three other Drosophila species: Drosophila suzukii, Drosophila subobscura, and Drosophila madeirensis (12). All members of the Sophophora subgenus contain an SP gene, albeit at distinct genomic locations within the melanogaster, obscura, and willistoni groups. Of three sequenced species in Drosophila subgenus, only the Drosophila virilis genome contains a SP-like gene. Extensive manual and BLAST search failed to identify any SP-related gene in Drosophila grimshawi, Drosophila mojavensis, or indeed in any other sequenced insect genomes. Dup99B, a peptide closely related to SP, appears to have an even more recent origin (10, 11). This gene is present in only a subset of melanogaster group, including D. melanogaster, Drosophila simulans, Drosophila sechellia, and Drosophila yakuba. Dup99B appears to be lacking in Drosophila erecta and more distant species (Fig. 1). The most parsimonious explanation for these observations is that SP first arose in a common ancestor of both the Sophophora and Drosophila subgenuses, and, during the early diversification of the melanogaster group, it was duplicated and translocated to generate Dup99B. SP may have then been lost within the mojavensis and grimshawi lineages, and Dup99B was lost in the erecta lineage.
In contrast to SP and DUP99B, SPR was found in almost all sequenced insect genomes, with the exception of the honey bee Apis melifera and the parasitoid wasp Nasonia vitripennis, both of which belong to hymenoptera order (Fig. 1). SPR is not only found in ecdysozoa, but also in lophotrochozoa, including Aplysia. The presence of SPR in many species that lack SP and DUP99B suggests that, at least in these species, SPR is likely to have other ligands.
Strategy for Identification of Other SPR Ligands.
We previously demonstrated the functional expression of both D. melanogaster and A. aegypti SPRs in mammalian CHO cells and showed that SPRs from both species respond to Drosophila SP. SP activates Drosophila SPR strongly, with an EC50 of approximately 1.3 nM, but Aedes SPR only weakly, with an EC50 of approximately 167 nM (5). Although SPR activation is normally coupled to cAMP signaling, in this assay it is linked to an increase in [Ca2+]i. This allows convenient detection of SPR activation using the Ca2+ reporter aequorin to produce luminescent light. We now used this assay to search for additional ligands for Aedes and Drosophila SPR. We followed two approaches in parallel. First, we took a candidate approach, based on the identification of myoinhibitory peptides (MIPs) as ligands for SPR in Bombyx mori (13). Second, we attempted to identify ligands in fractionated extracts of Aedes whole bodies (for results, see SI Text).
MIPs Are Potent SPR Agonists.
MIPs are a set of highly conserved peptides that have been identified or predicted in many different insect species (14–23). Interestingly, however, MIPs, like SPR, cannot be detected in the sequenced hymenoptera genomes (Fig. 1). Typically, five to seven mature MIPs are produced by processing of a single preprohormone precursor. Each of these MIPs has a signature W(X)6W-amide C-terminal motif, making them structurally distinct from SP and its close relatives (Table S2). MIPs have been shown to suppress the spontaneous contractions of the hindgut and the oviduct, leading to their designation as myoinhibitory peptides (15, 22). In B. mori, MIPs have also been found to suppress ecdysteroid production in the prothoracic gland and accordingly have also been named PTSPs (13, 16). The conserved physiological functions of the MIPs, however, remain obscure.
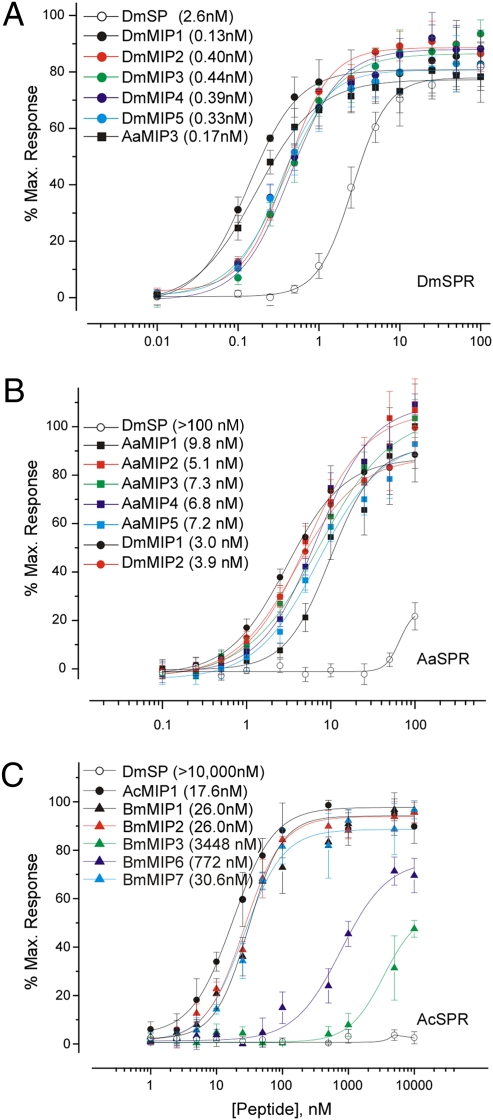
Based on the observation that Bombyx SPR is activated by Bombyx MIPs (13), we tested whether Drosophila SPR is also sensitive to MIPs. Indeed, Drosophila MIPs are potent agonists for Drosophila SPR, with subnanomolar EC50s (Fig. 2A). In these assays, MIPs were even more potent agonists for SPR than SP itself. MIPs from Bombyx and Aedes were also highly active on Drosophila SPR (Fig. S1), as anticipated from their high sequence conservation. Aedes MIP3, for example, activated Drosophila SPR with a similar potency to each of the Drosophila MIPs (Fig. 2A).
Fig. 2.
MIPs are highly potent agonists for SPRs from various taxa: (A) D. melanogaster, (B) A. aegypti, and (C) A. californica. Dose-response curves of CHO cells expressing SPR, aequorin, and a chimeric G protein, Gαqi, and treated with varying concentrations of MIPs and SP. Each data point is mean ± SEM (n = 4). Values in parenthesis are EC50s.
Similarly, we found that Aedes SPR is also strongly activated by both Aedes and Drosophila MIPs, with EC50s in the low nanomolar range (Fig. 2B), as has also been found for activation of Bombyx SPR by Bombyx MIPs (13). In contrast, and as previously reported (5), Drosophila SP activates Aedes SPR with approximately 100-fold lower potency (Fig. 2B). Moreover, we also found that insect MIPs strongly activate the Aplysia SPR (Fig. 2C and Fig. S2). Indeed, Bombyx MIP1 (EC50, 26.0 nM) was just slightly less potent than Aplysia MIP1 (EC50, 14.6 nM) in these assays (Fig. 2C). On the contrary, Drosophila SP failed to activate Aplysia SPR. We conclude that MIPs are conserved and highly potent agonists for SPR.
Structural Requirements of SP and MIP for SPR Activation.
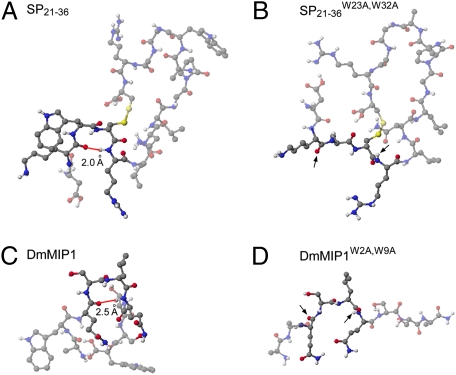
The primary amino acid sequences of SPs and MIPs suggest little if any similarity between the two classes of SPR ligands (Table S2 and Table 1). SP is a 36-amino acid polypeptide carrying a disulfide bridge, whereas MIPs are relatively short (9–12 amino acids) and do not have a disulfide bridge. The only shared features of the two peptides classes are a pair of tryptophan residues, separated by eight amino acids in SP and six amino acids in MIPs. To assess possible similarity in their secondary structures, we predicted the structures of low-energy conformers of SP and MIPs. Although the overall conformations of both ligand classes differed considerably, both classes are predicted to adopt a beta-turn conformation stabilized by the conserved tryptophans (Fig. 3).
Table 1.
EC50 of synthetic SP and MIP derivatives on Drosophila SPR expressed in CHO cells
| Name | Sequence | EC50, nM |
| SP | WEWPWNRKPTKFPIPSPNPRDKWCRLNLGPAWGGRC | 2.6 |
| SP12–36 | FPIPSPNPRDKWCRLNLGPAWGGRC | 5.0 |
| SP12–36C36A | FPIPSPNPRDKWCRLNLGPAWGGRA | 161.1 |
| DUP99B | *QDRNDTEWIQSQKDREKWCRLNLGPYLGGRC | 35.7 |
| SP21–36 | DKWCRLNLGPAWGGRC | 6.1 |
| SP21–36, reduced | DKWCRLNLGPAWGGRC | 6.4 |
| SP21–36W23A,C24A,W32A,C36A | DKAARLNLGPAAGGRA | >10,000† |
| SP21–36W23A,W32A | DKACRLNLGPAAGGRC | >10,000† |
| SP21–36C24A,C36A | DKWARLNLGPAWGGRA | 80.2 |
| 16AA3W,A12W | AAWAAAAAAAAWAAAA | >10,000† |
| 16AA3W,A4C,A12W,A16C | AAWCAAAAAAAWAAAC | >10,000† |
| DmMIP1 | AW.QSLQS.SW-a | 0.1 |
| DmMIP1, w/o amidation | AW.QSLQS.SW | 151.9 |
| DmMIP1W2A,W9A | AA.QSLQS.SA-a | >10,000† |
| DmMIP13-8A | AW.AAAAA.AW-a | 12.3 |
†>10,000, no activity at 10 μM concentrations. *Q, underlined P and underlined Cs indicate pyroglutamic acid, hydroxyproline, and disulfide bonds between Cs respectively.
Fig. 3.
Conserved Trp residues stabilize beta-turn conformation in both SP and MIPs. Secondary structures of (A) SP21–36, (B) SP21–36W23A,W32A, (C) DmMIP1, and (D) DmMIP1W2A,W9A are predicted by low energy conformers calculated with MM2 and MOPAC (Materials and Methods). (A and C) In wild-type peptides, the distances between carbonyl groups of i amino acids and amino groups of I + 3 amino acids are determined in the range of hydrogen bonding distance (red line, 1.9–2.8 Å), a key signature of beta-turn conformation. (B and D) Trp > Ala mutations abolished the beta-turn conformations completely. The corresponding carbonyl groups of i amino acids and NH groups of I + 3 amino acids in mutants were indicated by arrows. Amino acids participating in the beta-turn conformations and their corresponding ones in mutants are highlighted. Black, blue, red, and white balls indicate carbon, nitrogen, oxygen, and hydrogen atoms respectively.
To test the functional significance of the conserved tryptophan residues, we prepared mutant synthetic derivatives of both SP and MIP in which the tryptophan residues are replaced by alanine, predicted to disrupt the beta-turn conformations (Fig. 3). These mutant SP and MIP1 peptides (SP21–36W23A,W32A, MIP1W2A,W9A) were completely inactive even in 10 μM concentration (Table 1), suggesting that these residues, and by inference the turn conformations, are critical for SPR activation. Furthermore, an MIP mutant (DmMIP13-8A) carrying Ala-replacements in all residues except the conserved Trp residues retained a strong SPR agonist activity (Table 1). Together, these results suggest that SP and MIP share similar structural requirements for the SPR activation, possibly reflecting their interactions with a common binding site on the SPR.
MIPs Are Expressed in the CNS but Not Male Reproductive Organs.
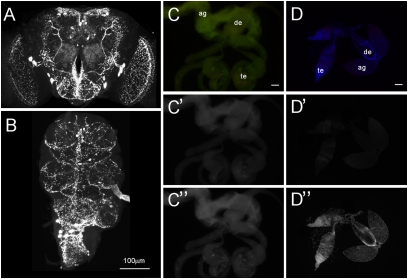
Because MIPs are highly active on SPR, we wondered whether they are involved in inducing postmating responses in Aedes and whether they might also contribute alongside SP to postmating responses in Drosophila. To address this, we first examined whether MIPs are present in male reproductive organs that produce the seminal chemicals essential for the postmating switch in both species (for review see ref. 24). We used MIP-antibody to detect mature MIP peptides in Aedes as well as in Drosophila. The MIP-antibody we used reportedly visualizes MIPs across diverse insect species, including D. melanogaster, Manduca sexta, B. mori, and Rhipicephalus appendiculatus (13, 25–27), and staining in these species coincides with the expression of MIP mRNA (13, 25, 26). We confirmed the strong MIP immunoreactivity in the CNS of both Drosophila and Aedes but could not detect any staining in the male reproductive organs of either species (Fig. 4). This is also consistent with the FlyAtlas database (28), which reports a lack of MIP transcript in male accessory gland but enrichment in CNS. We also failed to detect MIP immunoreactivity in the reproductive organs from various other insect species, including D. mojavensis, B. mori, and Tribolium castaneum.
Fig. 4.
MIPs are expressed in the CNS but not in reproductive organs. Antibody against MIP detected MIP-positive neurons and their arborizations in the brain (A) and the ventral nerve cord (B). In contrast, no MIP immunoreactivity was detected in male reproductive organs of Drosophila (C’) or Aedes (D’). GFP (C’’) and DAPI (D’’) were used for counterstaining in Drosophila (w;UAS-lamin-EGFP;Fru-GAL4) and Aedes respectively. Ag, accessory gland, de, ejaculatory duct, te, testis. (Scale bar, 100 μm.)
MIPs Are Not Required for Postmating Responses in Drosophila.
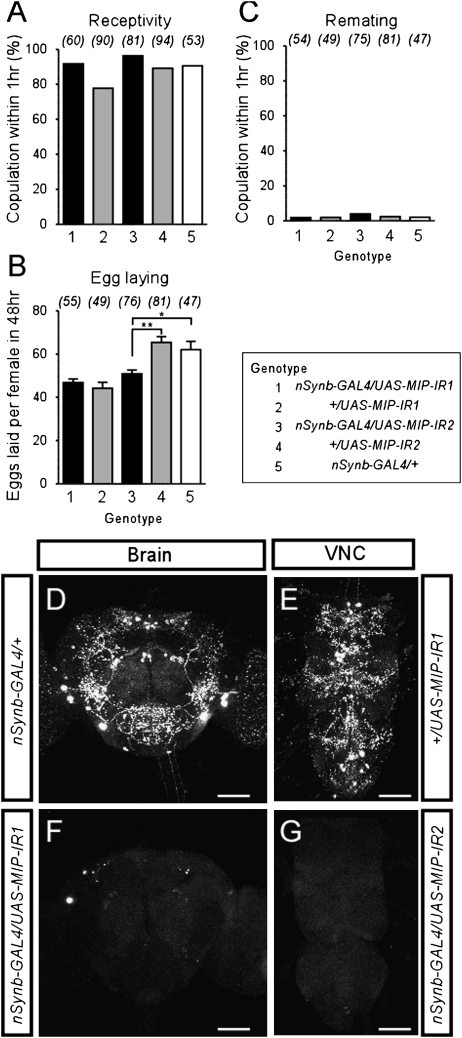
The lack of MIP expression in the male reproductive system argues against the model that MIPs, like SP, are transferred to females in the seminal fluid. Nonetheless, MIPs might still act centrally in the female to modulate reproductive behavior, possibly even within the cascade of events triggered by SP activation of SPR in the periphery. To test this hypothesis, we investigated female postmating responses upon knockdown of MIP expression in the nervous system, induced by pan-neuronal expression of RNAi transgenes targeting MIPs (nSynb-GAL4 UAS-MIP-IR, henceforth MIP RNAi). To control for potential off-targeting effects, two independent RNAi lines (UAS-MIP-IR1, UAS-MIP-IR2) were used. For behavioral assays, individual virgin females were first tested for receptivity toward a naïve male. Those females that mated were then allowed to lay eggs for 48 h before being retested for receptivity with a second naïve male. In all of these assays, there was little difference between MIP RNAi and the control females (Fig. 5 A–C). The only exception was a modest reduction in egg-laying with one of the RNAi lines. Staining of the brain and ventral nerve cord with anti-MIP antibody confirmed the efficient knockdown of MIP in both RNAi females (Fig. 5 D–G). We conclude that endogenous MIPs are not required for female postmating responses, at least in Drosophila.
Fig. 5.
MIPs are not required for the postmating switch. (A) Receptivity of virgin females, scored as the percentage of females that copulated with canton-S male within 1 h. Numbers in parentheses represent the number of samples, P > 0.01 for all comparisons against +/nSynb-GAL4 (genotype 5); 1-way ANOVA followed by Bonferroni test. (B) Number of eggs laid per female. Data are shown as mean ± SEM. * P < 0.01; ** P < 0.001. (C) Remating frequency, scored as the percentage of females that copulated with naïve canton-S male within 1 h. P > 0.01 for all comparisons against +/nSynb-GAL4 (genotype 5). (D–G). Brain and ventral nerve cord of nSynb-GAL4/+ (D; genotype 5), +/UAS-MIP-IR1 (E; genotype 2), nSynb-GAL4/UAS-MIP-IR1 (F; genotype 1), nSynb-GAL4/UAS-MIP-IR2 (G; genotype 3) stained with MIP-antibody. (Scale bars, 100 μm.)
MIPs Do Not Trigger Postmating Responses in Drosophila.
Although not required for postmating responses in vivo, the fact that MIPs are potent SPR agonists in vitro predicts that exogenous MIPs should nonetheless be capable of eliciting a postmating response in females provided they gain access to the relevant SPR-expressing neurons. We tested this prediction in two ways: first, by expressing MIPs in the male seminal fluid in place of SP, and, second, by direct injection of synthetic MIPs into the female haemolymph.
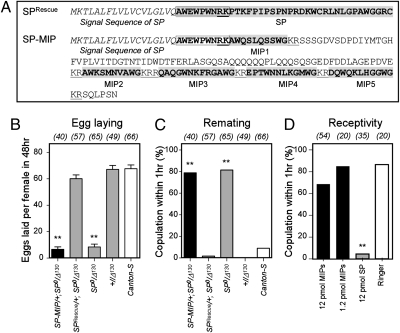
We expressed MIPs in the male seminal fluid by generating a transgene in which MIPs are expressed under the control of regulatory sequences from the SP locus. Because the N-terminal region of SP mediates sperm attachment and the gradual release required for a long-term response (29), we fused the SP signal peptide and N-terminal region to the MIP precursor protein (SP-MIP). This fusion protein is predicted to generate an SP-MIP1 fusion peptide that should bind to sperm, as well as unfused MIP2-5 peptides (Fig. 6A). The expression of MIP in the accessory gland was confirmed by anti-MIP staining (Fig. S3). Genital tract extracts from male flies carrying the SP-MIP transgene in an SP null mutant background showed elevated levels of SPR agonist activity in vitro, comparable to the increase obtained with a control SP transgene (Fig. S3). These assays confirm the expression of functional MIP proteins in the male reproductive tract. Surprisingly, however, wild-type females mated with SP-MIP males did not show any sign of the postmating responses (Fig. 6 B and C). Egg-laying rate and remating frequency of these females were indistinguishable from those of females mated with SP null (SP0/Δ130) males. In contrast, males carrying the control SP transgene (SPRsecue/+; SP0/Δ130) or a wild-type SP allele (+/Δ130) elicited a robust postmating responses from their partners (Fig. 6 B and C).
Fig. 6.
MIPs do not trigger postmating responses in vivo. (A) Predicted amino acid sequence of SP rescue and SP-MIP constructs. Bold, boxed, underlined sequences indicate a mature peptide, SP attachment site and a dibasic peptide cleavage site respectively. Males expressing an SP-MIP chimera (SP-MIP) instead of SP do not induce postmating responses (black bars), as measured by (B) egg-laying and (C) remating frequency. Canton-S virgin females were crossed individually with males from corresponding genotypes. (B) Numbers of eggs laid per females for 48 h after copulation. Data are mean ± SEM. Numbers in parentheses indicate the number of samples. Double asterisk, P < 0.0001 for comparisons against wild-type Canton-S; 1-way ANOVA followed by Bonferroni test. (C) Remating frequency, scored as the percentage of females that copulated with naïve canton-S male within 1 h. Double asterisk, P < 0.0001. (D) Receptivity of Canton-S virgin females assayed 5 h after injection with MIP mixture, SP or Ringer's solution alone. All injected females were virgins. ** P < 0.001 for comparisons against Ringer injection; 1-way ANOVA with Bonferroni correction.
One possible explanation for the inability of SP-MIP peptides to elicit a female postmating response is their inability to pass from the female reproductive tract into the haemolymph (30, 31). To test this possibility, we injected varying concentrations of synthetic MIPs into the hemocoel of virgin females. These females showed no sign of a postmating response, as measured by their receptivity, at any of tested doses (Fig. 6C). In contrast, and as reported previously (5, 32), virgins injected with SP became unreceptive and actively rejected courting males (Fig. 6C).
It is unclear why MIPs should be inactive in these in vivo assays, when they are so potent in vitro. We speculate that additional factors are required for SPR activation by SP in vivo and that these factors may discriminate between the two classes of ligands. Indeed, several proteins have been described that regulate the release, stability, and localization of SP within the female reproductive tract and are thought to be critical for coordinating reproduction (33, 34). One or more of these proteins may ensure that only SP can activate SPR to trigger the postmating changes in female reproductive behavior.
Conclusion
We have presented here evidence that MIPs are ligands for SPR. The evolutionary conservation of both MIPs and SPR across most of the invertebrates, including both ecdysozoa and lophotrochozoa, stands in striking contrast to the recent origin of SP, which can only be detected in the genomes of a limited number of Drosophila species. We also note that the sequenced hymenopteran genomes that lack SPR also lack MIPs. We therefore conclude that MIPs are the ancestral ligands for SPR. Because we cannot detect MIPs in the male reproductive organs of any species tested, and MIPs are neither required nor sufficient to trigger postmating changes in Drosophila females, we infer that the ancestral function for MIPs and SPR is most likely unrelated to the regulation of female reproductive behavior. Our genetic analysis in Drosophila has, however, not yet revealed any other functions for SPR that might be ascribed to MIPs. This may be due to functional redundancy with other putative MIP receptors, such as CG30106 (35) and its closely related homolog CG14593. In other insects, MIPs have been shown to suppress visceral muscle contractions (15, 22), regulate production of juvenile hormones (18) and ecdysteroid (13, 16), and modulate ecdysis behavior (25, 26). It remains to be seen whether any of these functions are mediated by SPR.
Nonetheless, our data do not formally exclude the possibility that SPR regulates female reproductive behavior in species other than Drosophila. We have also found that Aedes contains at least one other potent SPR ligand, presumably unrelated to both MIPs and SP (SI Text). It is possible that this ligand may function in Aedes in a manner analogous to SP in Drosophila.
Materials and Methods
CHO-K1 cells were transiently transfected, and luminescent signals were measured as described previously (36). Expression constructs for Drosophila SPR, Aedes SPR, and Bombyx SPR have been described previously (5). Aplysia SPR was acquired from Dr. J. Edwards, Columbia Genome Center Aplysia EST Project Site (Columbia University, New York, NY) (Clone ID, CNSN01-C-009396), and its ORF was subcloned into pcDNA3.1(+). The peptides used in this study are shown in Table 1 and Table S2. Peptides were synthesized using the FMOC strategy and solid-phase method on an ABI433A Peptide Synthesizer and were purified with HPLC.
To predict the secondary structure of MIP and SP, a computational analysis was performed using the CAChe program (BioMedCAChe Version 5.0, CAChe Scientific, Inc.). Each peptide was built, subjected to Beautify/Comprehensive, and saved as .cdw file. The saved structure was calculated by Augmented MM2 parameter, which enables minimization calculations for square planar, trigonal bipyramidal, and octahedral atoms. After calculating the structure's potential energy, Mechanics adjusts the atomic positions and calculates the potential energy of the new structure. This procedure is an iterative process until the energy change between iterations is within a specified limit. The obtained structure by Mechanics was calculated MOPAC 2002 using PM3 parameter, which determines bond strengths, atomic hybridizations, partial charges, and orbitals from the positions of the atoms and the net charge, to search the lowest energy conformer with comparisons of HF (heat of formation).
Mouse monoclonal MIP antibody (1A4) was a gift from Dr. A. Mizoguchi (Nagoya University, Nagoya, Japan) (13). Tissues were fixed overnight at 4 °C in 4% paraformaldehyde in PBS (or in some cases at 4 °C for 2 h). The tissues were incubated in primary antibody (1:1,500–1:2,000) for 48 h at 4 °C, and in secondary antibody for 24 h at 4 °C. Other antibodies used were: Alexa 488-conjugated goat anti-mouse and Alexa 568-conjugated goat anti-mouse (all 1:1,000–1:2,000; Molecular Probes). Images were acquired with a Zeiss LSM 510 (the CNS) and a Zeiss Axioscope (male reproductive organs) were processed in Adobe Photoshop.
UAS-MIP-IR1 and UAS-MIP-IR2 were obtained from the genome-wide transgenic RNAi library (37) maintained at the Vienna Drosophila RNAi Center (Stock numbers 5294 and 106076). nSynb-GAL4 (obtained from Dr. J. Simpson, Janelia Farm, Ashburn, VA) was used for the RNAi experiments together with UAS-Dcr-2 (37). SP-MIP was prepared by cloning the part of MIP precursor ORF (+196 to +633) between the 5′ (−873 to +81) and 3′ (+231 to +1020) regions of a SP genomic fragment. The control SP rescue construct contains the corresponding SP genomic region (−873 to +1020). These SP-MIP and SP genomic constructs were cloned into pKM20 vector (a custom designed vector modified from a standard GAL4 vector) (6) and inserted into a specific second chromosome site (VIE-72a) using the ΦC31 system (38). Other stocks used were SP0/TM3, Sb (9), Δ130/TM3, Sb (9) and Canton-S for the wild-type control. Peptide injections and behavioral tests for female reproductive behaviors were performed as described previously (5). All behavioral tests were repeated three times.
Supplementary Material
Acknowledgments
We thank G. Krssakova and M. Madalinski for synthesis of peptides used in this study, K. Humphreys for mosquito rearing, J. Simpson for nSynb-GAL4, A. Mizoguchi for MIP antibody and J.-H. Jung for cloning SP-MIP chimera and SP rescue constructs. Y.-J.K. was supported by an European Molecular Biology Organization postdoctoral fellowship, National Research Foundation of Korea (Grant 2009-0089247) and Gwangju Institute of Science and Technology (Dasan Project 2009). Basic research at the Institute of Molecular Pathology is funded by Boehringer Ingelheim GmbH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914764107/DCSupplemental.
References
- 1.Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E. The roles of co-transmission in neural network modulation. Trends Neurosci. 2001;24:146–154. doi: 10.1016/s0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho GB, Kapahi P, Anderson DJ, Benzer S. Allocrine modulation of feeding behavior by the Sex Peptide of Drosophila. Curr Biol. 2006;16:692–696. doi: 10.1016/j.cub.2006.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen PS, et al. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54:291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 4.Kubli E. Sex-peptides: Seminal peptides of the Drosophila male. Cell Mol Life Sci. 2003;60:1689–1704. doi: 10.1007/s00018-003-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yapici N, Kim YJ, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451:33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- 6.Häsemeyer M, Yapici N, Heberlein U, Dickson BJ. Sensory neurons in the Drosophila genital tract regulate female reproductive behavior. Neuron. 2009;61:511–518. doi: 10.1016/j.neuron.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Yang CH, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman T, et al. The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci USA. 2003;100:9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Kubli E. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc Natl Acad Sci USA. 2003;100:9929–9933. doi: 10.1073/pnas.1631700100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saudan P, et al. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur J Biochem. 2002;269:989–997. doi: 10.1046/j.0014-2956.2001.02733.x. [DOI] [PubMed] [Google Scholar]
- 11.Rexhepaj A, Liu H, Peng J, Choffat Y, Kubli E. The sex-peptide DUP99B is expressed in the male ejaculatory duct and in the cardia of both sexes. Eur J Biochem. 2003;270:4306–4314. doi: 10.1046/j.1432-1033.2003.03823.x. [DOI] [PubMed] [Google Scholar]
- 12.Cirera S, Aguadé M. Molecular evolution of a duplication: The sex-peptide (Acp70A) gene region of Drosophila subobscura and Drosophila madeirensis. Mol Biol Evol. 1998;15:988–996. doi: 10.1093/oxfordjournals.molbev.a026014. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka N, et al. Bombyx prothoracicostatic peptides activate the sex peptide receptor to regulate ecdysteroid biosynthesis. Proc Natl Acad Sci USA. 2010;107:2060–2065. doi: 10.1073/pnas.0907471107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackburn MB, Jaffe H, Kochansky J, Raina AK. Identification of four additional myoinhibitory peptides (MIPs) from the ventral nerve cord of Manduca sexta. Arch Insect Biochem Physiol. 2001;48:121–128. doi: 10.1002/arch.1064. [DOI] [PubMed] [Google Scholar]
- 15.Blackburn MB, et al. The identification of two myoinhibitory peptides, with sequence similarities to the galanins, isolated from the ventral nerve cord of Manduca sexta. Regul Pept. 1995;57:213–219. doi: 10.1016/0167-0115(95)00034-9. [DOI] [PubMed] [Google Scholar]
- 16.Hua YJ, et al. Identification of a prothoracicostatic peptide in the larval brain of the silkworm, Bombyx mori. J Biol Chem. 1999;274:31169–31173. doi: 10.1074/jbc.274.44.31169. [DOI] [PubMed] [Google Scholar]
- 17.Li B, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18:113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz MW, Kellner R, Hoffmann KH. A family of neuropeptides that inhibit juvenile hormone biosynthesis in the cricket, Gryllus bimaculatus. J Biol Chem. 1995;270:21103–21108. doi: 10.1074/jbc.270.36.21103. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz MW, Kellner R, Hoffmann KH, Gäde G. Identification of multiple peptides homologous to cockroach and cricket allatostatins in the stick insect Carausius morosus. Insect Biochem Mol Biol. 2000;30:711–718. doi: 10.1016/s0965-1748(00)00042-4. [DOI] [PubMed] [Google Scholar]
- 20.Predel R, Rapus J, Eckert M. Myoinhibitory neuropeptides in the American cockroach. Peptides. 2001;22:199–208. doi: 10.1016/s0196-9781(00)00383-1. [DOI] [PubMed] [Google Scholar]
- 21.Riehle MA, Garczynski SF, Crim JW, Hill CA, Brown MR. Neuropeptides and peptide hormones in Anopheles gambiae. Science. 2002;298:172–175. doi: 10.1126/science.1076827. [DOI] [PubMed] [Google Scholar]
- 22.Schoofs L, Holman GM, Hayes TK, Nachman RJ, De Loof A. Isolation, identification and synthesis of locustamyoinhibiting peptide (LOM-MIP), a novel biologically active neuropeptide from Locusta migratoria. Regul Pept. 1991;36:111–119. doi: 10.1016/0167-0115(91)90199-q. [DOI] [PubMed] [Google Scholar]
- 23.Williamson M, et al. Molecular cloning, genomic organization, and expression of a B-type (cricket-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Commun. 2001;281:544–550. doi: 10.1006/bbrc.2001.4402. [DOI] [PubMed] [Google Scholar]
- 24.Klowden MJ. The check is in the male: Male mosquitoes affect female physiology and behavior. J Am Mosq Control Assoc. 1999;15:213–220. [PubMed] [Google Scholar]
- 25.Kim YJ, Zitnan D, Galizia CG, Cho KH, Adams ME. A command chemical triggers an innate behavior by sequential activation of multiple peptidergic ensembles. Curr Biol. 2006;16:1395–1407. doi: 10.1016/j.cub.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, et al. Central peptidergic ensembles associated with organization of an innate behavior. Proc Natl Acad Sci USA. 2006;103:14211–14216. doi: 10.1073/pnas.0603459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simo L, Slovák M, Park Y, Zitnan D. Identification of a complex peptidergic neuroendocrine network in the hard tick, Rhipicephalus appendiculatus. Cell Tissue Res. 2009;335:639–655. doi: 10.1007/s00441-008-0731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 29.Peng J, et al. Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr Biol. 2005;15:207–213. doi: 10.1016/j.cub.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 30.Pilpel N, Nezer I, Applebaum SW, Heifetz Y. Mating-increases trypsin in female Drosophila hemolymph. Insect Biochem Mol Biol. 2008;38:320–330. doi: 10.1016/j.ibmb.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Lung O, Wolfner MF. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem Mol Biol. 1999;29:1043–1052. doi: 10.1016/s0965-1748(99)00078-8. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt T, Choffat Y, Klauser S, Kubli E. The Drosophila melanogaster sex-peptide: A molecular analysis of structure-function relationships. J Insect Physiol. 1993;39:361–368. [Google Scholar]
- 33.Ram KR, Wolfner MF. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc Natl Acad Sci USA. 2009;106:15384–15389. doi: 10.1073/pnas.0902923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfner MF. Battle and ballet: Molecular interactions between the sexes in Drosophila. J Hered. 2009;100:399–410. doi: 10.1093/jhered/esp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson EC, et al. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J Biol Chem. 2003;278:52172–52178. doi: 10.1074/jbc.M306756200. [DOI] [PubMed] [Google Scholar]
- 36.Kim YJ, et al. Corazonin receptor signaling in ecdysis initiation. Proc Natl Acad Sci USA. 2004;101:6704–6709. doi: 10.1073/pnas.0305291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 38.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moroz LL, et al. Neuronal transcriptome of aplysia: Neuronal compartments and circuitry. Cell. 2006;127:1453–1467. doi: 10.1016/j.cell.2006.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.