Abstract
Bacillithiol (BSH), the α-anomeric glycoside of L-cysteinyl-D-glucosamine with L-malic acid, is a major low-molecular-weight thiol in Bacillus subtilis and related bacteria. Here, we identify genes required for BSH biosynthesis and provide evidence that the synthetic pathway has similarities to that established for the related thiol (mycothiol) in the Actinobacteria. Consistent with a key role for BSH in detoxification of electrophiles, the BshA glycosyltransferase and BshB1 deacetylase are encoded in an operon with methylglyoxal synthase. BshB1 is partially redundant in function with BshB2, a deacetylase of the LmbE family. Phylogenomic profiling identified a conserved unknown function protein (COG4365) as a candidate cysteine-adding enzyme (BshC) that co-occurs in genomes also encoding BshA, BshB1, and BshB2. Additional evolutionarily linked proteins include a thioredoxin reductase homolog and two thiol:disulfide oxidoreductases of the DUF1094 (CxC motif) family. Mutants lacking BshA, BshC, or both BshB1 and BshB2 are devoid of BSH. BSH is at least partially redundant in function with other low-molecular-weight thiols: redox proteomics indicates that protein thiols are largely reduced even in the absence of BSH. At the transcriptional level, the induction of genes controlled by two thiol-based regulators (OhrR, Spx) occurs normally. However, BSH null cells are significantly altered in acid and salt resistance, sporulation, and resistance to electrophiles and thiol reactive compounds. Moreover, cells lacking BSH are highly sensitive to fosfomycin, an epoxide-containing antibiotic detoxified by FosB, a prototype for bacillithiol-S-transferase enzymes.
Keywords: Bacillus subtilis, glutathione, glutaredoxin, mycothiol, thioredoxin
Low-molecular-weight (LMW) thiols play critical roles in cell physiology. In most cells, the tripeptide glutathione (GSH) is the major LMW thiol. However, many bacteria lack GSH and instead synthesize alternative LMW thiols (1). In the mycobacteria, mycothiol (MSH) is the major LMW thiol (2), and in Staphylococcus aureus coenzyme-A (CoASH) may, at least partially, fulfill this role. Previously, Bacillus subtilis was thought to rely on Cys as the dominant LMW thiol (1). However, bacillithiol (BSH) was recently identified as a major thiol in B. subtilis and related Firmicutes (3). Organisms that contain BSH may also contain other abundant LMW thiols. This suggests that various LMW thiols have specialized functions in the cell and that BSH, in particular, may be adept at chelating metal ions (3).
The thiol group has exceptionally complex chemistry but is perhaps best known for its ability to form disulfides (RS-SR) under oxidizing conditions. Disulfide bond formation between cysteine thiols provides stability and determines the structure of extracytoplasmic proteins. However, in most cells the cytosol is reducing and protein thiols are largely in the reduced state. Under oxidative stress, such as elicited by exposure to peroxides, disulfide bond formation can occur either between protein thiols or between protein thiols and LMW thiols. For example, S-glutathionylation of active site Cys residues inactivates some enzymes (4). S-thiolation protects Cys from further oxidation, which could lead to irreversibly damaged proteins containing cysteine sulfinic and sulfonic acids. Reduction of protein disulfides is generally mediated by thiol:disulfide oxidoreductases (TDORs) including thioredoxins (Trx) and glutaredoxins (5). Oxidized glutaredoxins are reduced by GSH generating oxidized GSSG, which is in turn reduced by glutathione reductase at the expense of NADPH. Thioredoxins are directly reduced by Trx reductase.
In addition to disulfide bond formation, thiols can function as nucleophiles to form S-conjugates (6, 7). Thiols react rapidly with some alkylating agents (e.g., N-ethylmaleimide, iodoacetamide) and with electrophiles (formaldehyde, methylglyoxal, S-nitrosocompounds). LMW thiols provide protection against these types of compounds by, minimally, acting as a buffer and, in several cases, by generating adducts that can be enzymatically converted to less harmful compounds (8). The formation of S-conjugates by glutathione-S-transferases is a key step in the detoxification of many xenobiotics (9). Finally, thiols react avidly with thiophilic metals and metalloids and thereby buffer the potentially harmful impact of these elements on cytosolic chemistry.
BSH was characterized as a 398-Da unknown thiol in extracts of B. anthracis (10) and, independently, as the mixed disulfide with B. subtilis OhrR, an organic peroxide-sensing repressor (11). In retrospect, BSH likely corresponds to an unknown thiol (U16) detected earlier in S. aureus (12). BSH and Cys are present in B. subtilis in approximately equal concentrations, and either thiol can mediate protein S-thiolation, as observed with OhrR (11). The thiol-oxidixing agent diamide elicits S-cysteinylation of a large number of cellular proteins in both B. subtilis and S. aureus (13, 14). The extent to which some or all of these proteins may also be modified by S-bacillithiolation has yet to be determined. Further, it is not yet clear whether some protein thiols are preferentially modified with one or another LMW thiol.
Here, we define the biosynthetic pathway for BSH using both genetic and biochemical evidence. Analysis of mutant strains demonstrates that BSH plays a significant role in resistance to thiol-reactive compounds, alkylating agents, and electrophiles. Moreover, BSH null cells are highly sensitive to fosfomycin, which identifies FosB as a likely bacillithiol-S-transferase.
Results and Discussion
Overview of Biosynthetic Pathway for BSH Synthesis.
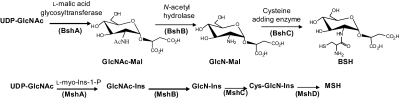
We predicted a likely pathway for BSH biosynthesis (Fig. 1) based on parallels to that described for MSH (6, 15). BshA catalyzes the formation of the first intermediate, GlcNAc-Mal, using L-malate and the glucosamine donor substrate UDP-N-acetylglucosamine (UDP-GlcNAc) as substrates. BshB then hydrolyzes the acetyl group from GlcNAc-Mal to generate GlcN-Mal. Subsequent addition of cysteine, catalyzed by BshC, generates BSH (Cys-GlcN-Mal).
Fig. 1.
Parallels between the predicted BSH biosynthetic pathway (Upper) and the pathway for MSH biosynthesis (Lower). BSH synthesis entails three steps catalyzed by BshA (glycosyltransferase), BshB (N-acetylhydrolase), and BshC (cysteine-adding enzyme). MSH biosynthesis follows a parallel logic, with the addition of a dephosphorylation step (MshA2) after the initial glycosyltransferase reaction and a final N-acetylation of cysteine (MshD).
BshA(YpjH) Is a Glycosyltransferase Essential for BSH Biosynthesis.
Bioinformatic analysis identified five candidate glycosyltransferases in B. subtilis with distant similarity (<22% identity) to MshA (16). We noted that one candidate was encoded by a gene (ypjH) immediately downstream from an mshB homolog (ypjG). On the basis of results shown herein, YpjH and YpjG are renamed BshA and BshB1, respectively. Significantly, B. anthracis BshA (BA1558; 64% identical to BshA) has been crystallized (17), and despite its low degree of identity with Mycobacterium tuberculosis MshA (≈20%), the active site structure is conserved (18). The binding site residues for the acceptor substrate of MshA (1-L-inositol-1-phosphate) are located predominantly in the N-terminal domain (18), which has no similarity to the N-terminal domain of BshA (which presumably binds L-malate).
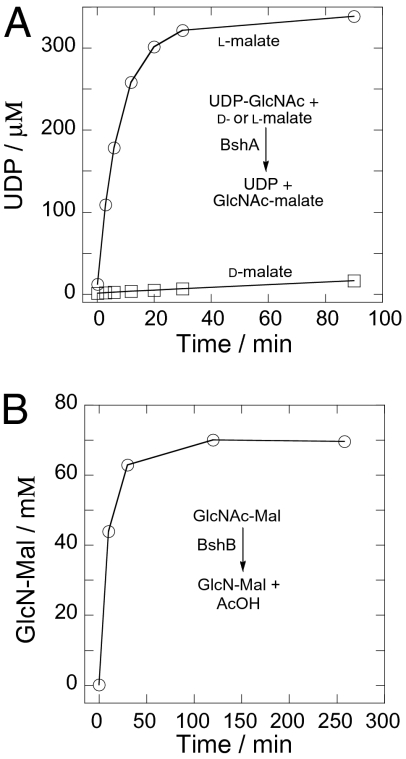
We constructed a bshA null mutant (bshA::mls) using an antibiotic resistance cassette oriented to allow transcription of down-stream genes. In this mutant, BSH was below the detection limit of our assays (Table 1). Complementation with an inducible copy of bshA restored production of BSH in both the bshA null mutant and in a strain in which the mgsA-bshB1-bshA region was replaced with an antibiotic cassette (Table 1). Verification that bshA encodes BshA activity was provided by direct enzymatic assay of the purified protein (Fig. 2A). Incubation with UDP-GlcNAc and L-malate led to the production of GlcNAc(α1→2)L-malate (D-malate was a much poorer substrate). BshA and its orthologs represent a unique class of glycosyltransferase and are present in all species known to synthesize BSH (Fig. S1A).
Table 1.
Genetic determinants of BSH biosynthesis
| Strain and relevant genotype | BSH | Cys | CoA |
| CU1065 wild type | 1.0 ± 0.1 | 0.58 ± 0.14 | 1.1 ± 0.1 |
| HB11002 bshA | <0.007 | 0.50 | 0.59 |
| HB11052 bshA amyE::PxylAbshA | <0.03 | 0.31 ± 0.13 | 1.0 ± 0.15 |
| HB11052 bshA amyE::PxylAbshA + 2% xylose | 0.52 ± 0.07 | 0.49 ± 0.15 | 0.65 ± 0.12 |
| HB11000 bshB1 | 0.42 ± 0.06 | 0.40 ± 0.14 | 0.83 ± 0.15 |
| HB11042 bshB2 | 1.6 ± 0.2 | 0.28 ± 0.14 | 0.85 ± 0.11 |
| HB11053 bshB1 bshB2 | <0.02 | 0.29 ± 0.1 | 0.71 ± 0.06 |
| HB11051 mgsA bshB1 bshA amyE::PxylAbshA | <0.03 | 0.29 ± 0.17 | 1.0 ± 0.1 |
| HB11051 mgsA bshB1 bshA amyE:: PxylAbshA 2% xylose | 1.1 ± 0.03 | 0.51 ± 0.1 | 0.9 ± 0.1 |
Values are μmol/g residual dry weight. Cells cultured to OD600 = 0.8–1.2. Triplicates analyses from single cultures in trypticase soy broth.
Fig. 2.
Enzymatic activities of BshA and BshB. (A) Production of UDP from 1 mM UDP-GlcNAc with 300 μM D- or L-malic acid in the presence of 6.3 μg/mL of B. subtilis BshA. (B) Analyses for monitoring of the preparative reaction used to generate GlcN-Mal from 70 mM GlcNAc-Mal in the presence of 220 μg/mL of B. anthracis Ba1557 BshB1.
Either BshB1(YpjG) or BshB2(YojG) Is Essential for BSH Biosynthesis.
The BshB1 deacetylase is encoded immediately upstream of bshA (Fig. S2). BshB1 is 62% identical in sequence to BcZBP, a zinc-binding protein from B. cereus that has a 1.8-Å resolution structure (19). BcZBP is similar in structure in its N-terminal domain to MshB (rmsd = 1.6 Å for 127 aa) despite relatively low overall identity (26%). Analysis of a bshB1 null mutant revealed an ≈2-fold reduction in BSH levels relative to wild-type. Thus, this gene is clearly not essential for BSH biosynthesis (Table 1). This is reminiscent of prior results for MSH biosynthesis: the mycothiol conjugate amidase (Mca) also displays significant deacetylase activity (20). Mca cleaves MSH S-conjugates at the amide bond linking the N-acetylCys conjugate (mercapturic acid) to glucosamine, a reaction mechanistically and positionally identical to the deacetylation reaction, which likely accounts for the overlap in function (6, 15). We therefore postulated that there was a second deacetylase that partially overlaps in function with BshB1. A candidate for such a function is BshB2(YojG), a paralog of BshB1 (≈25% identity) with predicted GlcNAc deacetylase activity (21). A bshB2 null mutant had normal levels of BSH, but in a bshB1 bshB2 double mutant BSH was undetectable (Table 1). Thus, both bshB1 and bshB2 encode enzymes with sufficient deacetylase activity to allow BSH synthesis, although BshB1 seems to play the major role.
To demonstrate that BshB1 can effectively deacetylate GlcNAc-Mal, we overproduced the B. anthracis ortholog (BA1557). When incubated with GlcNAc-Mal (prepared using BshA as described in SI Materials and Methods), BA1557 rapidly deacetylated the substrate to yield the expected product (Fig. 2B and Fig. S3). Thus, BshB1 is a GlcNAc-Mal deacetylase competent to catalyze the second step of BSH biosynthesis. Significantly, BshB1 and BshB2 homologs are conserved in a wide variety of species known to produce BSH, although with some variations. In S. aureus, BshB1 seems to be missing, and there is a single BshB2 homolog. Conversely, B. anthracis encodes BshB1 and two paralogous BshB2 enzymes (Fig. S1 B and D).
BshC Is a Candidate for a Unique Cysteine-Adding Enzyme.
The MshC cysteine ligase is a homolog (34% amino acid identity) of tRNA-cysteinyl synthetase (CysS) (22). A similar homolog is lacking in Firmicutes, and purified S. aureus CysS lacks detectable BshC activity (SI Materials and Methods). This suggests that BshC function is provided by a previously uncharacterized enzyme.
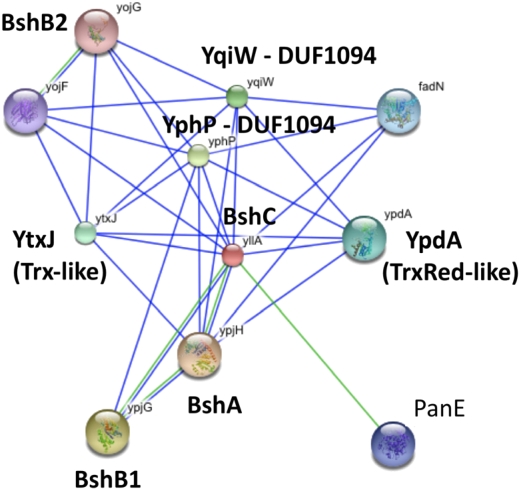
We reasoned that the BshC would likely be encoded by a gene present in those organisms synthesizing BSH, and therefore also having bshA and bshB, and absent from other genomes. To identify candidate loci, we used EMBL STRING (23) using bshA as query. This search tool identifies interaction partners according to a variety of criteria, including colocalization in one or more genomes (e.g., operon structures), co-occurrence across genomes (phylogenomic profiling), and, where available, correlated expression data and/or literature citations. Phylogenomic profiling identified YllA, a 539-aa unknown function protein (COG4365) with no recognizable domains, as a BshC candidate present in genomes that also encode BshA, BshB1, and BshB2. Orthologs include the B. anthracis BA4058 and S. aureus (Newman) NWMN_1087 proteins (Fig. S1 C and D). This analysis further revealed that Myxococcus xanthus, a delta-proteobacterium not previously suspected of containing BSH, contains a likely operon with gene order bshC(yllA)-bshB1-bshA. A similar arrangement is present in Natranaerobius thermophilus, a halophilic alkalithermophile. Colocalization of these three genes provides additional support for the notion that their products define a biosynthetic pathway.
To determine whether yllA encodes a function needed for BSH biosynthesis, we generated a null mutant. The resulting strain lacks BSH (Table 2). Therefore, we tentatively assign yllA (and its orthologs) as bshC. If BshC functions as a cysteine-adding enzyme, we reasoned that the null mutant might accumulate the GlcN-Mal intermediate. Indeed, HPLC analysis of the bshC null mutant revealed a complete absence of BSH and a ≈5-fold increased accumulation of GlcN-Mal relative to wild type. Complementation by ectopic expression of bshC led to a reduction in GlcN-Mal and a restoration of BSH synthesis (Table 2). Thus, BshC is required for BSH synthesis and seems to function in the cysteine coupling reaction.
Table 2.
Characterization of bshC mutant
| Strain and relevant genotype | BSH | Cys | CoA | GlcN-Mal |
| CU1065 wild type | 1.9 ± 0.3 | 0.38 ± 0.05 | 0.53 ± 0.16 | 0.24 ± 0.03 |
| HB11079 bshC | <0.01 | 0.29 ± 0.02 | 0.60 ± 0.06 | 1.2 ± 0.1 |
| HB11091 bshC amyE::PxylAbshC | <0.01 | 0.31 ± 0.06 | 0.45 ± 0.04 | 0.87 ± 0.01 |
| HB11091 bshC amyE:: PxylAbshC + 2% xylose | 1.1 ± 0.1 | 0.30 ± 0.03 | 0.53 ± 0.06 | 0.58 ± 0.04 |
Values are μmoles/g residual dry weight. Cells cultured in trypticase soy broth to OD600 = 0.8. Triplicate samples from a single culture (n = 3).
Phylogenomic Profiling Identifies Four Putative BSH-Related TDORs.
In addition to BshC, four additional proteins are highly correlated in their occurrence with BSH biosynthetic functions (Fig. 3). These include YpdA, YqiW, YphP, and YtxJ. Remarkably, all of these proteins are likely TDORs. YpdA is a member of COG0492 (Trx reductase family) with 24% identity to B. subtilis TrxB. YqiW and YphP are paralogs (53% identity), and each contain an unusual CxC redox motif. Recently, the structure of YphP was reported, and this protein was shown to function as a disulfide isomerase (24). YqiW contains redox active Cys residue(s) visible in redox proteomics (25). YtxJ is a Trx family member that contains a single conserved Cys residue in a motif (TCPIS) reminiscent of that found in monothiol glutaredoxins. Collectively, these proteins define a group of unlinked genes that cooccur in those species predicted to synthesize BSH. Therefore, we suggest that these proteins likely reduce intra- or intermolecular disulfides, including mixed disulfides with BSH. Some or all of YqiW, YphP, and YtxJ may be reduced enzymatically by a Trx-reductase type enzyme (perhaps YpdA). Alternatively, they may be reduced directly by BSH, much like glutaredoxins are reduced by GSH. Recently, an MSH-dependent mycoredoxin was identified in Corynebacterium glutamicum (26). By analogy, it is reasonable to anticipate the existence of BSH-coupled reductases for which we propose the term bacilliredoxins.
Fig. 3.
Phylogenomic profiling of BSH-related functions. The interaction network (as displayed by EMBL STRING) for genetically interacting proteins possibly related in function with B. subtilis BshC (YllA) is shown. Green lines indicate colocalization in genomes (likely operon structures), and blue lines indicate statistically significant co-occurrence across multiple genomes.
BSH Biosynthesis Genes Are Clustered with Functions Required for Coenzyme-A Biosynthesis.
Both genome arrangement and evidence from high-density tiling array transcriptome experiments (27) suggest that the BSH biosynthetic genes are expressed as members of three operons (Fig. S2). The bshA and bshB1 enzymes are encoded as part of an apparent heptacistronic ypjD operon together with dihydrodipicolinate reductase (dapB), methylglyoxal synthase (mgsA), CCA tRNA nucleotidyltransferase (cca), and the biotin repressor (birA). Immediately downstream of the ypjD operon lies panBCD, encoding three enzymes of pantothenate biosynthesis. Transcriptomic data suggest that there is significant readthrough from the ypjD operon into panBCD (Fig. S2). The bshC gene is in an apparent operon with ylbQ, which encodes a predicted 2-dehydropantoate 2-reductase (PanE). Together, these genomic colocalization and transcriptomics results suggest a coordination of BSH and pantothenate biosynthesis. Pantothenate is a precursor of CoA (CoASH), which like BSH is a major LMW thiol in low GC Gram-positive bacteria (3). The bshB2 gene is encoded as part of the ypyC-yojF-bshB2 operon. Regulation of these operons has yet to be elucidated, but bshB2 is up-regulated as part of the disulfide stress responsive Spx regulon (28).
BSH Is Not Essential for Maintaining Protein Thiols in a Reduced State.
We used a previously described 2D gel fluorescence-based thiol-modification assay (25) to identify reversibly oxidized proteins both before and after treatment with diamide, a thiol-oxidizing agent that catalyzes the formation of disulfide bonds (Fig. S4). The major fluorescently labeled proteins (those that contain Cys residues that become accessible for labeling only upon reduction) are identical in the three strains (wild-type, bshA, and bshB1bshB2) both before and after diamide treatment. These include AhpC, Tpx, and IlvC that are partially oxidized under control conditions in all strains. Consistent with previous results (25), there is a large increase of reversibly oxidized proteins in wild-type and BSH null cells after diamide treatment, including TrxA, AccB, CysC, GapA, GuaB, LeuC, MetE, PurL, and TufA. Thus, the protein redox status in the cytosol is not grossly perturbed in a BSH null cell. Because BSH and Cys are both abundant LMW thiols in B. subtilis (Table 1), some redundancy in function is expected.
BSH Is Not Essential for Activating Oxidative and Disulfide Stress Responses.
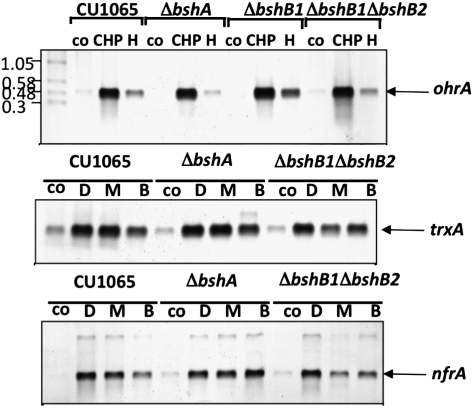
Both Cys and BSH form mixed disulfides in vivo with OhrR, a cysteine-based peroxide sensor (11). In vitro, S-thiolation of OhrR with Cys is sufficient for regulation. To determine whether S-bacillithiolation is required for redox regulation of OhrR, we performed Northern blot analyses of the OhrR-repressed ohrA gene in wild-type and BSH null cells (Fig. 4). As expected, ohrA is strongly induced by cumene hydroperoxide and only very weakly by H2O2. Significantly, this pattern of induction is not overtly affected in BSH null strains (bshA and bshB1bshB2 mutants). Thus, BSH is not required for the regulation of gene expression by OhrR.
Fig. 4.
Northern blots of gene regulation in wild-type vs. BSH null cells. RNA samples were probed with the ohrA gene (to detect derepression by oxidation of OhrR) and with the trxA and nrfA genes (for regulation by Spx). co, untreated control cells; CHP, 100 μM cumene hydroperoxide; H, 100 μM hydrogen peroxide; D, 1 mM diamide; M, 0.5 mM methylhydroquinone; B, 5 μM benzoquinone.
In parallel with this analysis, we also tested the effects of oxidants on Spx, a disulfide-stress sensor activated by formation of an intramolecular disulfide (29, 30). We monitored expression of two Spx target genes, trxA and nfrA, under control and electrophile stress conditions (Fig. 4, Middle). These genes are expressed at low levels in both wild-type and BSH null cells in the absence of stress. This suggests that BSH null cells are not experiencing disulfide stress, consistent with redox proteomics. Moreover, both Spx-regulated genes are normally induced in both wild-type and BSH null cells by the electrophilic quinones and diamide. Thus, both the OhrR and Spx regulatory systems function normally independent of BSH.
Phenotypic Survey of BSH Null Mutants.
To investigate the functions of BSH, we have compared the growth properties and chemical sensitivity of wild-type and mutant strains lacking BSH (BSH null strains). Overall, the growth rate of the BSH null strains is comparable to that of wild-type in both rich and minimal medium, there are no additional auxotrophies, and the mutant strains are competent for DNA transformation. However, a broad survey of growth phenotypes revealed several striking differences. First, the BSH null cells sporulate with ≈100-fold reduced efficiency relative to wild type. Second, the mutant strains displayed a greatly increased sensitivity to high salt: growth was drastically reduced in LB medium amended with 0.9 M NaCl, whereas the growth rate of wild type was little affected. Third, the BSH null cells were sensitive to acid stress and grew slower at pH 5.8 when compared with wild type.
To survey the effects of BSH deficiency on sensitivity to a wide range of chemicals, we used plate sensitivity (zone of growth inhibition) tests in combination with growth analyses (using BioScreen C). Cells lacking BSH have a significantly increased sensitivity to thiol alkylating agents (monobromobimane, N-ethylmaleimide, and iodo-acetamide; Table 3). They also display a modest increase in sensitivity to diamide and methylglyoxal, an endogenously produced thiol-reactive electrophile (Table 3).
Table 3.
Phenotypes of BSH null cells
| Compound | Wild type* | bshA* |
| mBBr 1.5 mmol | 2.15 ± 0.07 | 2.55 ± 0.06 |
| Diamide 15 mmol | 2.13 ± 0.1 | 2.4 |
| H2O2 150 μmol | 2.8 ± 0.14 | 2.9 |
| Methylglyoxal 27.5 mmol | 3.45 ± 0.07 | 3.95 ± 0.06 |
| N-ethylmaleimide 0.5 mmol | 2.87 ±0.15 | 3.37 ±0.06 |
| Iodoacetamide 0.25 mmol | 1.97 ±0.06 | 2.5 ± 0.1 |
| Fosfomycin 500 mg | 2.0 ± 0.1 | 4.3 ±0.06 |
| Penicilllin G 2mg | 3.0 ± 0.1 | 3.4 ± 0.2 |
| Rifampin 1mg | 2.4 | 2.5 |
| Nitrofurantoin 0.5 mmol | 2.0 | 1.7 |
| Bacitracin 0.75 mmol | 1.15 ± 0.07 | 1.25 ± 0.07 |
*Diameter (cm) of the zone of growth inhibition around a 0.6-cm filter paper disk containing the indicated compounds. Values shown are averages ± SD (n ≥ 3), and where no SD is indicated all measurements were identical within the precision of the assay (±0.05 cm).
BSH Is Important for Resistance to Fosfomycin.
GSH (and presumably MSH) are substrates for cognate S-transferases that are involved in detoxification of xenobiotics, including many antibiotics (7, 9). We screened BSH nulls for sensitivity to antibiotics using plate sensitivity assays (Table 3) and confirmed positive results using growth experiments. We noted a slight increase in sensitivity for penicillin G and rifampin and a dramatic increase in fosfomycin sensitivity. In B. subtilis, fosfomycin resistance depends on FosB, previously shown to function as a thiol-dependent S-transferase mechanistically related to the FosA glutathione-S-transferase (31). Unlike FosA, FosB does not use GSH, and although L-Cys supports catalysis in vitro, FosB has a low affinity for this cosubstrate (KM of ≈35 mM). We therefore hypothesized that BSH might be an obligate cosubstrate for FosB. Indeed, BSH null cells are as sensitive to fosfomycin as fosB null mutants. Moreover, a fosBbshA double mutant is no more sensitive than either single mutant. We therefore propose that FosB is a prototype bacillithiol-S-transferase.
Concluding Remarks.
Previous chemical analyses identified BSH (Cys-GlcN-Mal) as a major LMW thiol in B. subtilis and a variety of other bacterial species (3). Here, we have identified genes required for BSH biosynthesis and confirmed the predicted enzymatic activity for the first two biosynthetic enzymes, BshA and BshB1. Using phylogenomic profiling, we have also identified a gene, bshC(yllA), that is required for the final step in BSH synthesis: the coupling of Cys to GlcN-Mal. Further studies will be required to determine the nature of this final biosynthetic step. Phylogenomic analysis also identifed several other proteins, predicted to function as TDORs, that co-occur in those genomes encoding BSH.
Finally, we present the results of an initial survey to determine the phenotypic consequences of a loss of BSH in the model organism, B. subtilis. There were no gross changes relative to wild type in the redox status of cytosolic proteins under either nonstressed or diamide-treated conditions. Moreover, two well-characterized Cys-dependent transcription factors, OhrR and Spx, both seem to function normally in cells lacking BSH. However, BSH null cells are significantly altered in a number of phenotypes, including acid and salt resistance, sporulation, and resistance to antibiotics. BSH null mutants are greatly increased in their sensitivity to fosfomycin, and genetic evidence indicates that this is due to a requirement for BSH in the detoxification reaction catalyzed by the FosB resistance enzyme. Thus, FosB represents a prototype for bacillithiol-S-transferases.
Materials and Methods
Strain Construction and Growth Conditions.
All genetic and physiologic studies were conducted in B. subtilis 168 strain CU1065 (trpC2 attSPβ) and isogenic derivatives (Table S1). Disruption of the bshA, bshB1, bshB2, and bshC genes was achieved by transformation with PCR products constructed using oligonucleotides (Table S2) to amplify DNA fragments flanking each target gene and an intervening antibiotic cassette as described previously (32). Double mutants were constructed by chromosomal transformation. For complementation, the coding region of each gene was cloned into the xylose-inducible pSWEET plasmid (33). B. subtilis strains were assayed for growth and chemical sensitivity using standard zone of growth inhibition assays and by monitoring growth in a Bioscreen C as described in SI Materials and Methods.
Measurement of LMW Thiols.
LMW thiols were measured by HPLC analysis of fluorescent thiol adducts with monobromobimane as described previously (3). Cells were cultured in tryptone soy broth to OD600 = 0.8, and triplicate samples were analyzed from a single culture.
Expression, Purification, and Assay of BshA, BshB1(Ba), and BshC.
Proteins were purified after overproduction in Escherichia coli BL21/DE3(pLysS) strains by induction of T7 RNAP and assayed for the predicted enzyme activities as detailed in SI Materials and Methods.
Proteome and Thiol-Redox Proteome Analysis.
The redox proteome analysis was performed as described previously (25) for wild-type and BSH null cells with and without treatment with diamide for 15 min. Details are in SI Materials and Methods.
Northern Blot Experiments.
Northern blot analyses were performed using RNA isolated from type, bshA, bshB2, and bshB1bshB2 mutants before (control) and 10 min after treatment with 5 μM benzoquinone, 0.5 mM methylhydroquinone, 1 mM diamide, 5.6 mM methylglyoxal, 100 μM H2O2, or 100 μM cumene hydroperoxide. Details of probe construction and hybridization are in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grants MCB0640616 (to J.D.H.) and MCB0235705 (to R.C.F.), National Institutes of Health Grants GM47466 (to J.D.H.), GM061223 (to M.R.), GM35394 (to A.C.), and AI72133 (to R.C.F.), and Deutsche Forschungsgemeinschaft Grant AN746/2-1 (to H.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
1A.G. and G.L.N. contributed equally to this work.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000928107/DCSupplemental.
References
- 1.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol. 2001;55:333–356. doi: 10.1146/annurev.micro.55.1.333. [DOI] [PubMed] [Google Scholar]
- 2.Newton GL, et al. Distribution of thiols in microorganisms: Mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton GL, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 6.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72:471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawat M, Av-Gay Y. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol Rev. 2007;31:278–292. doi: 10.1111/j.1574-6976.2006.00062.x. [DOI] [PubMed] [Google Scholar]
- 8.Antelmann H, Hecker M, Zuber P. Proteomic signatures uncover thiol-specific electrophile resistance mechanisms in Bacillus subtilis. Expert Rev Proteomics. 2008;5:77–90. doi: 10.1586/14789450.5.1.77. [DOI] [PubMed] [Google Scholar]
- 9.Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 10.Nicely NI, et al. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: Implications for coenzyme A-dependent redox biology. Biochemistry. 2007;46:3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lithgow JK, Hayhurst EJ, Cohen G, Aharonowitz Y, Foster SJ. Role of a cysteine synthase in Staphylococcus aureus. J Bacteriol. 2004;186:1579–1590. doi: 10.1128/JB.186.6.1579-1590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hochgräfe F, et al. S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem. 2007;282:25981–25985. doi: 10.1074/jbc.C700105200. [DOI] [PubMed] [Google Scholar]
- 14.Pöther DC, et al. Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jothivasan VK, Hamilton CJ. Mycothiol: Synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat Prod Rep. 2008;25:1091–1117. doi: 10.1039/b616489g. [DOI] [PubMed] [Google Scholar]
- 16.Newton GL, et al. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA) J Bacteriol. 2003;185:3476–3479. doi: 10.1128/JB.185.11.3476-3479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruane KM, Davies GJ, Martinez-Fleites C. Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins. 2008;73:784–787. doi: 10.1002/prot.22171. [DOI] [PubMed] [Google Scholar]
- 18.Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: Substrate-assisted catalysis. J Biol Chem. 2008;283:15834–15844. doi: 10.1074/jbc.M801017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fadouloglou VE, et al. Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. FEBS J. 2007;274:3044–3054. doi: 10.1111/j.1742-4658.2007.05834.x. [DOI] [PubMed] [Google Scholar]
- 20.Newton GL, Av-Gay Y, Fahey RC. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 2000;39:10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka T, Fukui T, Fujiwara S, Atomi H, Imanaka T. Concerted action of diacetylchitobiose deacetylase and exo-beta-D-glucosaminidase in a novel chitinolytic pathway in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Biol Chem. 2004;279:30021–30027. doi: 10.1074/jbc.M314187200. [DOI] [PubMed] [Google Scholar]
- 22.Sareen D, Steffek M, Newton GL, Fahey RC. ATP-dependent L-cysteine:1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41:6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 23.Jensen LJ, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derewenda U, et al. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry. 2009;48:8664–8671. doi: 10.1021/bi900437z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochgräfe F, Mostertz J, Albrecht D, Hecker M. Fluorescence thiol modification assay: Oxidatively modified proteins in Bacillus subtilis. Mol Microbiol. 2005;58:409–425. doi: 10.1111/j.1365-2958.2005.04845.x. [DOI] [PubMed] [Google Scholar]
- 26.Ordóñez E, et al. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J Biol Chem. 2009;284:15107–15116. doi: 10.1074/jbc.M900877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano S, Küster-Schöck E, Grossman AD, Zuber P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc Natl Acad Sci USA. 2003;100:13603–13608. doi: 10.1073/pnas.2235180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]
- 31.Cao M, Bernat BA, Wang Z, Armstrong RN, Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol. 2001;183:2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: Conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.