Abstract
Papillomaviruses, members of a group of dsDNA viruses associated with epithelial growths and tumors, have compact capsids assembled from 72 pentamers of the protein L1. We have determined the structure of bovine papillomavirus by electron cryomicrosopy (cryoEM), at ∼3.6 Å resolution. The density map, obtained from single-particle analysis of ∼4,000 particle images, shows the trace of the L1 polypeptide chain and reveals how the N- and C-terminal “arms” of a subunit (extensions from its β-jelly-roll core) associate with a neighboring pentamer. Critical contacts come from the C-terminal arm, which loops out from the core of the subunit, forms contacts (including a disulfide) with two subunits in a neighboring pentamer, and reinserts into the pentamer from which it emanates. This trace corrects one feature of an earlier model. We discuss implications of the structure for virion assembly and for pathways of infectious viral entry. We suggest that it should be possible to obtain image reconstructions of comparable resolution from cryoEM images of asymmetric particles. From the work on papillomavirus described here, we estimate that such a reconstruction will require about 1.5 million images to achieve the same number of averaged asymmetric units; structural variability will increase this number substantially.
Keywords: virus assembly, virus structure, icosahedral symmetry
The papillomaviruses are a group of small, DNA viruses associated with benign and malignant epithelial growths (1). Human papillomavirus (HPV) isolates fall into over 100 “types,” initially classified serologically and now by DNA sequence variations that primarily reflect differences in the amino acid sequences of several prominent loops on the major structural protein L1. Infection with “high risk” types, e.g., HPV16 and HPV18, predispose to cervical cancer, prompting development of the recently introduced recombinant vaccines (2). The immunogens in these vaccines are virus-like particles (VLPs), obtained by expressing L1 in eukaryotic cells.
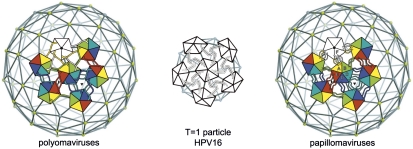
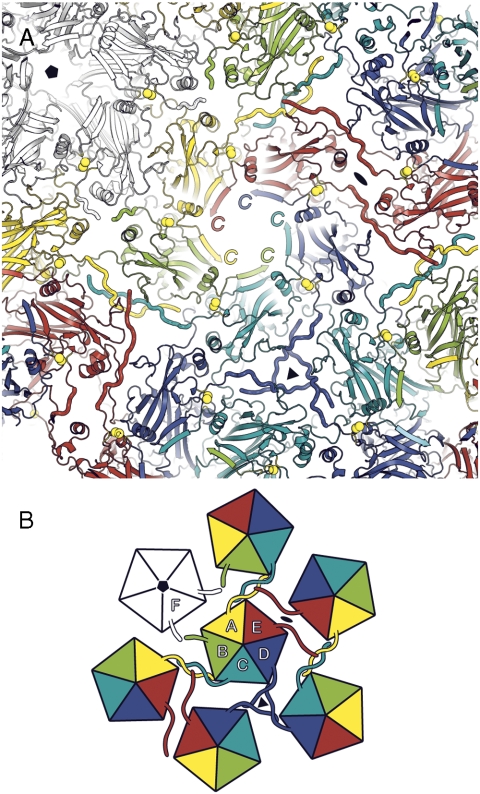
Seventy-two pentamers of L1 assemble to form the outer shell of a papillomavirus particle (3), which packages the closed circular dsDNA genome and associated nucleosomal histones. Twelve of the L1 pentamers lie on 5-fold axes; the other 60, at general positions in the icosahedral surface lattice, are 5-fold symmetric structures in a 6-coordinated surround (Fig. 1). Capsids of the polyomaviruses (in particular, those of SV40 and murine polyomavirus, studied crystallographically) have a closely related, 72-pentamer architecture (4–6). Crystal structures of recombinant L1 pentamers from HPV16 (7) and from three other HPV types (8) have shown as expected that they resemble the major capsid protein (VP1) pentamers from polyomaviruses. The core of the L1 subunit, like those of polyomavirus VP1, is a β-jelly roll domain, with N- and C-terminal extensions (“arms”). In the polyomavirus structures, the C-terminal arms mediate nearly all the interpentamer contacts. They “invade” a neighboring pentamer, forming part of its folded structure; bound Ca2+ ions stabilize the invading conformation of the arm, and inter- or intrapentamer disulfide bonds further lock the assembly together (5, 6).
Fig. 1.
Diagrams of pentamer packing and C-terminal arm interchange in polyoma and papillomaviruses. In the full-size capsids of viruses from both groups, 72 pentamers of the major capsid protein (called VP1 in the polyomaviruses and L1 in the papillomaviruses) are arrayed at the vertices of a T = 7 d icosahedral lattice. Twelve pentamers (white) are centered on 5-fold symmetry axes, whereas the 60 others (6 of which are shown, colored) are at 6-coordinated positions. In a 12-pentamer (T = 1) icosahedral particle, obtained with N-terminally truncated L1 from HPV16, all 12 pentamers are on icosahedral 5-fold symmetry axes. Left: In the polyomaviruses, the VP1 C-terminal arm “invades” a neighboring pentamer, in the pattern shown, and “inserts” into its structure. Right: In the papillomaviruses, the L1 C-terminal arm invades a neighboring pentamer but turns around and inserts back into its pentamer of origin. The T = 1 shell has arm–arm interactions distinct from those in the virion.
When the HPV16 L1 structure was used to interpret a moderate-resolution electron cryomicrosopy (cryoEM) reconstruction of bovine papillomavirus (BPV), the best fit to the density suggested that the L1 C-terminal arms mediate interpentamer contacts with a pattern of invasion very similar to the one seen in SV40 and polyoma (9). That picture was consistent with protein-chemical evidence for an interpentamer disulfide between a cysteine in the C-terminal arm (BPV Cys426) and one within the jelly roll β-barrel (BPV Cys171) (10, 11). We have now obtained a near-atomic resolution cryoEM reconstruction of BPV that allows us to follow the full polypeptide chain (with the exception of a few, inward-projecting C-terminal residues). The long, C-terminal arms indeed interact with neighboring pentamers, forming disulfide bonds in the pattern previously suggested. The full model corrects a feature of the earlier one, however, and thereby alters one principal conclusion of the previous work. Unlike the invading arms of the polyomaviruses, which terminate within the target subunit, those of BPV L1 reinsert into the core of the pentamer from which they emerge. That is, they are essentially elaborate loops, which extend away from the jelly roll core to create the interpentamer contacts (including the disulfide bond), while retaining a C-terminal anchor within their pentamer of origin. The L1 C-terminal arms in the recombinant HPV16 L1 crystal structure also reinsert in this way. Our map thus shows that this reinsertion is a conserved property of the L1 pentamer rather than (as proposed in connection with the earlier cryoEM fit) a specific consequence of truncations needed to obtain the HPV16 L1 crystals.
The complete polypeptide chain trace shows that the N-terminal extensions (about 20 residues) also participate in contacts among pentamers. These interactions are incompatible with formation of the smaller T = 1 shell (12 pentamers) that is seen in the crystals of N-terminally truncated L1 (7). They appear to be one of the switching elements that govern accurate assembly of a 72-pentamer capsid. Comparison of the BPV and HPV16 L1 structures and sequences allows us to identify a number of conserved motifs, including an additional disulfide bond in a subset of papillomavirus types.
Results
Structure Determination.
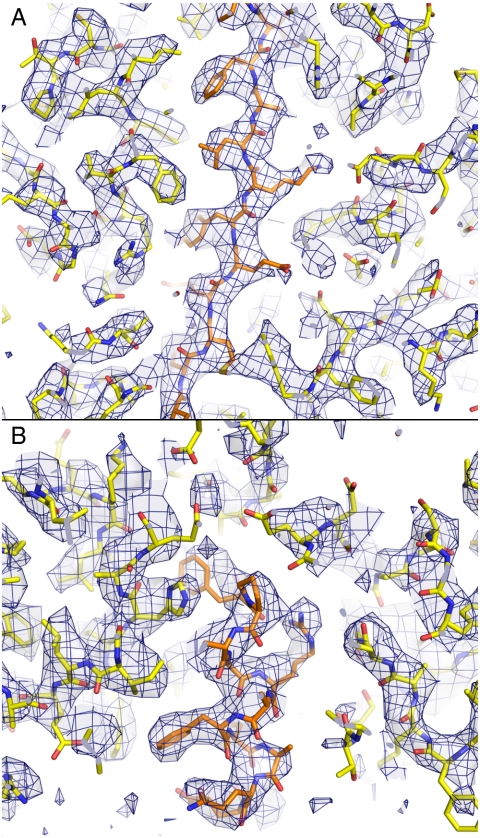
We have obtained a three-dimensional density map of BPV type 1 by single-particle reconstruction from electron micrographs of individual virus particles in vitreous ice. Images of about 4,000 particles from 49 different films contributed to the reconstruction. The definition of features in the maps corresponds to an experimental x-ray crystallographic density map at about 3.5 Å resolution (Fig. 2). The data were acquired as described in Materials and Methods. We could follow nearly all the L1 chain, including previously undescribed segments, by using the icosahedrally symmetrized map. Additional 6-fold “noncapsid symmetry” (NCS) averaging, superposing the core domains of the six subunits in the icosahedral asymmetric unit, showed detail at the level of amino acid side chains. An atomic model of the icosahedral asymmetric unit (six L1 chains) has a conventional R factor, after energy-minimization refinement, of 0.385 (0.570 in the shell at 3.6 Å resolution), calculated against “Fobs” computed from the final icosahedral map (not NCS-averaged) by embedding it in a P1 cell (see Materials and Methods).
Fig. 2.
Portions of the BPV cryoEM density map, in the 6-fold NCS-averaged region, with the refined molecular model superposed. The map is contoured at 3.2σ above the average density within the L1 β-barrel core. Side-chain densities in an α-helix (A) and a β-strand (B) lead to clear assignment of rotamers in many cases.
Interpentamer Contacts: C-Terminal Arms.
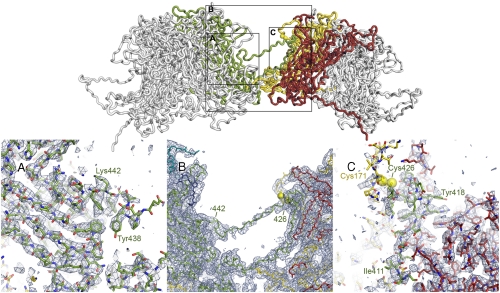
Cross-bridges formed by the long C-terminal extensions (residues 404–495) of the core β-jelly roll link the L1 pentamers, in a pattern of interchange shown in Fig. 1 Right. The polypeptide chain emerges from the jelly-roll domain at its base and extends toward the neighboring pentamer. A segment with a conserved LxDtYR(Y/F) amino acid sequence (residues 412–418 in BPV1) augments (at the B-strand edge) the sheet with strands B, I, D, and G of one subunit in the target pentamer, and the conserved cysteine a few residues C-terminal to this segment (Cys426) forms a disulfide bond with a similarly conserved cysteine (Cys171) in the elaborate EF loop of a second subunit in the target pentamer. The interactions of residues in the LxDxYR(Y/F) motif account for their conservation and for the conservation of their partners: Asp414 has a salt bridge with Arg40; Tyr416 packs against Leu42; Arg417 has a salt bridge with Asp193 and packs against Trp444 (Fig. 3).
Fig. 3.
The L1 C-terminal arm. Top: The interface between two 6-coordinated pentamers. Polypeptide chains are in “worm” representation. The arm of the “green” subunit in the left-hand pentamer (the other subunits are in gray, for clarity) invades the right-hand pentamer, where it augments a β-sheet in the red subunit, forms a disulfide bond (yellow spheres) with the yellow subunit, and extends back to reinsert into its pentamer of origin. The N-to-C direction of the green arm is left-to-right at the “lower” part of the interface shown and right-to-left in the contact-free segment that retraverses the interpentamer gap. The outlined rectangles indicate regions shown in the lower part of the figure. Bottom: Details of the density map to show unambiguous tracing of the L1 arm. In (A) and (C), the 6-fold NCS-averaged map is contoured at 3.0σ above mean density. In (B), a region in which the NCS symmetry does not hold, the map is the final icosahedrally symmetrized reconstruction, contoured at 2.5σ. (A) Reinsertion of the arm into the left-hand pentamer. The PYAGFKF sequence (residues 437–443) where the arm reinserts can be assigned unambiguously, as can residues following it. (B) Connection between the disulfide (Cys426) and the PYAGF segment in (A). The sequence in this bridge is proline-rich in most papillomaviruses. (C) Invasion of the red subunit of the target pentamer (augmentation of the BIDG sheet) and disulfide bond between Cys426 in the arm and Cys171 in the yellow subunit. Assignment of sequence to the augmenting (green) strand [LxDxYR(Y/F) motif, from 412–418] is unambiguous. Cys171 is in a surface loop, less rigid than the core β-jelly roll, and the density of the yellow chain in the vicinity of Cys171 is correspondingly weaker than the core density of the red subunit.
The charged and proline-rich segment immediately C-terminal to Cys426 returns the polypeptide chain to the core of the subunit from which it originated. This fully extended, 11-residue segment bridges the gap between pentamers, with essentially no contacts. It reinserts into the “parent” subunit as a strand (residues 444–449), which augments the sheet with strands C, H, E, and F at the C-strand edge. The tenuous density for the bridging connection was not evident in the lower-resolution map used to build the earlier model (9), and a link was instead suggested to residue 444 in the first subunit of the target pentamer. Except for this connection, which assigned residues from 438 to the C terminus to the wrong polypeptide chain (and the wrong pentamer), and for some conformational details of the segment between 405 and 427, the previous model (9) was correct. In particular, the disulfide was properly identified. The C-terminal arm is thus effectively a large loop: Residues 401–437 project away from the subunit, presenting Cys426 to Cys171 on a subunit of the neighboring pentamer, whereas residues 438–495 reinsert to hold the terminus of the loop within the pentamer from which it emerges.
The C-terminal part of the arm lies within the conical hollow around the pentamer axis (Fig. 3A). A short helix (residues 461–470) is followed by a strand (residues 478–484), which augments the G-strand edge of the BIDG sheet in the neighboring subunit (in a clockwise relationship, looking along the pentamer axis from outside the virion). A cysteine at position 473 pairs in a disulfide with a cysteine at neighboring-subunit position 22. The disulfide, for which there is clear density, thus links the C-terminal part of the L1 polypeptide chain with the N-terminal part of the adjacent chain. The two residues at the corresponding positions are both cysteines in several other papillomavirus types (see the HPV sequence database: http://hpv-web.lanl.gov/), but most types lack this disulfide. The disulfide is clearly not essential for anchoring the C-terminal segment, however, as there are also extensive noncovalent interactions. We can trace about ten residues following Cys473 in the polypeptide chain: They extend into a strand that augments the BIDG sheet of the neighboring subunit at the G-strand edge. The last ten residues of the chain, of which six are lysine, appear to project inward, probably contacting the minichromosome.
Conformational Variability in the C-Terminal Arm.
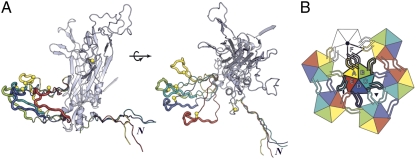
In the assembled virion, the projecting portion of the C-terminal arm must adopt 6 distinct conformations, to adapt to the 6 distinct packing relationships within an icosahedral asymmetric unit. Fig. 4 shows the range of variation within the BPV structure. The distal part of the loop, which contributes the augmenting strand J to the BIDG sheet in one subunit of the target pentamer and Cys426 to a disulfide bond with Cys171 of another subunit, has a uniform structure, determined by those interactions, and conformational variability is restricted to the outgoing and returning segments, residues 403–411 and 427–435 (Fig. 4A). In the T = 1 shell that assembles at low pH from N-terminally truncated HPV16 L1 pentamers (the small VLP), the projecting C-terminal arm also mediates interpentamer contacts, but the segment that would form strand J and the disulfide coils up as an α-helix (α4) instead (7).
Fig. 4.
Conformations of the BPV L1 arm. (A) Subunits A–E, superposed so that their core β-jelly rolls coincide (only one is shown, in gray), with N-terminal arms and extended part of C-terminal arms in color. The core of the subunit is in standard ribbon representation; the arms are in worm representation, in colors corresponding to those in the diagram in B and in Figs. 1,3, and 5. The gray arm, which has the same conformation as the yellow arm, is not shown. The parts of the C-terminal arms that contact subunits of the neighboring pentamer are shown as thicker worms; the conformation of these segments is the same for all subunits, because it is determined by conserved interactions (including the disulfide) with the target subunits. Yellow balls: The cysteines of the interpentamer disulfide. The N-terminal arms all have the same conformation C-terminal to residue 14 (the position at which the superposed arms splay apart). The two parts of A are related by a 90° rotation, as shown. (B) The icosahedral asymmetric unit, viewed from outside the particle, showing the pattern of C-terminal arm contacts.
Interpentamer Contacts: N-Terminal Arms.
The much shorter N-terminal extension (residues 1–20) also participates in a set of interpentamer contacts, largely with other N-terminal arms. One such interaction pairs the arms of chains A and C from two 6-coordinated pentamers related by an icosahedral 5-fold axis (yellow and light blue in Fig. 5). A second is a cyclic set of contacts among the three D-subunit arms related by a 3-fold axis (dark blue in Fig. 5). The third pairs the arm of one E subunit with the CD loop of another, 2-fold-related E subunit and with a short segment of its C-terminal arm (red in Fig. 5). The tip of the E arm also contributes to an A–C arm contact. Arms B and F are disordered up to residue 15. Despite the diversity of these interactions, the extended conformation of residues 1–14 is essentially invariant among the 4 ordered copies within an icosahedral asymmetric unit. A stretch of conserved residues (V/L-Y-L/V-P-P, 10–14 in BPV1) and a conserved tryptophan (residue 4 in BPV1) participate in these varied contacts: A similar N-terminal “scaffold” is therefore likely to be present in other papillomavirus particles.
Fig. 5.
View of BPV capsid from inside, looking outwards, centered along the axis of a 6-coordinated pentamer. (A) Ribbon representation, with N-terminal arms in worm representation, to distinguish them. Note the augmentation of a β-sheet in each subunit by the C-terminal segment of the neighboring subunit in the pentamer. (B) Icosahedral asymmetric unit, viewed from within the capsid. The pattern of N-terminal arm contacts is illustrated by the worm-like projections from each subunit. Note that N-terminal arms are ordered only on subunits A, C, D, and E.
Discussion
Virion Structure.
The central problem for the molecular architecture of a 72-pentamer, icosahedral structure is how to adapt a 5-fold symmetric assembly unit to a 6-coordinated surround. The solution found in capsids of polyomaviruses and papillomaviruses depends on the properties of adaptable subunit arms emanating from a conserved pentamer core. In capsids of both groups of viruses, the long C-terminal arms can have a variety of orientations and degrees of extension, but the segment that interacts with its target site in a neighboring pentamer has an invariant conformation and invariant set of contacts. Moreover, in both cases, disulfide bonds strengthen these contacts (5, 11), requiring that for productive infection the virus pass through a compartment in which these bonds are broken. The BPV structure shows that an important difference between capsids in the two groups of viruses—and a revision to the earlier model for the papillomavirus structure (9)—is whether the C-terminal arm inserts for its entire length into the target pentamer (in the polyomaviruses) or it inserts back into its pentamer of origin (in the papillomaviruses, as shown here). The BPV C-terminal arm emerges from the core of the subunit at residue 402. Residues 411–426 contact the target pentamer, to which Cys426 contributes a disulfide; residues 427–435 bridge back to the pentamer from which the chain originated. The new chain trace provides a more plausible relationship between the contacts in the T = 1 small VLP and the contacts in the virion. The C terminus of the L1 polypeptide chain is anchored identically in both assemblies, creating a long, flexible loop between residues 402 and 435. The tip of this loop contains the conserved LxDxYR(Y/F) motif (residues 412–418) and Cys426 and docks in a uniform way against the target pentamer in all 6 NCS-related contacts in the virus. The small VLP forms only when residues 1–10 are deleted (7). The BPV structure shows that the presence of these residues, which are a large part of the N-terminal arm interaction, is incompatible with the sharp curvature needed to generate the smaller icosahedral particle.
The atomic model accounts for several sequence motifs conserved among the large number of different papillomavirus types (see HPV sequence database: http://hpv-web.lanl.gov/). One of the most unusual features of the structure—the essentially contact-free, “strung out” conformation of residues 429–435—also correlates with conserved sequence characteristics (prolines and polar residues). These conservations are strong evidence that the same sets of L1 capsid contacts are present throughout the papillomavirus family.
Particle Assembly.
L1 pentamers (like polyomavirus VP1 pentamers) have such intricate and extensive interactions between adjacent subunits that soluble monomers are not present in detectable quantity and the pentameric “capsomere” is clearly the fundamental assembly unit of the virion. One copy of the internal protein L2 can associate with an L1 pentamer, through contacts formed within its inward-facing conical hollow (as seen at low resolution in cryoEM reconstructions of pseudovirions with high L2 occupancy) (12, 13). A set of hydrophobic residues toward the C terminus of L2 is essential for L1 binding (12). The resulting picture is similar to the one derived from crystallographic studies of a polyomavirus VP1:VP2/3 complex (14): The C-terminal part of L2 plugs the conical hollow in a hairpin-like conformation, with the hydrophobic segment in contact with one or more L1 subunits and the bulk of the L2 polypeptide chain projecting inward. Estimates of the number of L2 copies per virion vary, but in any case only a small fraction of the L1 pentamers bear this inward-projecting appendage. There is no evidence in our reconstruction for L2 density (or for other connections between L1 and the minichromosome), and we can therefore conclude that L2 is unlikely to be incorporated selectively into the 5-coordinated pentamers. L2 is critical both for DNA packaging and for entry (15).
A first step in particle assembly must be formation of a pentamer–pentamer interface. The contact between a 6- and a 5-coordinated pentamer is the only one that includes interactions other than those with N- or C-terminal arms. The CD loops in the jelly-roll cores of the B and F subunits abut, in a particularly tight contact. This interface is likely to be the one that forms when two pentamers associate, as suggested for polyomaviruses (16). Subsequent steps may also conform to the general considerations outlined for polyomavirus capsid formation. The disulfide bonds between cysteines 426 in the bridging arms and cysteines 171 in their target pentamers, shown to be required for virion stability, lock the capsid structure into place. During intranuclear assembly, however, the cysteines presumably remain reduced, forming disulfides only after the cell has lysed and the virus particles have entered an oxidizing environment. Indeed, pseudovirions or capsids that assemble in cells transfected with plasmids encoding L1, L2, and (for pseudovirions) packageable DNA have few if any disulfide bridges when freshly isolated from culture supernatants, and the particles collapse and flatten when negatively stained with uranyl acetate for electron microscopy (17). If the preparation is allowed to “mature” overnight, specific disulfides form, and particle images have the familiar round, rigid appearance of purified virus particles.
Virus Entry.
The primary cell-attachment receptors for many types of papillomavirus are thought to be heparan-sulfate proteoglycans (18), but specific interactions have yet to be visualized experimentally (19). Following attachment, virions undergo a conformational transition that allows the N-terminal part of L2 to become accessible to cell-surface furin (20). This transition may be related to an in vitro expansion detected by electron microscopy in samples imaged at low ionic strength or low pH (21). Furin cleavage (at a conserved site around residue 10), which is required for infectivity, in turn exposes an L2 epitope (residues 17–36 in HPV16) that can elicit neutralizing antibodies against multiple papillomavirus types (22). There is evidence that transfer to a secondary receptor follows the primary-receptor induced conformational transition and that association with the secondary receptor leads to endocytic uptake (23). A hydrophobic/basic sequence near the C terminus of the L2 polypeptide chain is essential for escape from endosomes (24).
The BPV shell has only rather restricted “holes” through which the N terminus or C terminus of L2 could emerge. The opening along the 5-fold axis of L1 has a van der Waals diameter of 10–12 Å, and a gap to either side of the 2-fold contact has about the same size. A small expansion that retained the contacts within a pentamer would leave the former opening unchanged but substantially increase the latter. For example, the 7% increase in diameter reported for the “open” form of cottontail rabbit papillomavirus (CRPV) (21) would increase the interpentamer distance by about 10 Å, essentially all of which would be an enlargement of the gaps between them. Such an expansion could be accommodated by conformational changes in the segments of the C-terminal arm that cross from one pentamer to another and back (residues 403–411 and 427–435), without breaking the Cys171-Cys426 interpentamer disulfide or disrupting its surrounding noncovalent contacts. Reduction of the disulfides in BPV also leads to expansion, accompanied by exposure of the L1 C-terminal arms (10), but reduction is probably a later event, after the particle has entered the cytosol.
High-Resolution cryoEM.
The clarity and resolution of the density map, from which we have derived an atomic model for BPV, are products of many incremental improvements in experimental and computational cryoEM methods. The virus preparation itself was exceptionally homogeneous, and the BPV particles are unusually robust. Merging the data may have been facilitated by the use of images all collected within a few hours from a single grid square, without major changes in beam geometry or eucentric height. The size of the reconstruction required a 64-bit processor for FREALIGN computation (see Materials and Methods). Sampling was adequate at 1.27 Å/pixel for the resolution obtained (2.9-fold oversampling, Nyquist limit at 2.54 Å) and required a relatively large reconstruction volume (8803 voxels). Accurate determination of defocus for each particle, on the basis of micrograph tilt and calibrated magnification, was essential (a reconstruction without individual defocus adjustment had 4.5 Å resolution). We also paid careful attention to scaling and weighting of the amplitudes for the reconstructed density, while avoiding oversharpening (see Materials and Methods). “Bad” particles were excluded on the basis of a projection-reference correlation criterion. The additional NCS averaging within the icosahedral asymmetric unit further improved the signal-to-noise ratio and extended the resolution from 4.2 to 3.6 Å. The reconstruction was computed from about 4,000 “good” particles. Because each view is icosahedrally equivalent to 59 others, the effective number of projections was about 240,000. The 6-fold NCS averaging implies that a better number, for comparison with required views from an asymmetric object, might be about 1.5 million. These estimates suggest that it may be possible to achieve similar resolution with asymmetric particles, if there is enough low-resolution contrast to avoid orientation ambiguities and if the objects themselves are as homogenous and rigid as BPV. Successful alignment will depend on the presence of clear particle features that help identify orientations. Structural variability of the particles can lead to misalignment and loss of resolution in the variable regions, and images will therefore need to be sorted into subsets of structurally equivalent particles. This sorting will increase the total number of required images, depending on the number of subsets.
Materials and Methods
Image Acquisition.
Data from vitrified samples of BPV, prepared as described (10), were collected on an FEI Tecnai F30 electron microscope (300 kV). Forty-nine micrographs, acquired on film during a single session at 25 e-/Å2, nominal magnification 59,000× (56,588× calibrated), defoci between -1.8 to -2.9 μm, were digitized at 7 µm (1.237 Å/pixel at the specimen). For further details, see SI Text.
Data Processing.
BPV images (6,015 particles) were selected from scanned micrographs with SIGNATURE (25), boxed, and padded. Defocus, tilt, and astigmatism of each micrograph were determined, and individual particle defocus adjusted, with CTFTILT (26). Details of these steps and of the determination and refinement of particle orientations are described in SI Text. Ewald-sphere correction (27) enabled during the final rounds of refinement and reconstruction had only a marginal influence on the final reconstruction (3,977 particles).
Resolution.
We estimated resolution by Fourier shell correlation (FSC) between two half-set reconstructions and by R factor with the final model. Resolution for the icosahedral map was 4.9 Å at FSC = 0.5 and 4.2 Å at FSC = 0.143. The R factor with the refined model reached 0.55 at a resolution of 4.1 Å, in accord with the FSC = 0.143 criterion (28). We estimated the influence of NCS on resolution by 6-fold averaging each half-set reconstruction within an asymmetric unit, by using a soft-edged spherical mask within the boundary of the binary NCS mask. The resolution after NCS averaging was 4.2 Å at FSC = 0.5 and 3.6 Å at FSC = 0.143 (SI Text).
Model Building, NCS Averaging, and Coordinate Refinement.
Procedures for initial model building, magnification calibration, sharpening, and NCS operator determination are described in SI Text. Refinement in CNSsolve (29) was carried out by placing the reconstruction in a cubic P1 cell (512 pixels on a side; 1.237 Å sampling), calculating structure factors and figures of merit (see SI Text), and applying standard procedures for crystallographic refinement within the resolution range 15–3.6 Å: isotropic B-factor correction, automatic bulk-solvent parameter search, bulk-solvent correction, coordinate refinement by energy minimization, restrained individual B-factor refinement, and unrestrained group B-factor refinement. We used icosahedral constraints plus 6-fold NCS restraints for the invariant cores of the 6 L1 protomers in each icosahedral asymmetric unit. See SI Text for details. Refinement statistics are in Table 1. Because the refinement target was the transform of the icosahedrally symmetrized map without local NCS averaging, the calculated R factor does not reflect the enhanced resolution of the NCS-averaged density. Unlike full crystallographic refinement, our procedures adjust the model to fit the final image reconstruction, subject to restraints, but do not iteratively correct the map itself. Thus, they are roughly equivalent to an implementation in reciprocal space of a round of real-space refinement.
Table 1.
Refinement statistics
| Number of “structure factors” | 16,264,110 |
| Constraints (icosahedral operators) | 60 |
| NCS restraints (local NCS operators) | 6 |
| NCS groups | 4 |
| Polypeptide chains in icosahedral a.u. | 6 |
| Number of residues (icosahedral a.u.) | 2,862 |
| Number of atoms in full assembly | 1,357,200 |
| Resolution range (Å) | 15–3.6 |
| R factor | 37.5 |
| Ramachandran plot: | |
| Residues in allowed regions | 100% |
| Residues in most favored region | 80.80% |
| rmsd bond lengths (Å) | 0.01 |
| rmsd bond angles (°) | 1.63 |
| Average B factor, all atoms (Å2) | 131 |
| Average B factor, backbone (Å2) | 126 |
Supplementary Material
Acknowledgments.
We thank Patti Estes for technical assistance and SBGrid for computational support. The work was supported by National Institutes of Health Grants R01 CA13202 (to S.C.H.), R01 CA37667 (to R.L.G.), and P01 GM62580 (to S.C.H. and N.G.). S.C.H. and N.G. are Investigators in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and maps have been submitted to EMDataBank-Research Collaboratory for Structural Bioinformatics with the deposition numbers EMD-5155 for the icosahedrally symmetric final reconstruction, EMD-5156 for the NCS-averaged subunit density, and 3IYJ for the coordinates.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914604107/DCSupplemental.
References
- 1.Howley PM, Lowy DR. Papillomaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2299–2354. [Google Scholar]
- 2.Shi L, et al. GARDASIL: Prophylactic human papillomavirus vaccine development—From bench top to bed-side. Clin Pharmacol Ther. 2007;81:259–264. doi: 10.1038/sj.clpt.6100055. [DOI] [PubMed] [Google Scholar]
- 3.Baker TS, et al. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayment I, Baker TS, Caspar DL, Murakami WT. Polyoma virus capsid structure at 22.5 A resolution. Nature. 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liddington RC, et al. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 6.Stehle T, Yan Y, Benjamin TL, Harrison SC. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen XS, Garcea RL, Goldberg I, Casini G, Harrison SC. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol Cell. 2000;5:557–567. doi: 10.1016/s1097-2765(00)80449-9. [DOI] [PubMed] [Google Scholar]
- 8.Bishop B, et al. Crystal structures of four types of human papillomavirus L1 capsid proteins: Understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 9.Modis Y, Trus BL, Harrison SC. Atomic model of the papillomavirus capsid. EMBO J. 2002;21:4754–4762. doi: 10.1093/emboj/cdf494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Beard P, Estes PA, Lyon MK, Garcea RL. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol. 1998;72:2160–2167. doi: 10.1128/jvi.72.3.2160-2167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapp M, Fligge C, Petzak I, Harris JR, Streeck RE. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J Virol. 1998;72:6186–6189. doi: 10.1128/jvi.72.7.6186-6189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnen RL, Erickson KD, Chen XS, Garcea RL. Interactions between papillomavirus L1 and L2 capsid proteins. J Virol. 2003;77:4818–4826. doi: 10.1128/JVI.77.8.4818-4826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buck CB, et al. Arrangement of L2 within the papillomavirus capsid. J Virol. 2008;82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen XS, Stehle T, Harrison SC. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998;17:3233–3240. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J Virol. 2005;79:3938–3948. doi: 10.1128/JVI.79.7.3938-3948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stehle T, Gamblin SJ, Yan Y, Harrison SC. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 17.Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of papillomavirus capsids. J Virol. 2005;79:2839–2846. doi: 10.1128/JVI.79.5.2839-2846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knappe M, et al. Surface-exposed amino acid residues of HPV16 L1 protein mediating interaction with cell surface heparan sulfate. J Biol Chem. 2007;282:27913–27922. doi: 10.1074/jbc.M705127200. [DOI] [PubMed] [Google Scholar]
- 19.Sapp M, Bienkowska-Haba M. Viral entry mechanisms: human papillomavirus and a long journey from extracellular matrix to the nucleus. FEBS J. 2009;276:7206–7216. doi: 10.1111/j.1742-4658.2009.07400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci USA. 2006;103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belnap DM, et al. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day PM, Gambhira R, Roden RB, Lowy DR, Schiller JT. Mechanisms of human papillomavirus type 16 neutralization by l2 cross-neutralizing and l1 type-specific antibodies. J Virol. 2008;82:4638–4646. doi: 10.1128/JVI.00143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selinka HC, et al. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol. 2007;81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamper N, et al. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol. 2006;80:759–768. doi: 10.1128/JVI.80.2.759-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 27.Wolf M, DeRosier DJ, Grigorieff N. Ewald sphere correction for single-particle electron microscopy. Ultramicroscopy. 2006;106:376–382. doi: 10.1016/j.ultramic.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.