Abstract
Bluetongue virus (BTV) is transmitted by blood-feeding insects (Culicoides sp.) and causes hemorrhagic diseases in livestock. BTV is a nonenveloped, double-stranded RNA (dsRNA) virus with two capsids: a well-studied, stable core enclosing the dsRNA genome and a highly unstable, poorly studied coat responsible for host cell attachment and entry. Here, based on cryo-electron microscopy (cryoEM), we report a 7-Å resolution structure of the infectious BTV virion, including the coat proteins. We show that unlike other dsRNA viruses, the VP2 attachment trimer has a triskelion shape composed of three tip domains branching from a central hub domain. We identify three putative sialic acid-binding pockets in the hub and present supporting biochemical data indicating sugar moiety binding is important for BTV infection. Despite being a nonenveloped virus, the putative VP5 membrane penetration trimer, located slightly inward of the VP2 attachment trimer, has a central coiled-coil α-helical bundle, similar to the fusion proteins of many enveloped viruses (e.g., HIV, herpesviruses, vesicular stomatitis virus, and influenza virus). Moreover, mapping of the amino acid sequence of VP5 to the secondary structural elements identified by cryoEM locates 15 amphipathic α-helical regions on the external surface of each VP5 trimer. The cryoEM density map also reveals few, weak interactions between the VP5 trimer and both the outer-coat VP2 trimer and the underlying core VP7 trimer, suggesting that the surface of VP5 could unfurl like an umbrella during penetration and shedding of the coat to release the transcriptionally active core particle.
Keywords: cryo-electron microscopy, dsRNA virus structure, membrane penetration protein, sialic acid-binding protein
Bluetongue virus (BTV) is a segmented double-stranded RNA (dsRNA) virus in the Orbivirus genus of the Reoviridae family. It infects both ruminants and blood-feeding insects of the Culicoides genus that vector the virus between ruminant hosts. BTV has recently emerged in European countries with severe economic consequences (1, 2), possibly due to climate change and the increased distribution of insect vectors (3).
The virus has four major structural proteins, two (VP2 and VP5) in the coat and two (VP3 and VP7) in the core. The virus also contains an RNA polymerase (4), a helicase (5), an mRNA capping enzyme (6), and the genome composed of 10 linear dsRNA molecules (7). In contrast to other members of the Reoviridae family, the coat of BTV is highly fragile. Upon entry into the cytoplasm, the unstable BTV coat is shed to release a stable core particle. High-resolution structures (3.5 Å) (8) are therefore available for the two core proteins, but only low-resolution structures (24 Å) (9) exist for the proteins that make up the unstable coat and that mediate attachment (10) and entry (11). Although the low-resolution structure places coat proteins VP2 and VP5 in sites consistent with their functions—VP2 (attachment) protruding outward and VP5 (membrane penetration) in a slightly more inward location (9)—the structural bases for BTV attachment and entry into host cells remain unresolved.
Here, we used a purification protocol that afforded complete BTV virions for cryo-electron microscopy (cryoEM) and obtained a 7-Å resolution structure of the whole virus, thus resolving its coat proteins VP2 and VP5 as well as its core proteins VP3 and VP7. These results suggest that VP2 has two sites for binding to the plasma membrane, an outer site and an inner site, the latter binding sialic acid. In addition, even though BTV lacks a lipid envelope, the VP5 trimer has a central coiled-coil helical bundle in this respect resembling the fusion proteins found in many enveloped viruses, including HIV, herpesviruses, vesicular stomatitis virus, and influenza virus. Moreover, by mapping VP5’s amino acid sequence to its structure, we locate 15 amphipathic α-helical regions on the external surface of the VP5 trimer. We suggest that these amphipathic helices play a critical role in penetration of the endosomal membrane and release of the core into the host cell cytoplasm.
Results
The VP2 Attachment Trimer of BTV Has an Exposed Receptor-Binding Tip Domain and an Internal Sialic Acid-Binding Pocket.
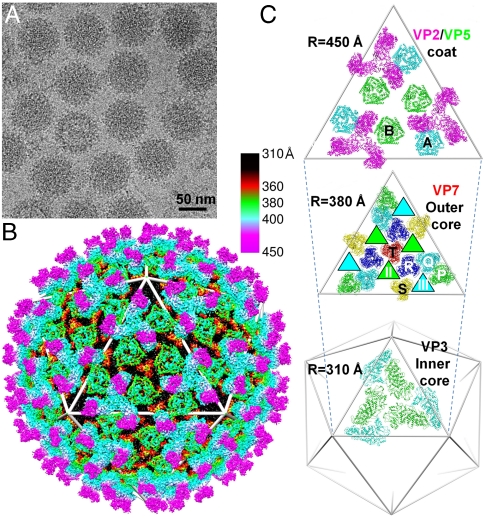
As detailed in Materials and Methods, the purification protocol provided complete BTV virions for cryoEM (Fig. 1A). The 7-Å resolution structure for BTV (Fig. 1B, Fig. S1, and Movie S1) resolved its two coat proteins and its two core proteins (Fig. 1C). We were able to identify the secondary structural elements and their topological arrangements, i.e., the folds, of all four structural proteins, totaling 1,440 molecules in each virion (SI Text). The excellent agreement of the folds of core proteins VP3 and VP7 with those determined by atomic resolution x-ray crystallography (Fig. S1 B and C) provides an internal validation of our structure. In addition, the respective cross-correlation coefficients between the cryoEM maps (at a contour level of 1.3 standard deviations above the mean) and the x-ray models filtered to 7 Å for the cyan VP3 monomer (Fig. 1C) and the VP7.R trimer are 0.54 and 0.59 as calculated with Situs (12).
Fig. 1.
Structure of BTV-1 by cryoEM at 7-Å resolution. (A) CryoEM pictures of BTV. (B) 3D reconstruction of BTV at 7-Å resolution, color coded by radial position: outer-coat VP2 (magenta and cyan), inner-coat VP5 (green), outer core VP7 (red and black), and inner core VP3 (not visible). See also Movie S1. (C Top) Outer-coat proteins VP2 (T = 3) (magenta) and VP5 (T = 6) (B and A conformers in green and cyan, respectively). (Middle) Outer core protein VP7 (T = 13) with channels II and III (green- and cyan-filled triangles underneath B and A conformers of VP5). VP7 trimers are designated as P, Q, R, S, and T based on their locations in the asymmetric unit (8). (Bottom) Two inner core protein VP3 conformers colored cyan and green.
The outer-coat VP2 triskelion (magenta in Fig. 1B and C) is assembled from three VP2 monomers, each contributing a hub domain and a tip domain (top view of density map in Fig. 2A; side view in Fig. 2B; Movie S2). The density map is clearly resolved for unambiguous assignment of secondary structural elements such as α-helices and β-sheets (side view in Fig. 2C) and reveals an arrangement of protein domains unlike that found in the coat of other dsRNA viruses; hence, they are unique to BTV. In the side view (Fig. 2B), the top of the tip domain projects upward from the surface of the virion, while the base of the tip domain sits atop VP7 trimers.
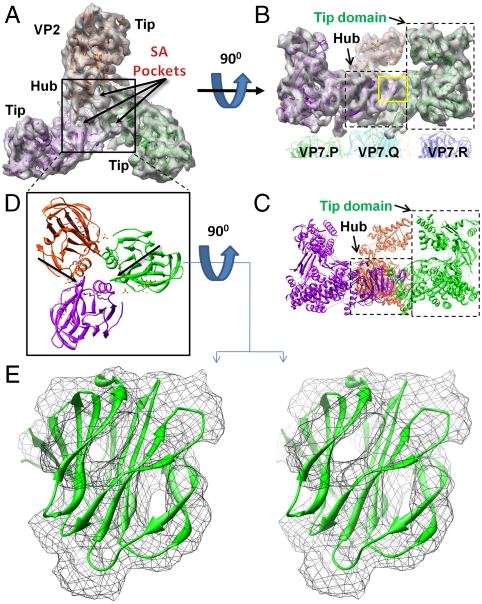
Fig. 2.
Fold of VP2 and identification of SA-binding pocket. Top (A) and side (B) views of the density map of a VP2 triskelion after 3-fold averaging, with three tip domains and a hub (black box) comprised of three hub domains. See also Movie S2. Each hub domain harbors a SA-binding pocket. The hub sits on a VP7.Q trimer, and each of the three VP2 tip domains sits atop its own VP7 trimer (either P, R, or S). (C) Ribbon model of (B), rich in α-helices and β-sheets. (D) Top view of atomic-resolution ribbon models of three VP8 SA-binding domains of rotavirus with positions determined by docking to the density map of VP2 of BTV. (E). Stereo view of the ribbon model of the VP8 SA-binding domain from rotavirus docked into the density map (threshold = 1.5σ) of BTV’s VP2 hub from the yellow box in (B).
The secondary structure of the tip domain is rich in both α-helices and β-sheets, represented by orange helices and blue arrows in Fig. S2 A–D. The density map shows the protruding nature of the tip domain and its possession of a cavity lined with β-sheets. The outermost location of this surface rich in β-sheets suggests that the tip domain is well positioned for the initial encounter of the virus with the host cell plasma membrane (13).
By contrast, the density map of the hub (dashed box labeled “Hub” in Fig 2B) reveals a β-barrel fold. A similar fold has been observed in the sialic acid (SA)-binding domain on the outermost region (called VP8) of the rotavirus VP4 spike (PDB ID code 1KQR) (14). When the ribbon model of the rotavirus SA-binding domain (shown in triplicate in Fig. 2D) is docked into the VP2 density map (Fig. 2E: enlarged view of the small yellow box in the Fig. 2B side view), we find an excellent fit between the two. Indeed, SA fits neatly into a pocket of this putative SA-binding site of VP2 (Fig. S2E). This docking dictates placement of one rotavirus β-barrel into each of the three hub domains that make up a VP2 hub (Fig. 2D) and also shows extensive interaction—revealed in the density map—between the three binding domains.
Effect of Sialic Acid-Binding on BTV Replication.
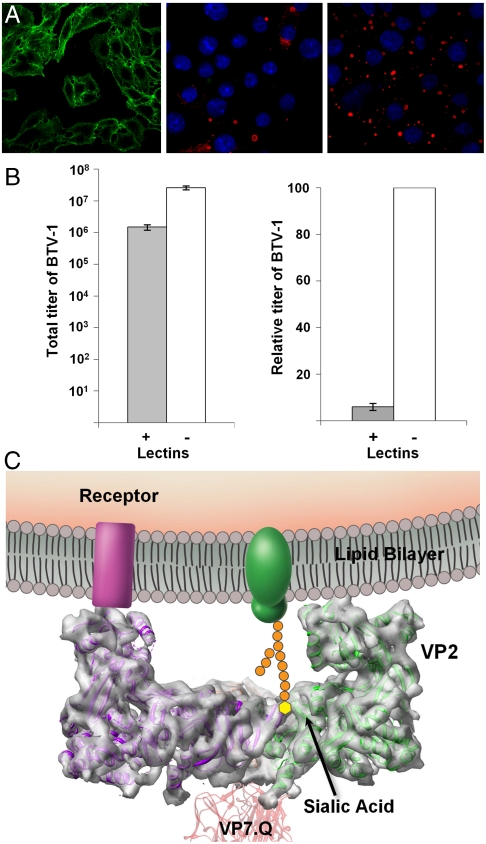
Even though there is no sequence homology between the rotavirus VP8 and the BTV VP2, our above structural findings suggest that VP2 binds SA, and this assignment was further investigated using biochemical approaches. BTV particles agglutinate erythrocytes of ruminants (15), and VP2 alone is responsible for this activity (13). To determine whether SA is available on the plasma membrane of permissive cells for BTV infection, we stained HeLa cells with fluorescently labeled wheat germ agglutinin (WGA), a lectin that binds SA and N-acetylglucosamine sugar residues (16) (Fig. 3A Left). We further monitored the expression of nonstructural protein 2 (NS2) at 16 h after BTV infection of HeLa cells in the presence (Fig. 3A Center) and absence (Fig. 3A Right) of WGA. The presence of WGA clearly reduced NS2 expression and reduced the amount of infectious BTV produced by over 10-fold (Fig. 3B). These results indicate that SA might play a functionally important role during infection of host cells and are consistent with the structural evidence of the existence of an SA-binding site in VP2.
Fig. 3.
Sialic acid-binding activities in BTV infection. (A) HeLa cells stained with (green) FITC-labeled WGA that binds SA (Left). Nonstructural protein 2 (red) was stained by indirect immunofluorescence, and nuclei (blue) were stained with Hoescht 33258, as described in Material and Methods, 16 h after infection with BTV in the presence (+WGA) (Center) or absence (-WGA) (Right) of excess competing WGA. (B) Total (Left) and relative (Right) viral titer 16 h after infection with (+) and without (-) competing WGA. Bars represent standard errors of four replicates (SI Text). (C) Schematic illustration of the membrane attachment of VP2 by its tip domain to a cell surface receptor and by its SA-binding domain to a cell surface glycoprotein.
The VP5 Trimer is a Globular Complex with a Unique Helical Structure.
Previous biochemical studies have shown that VP5 causes membrane leakiness and that its expression at the cell surface induces cell-cell fusion to produce syncytia upon exposure to low pH, similar to that found in the endosomal milieu (11). These data indicate that VP5 trimer is the membrane penetration protein of BTV. Our cryoEM analysis at 7-Å resolution reveals that the VP5 trimer is a globular complex (Fig. 4A). Furthermore, the density map reveals secondary structures demonstrating that VP5 contains many helices but only one β-sheet (Fig. 4A and Movie S3), in stark contrast to the myristoylated μ1 penetration protein of mammalian orthoreovirus with all β-sheets in a jelly-roll domain at its top surface (17). In parallel, we used Jpred software (18) to predict secondary structural regions of α-helices, loops, and β-strands from the amino acid sequence of VP5 (Fig. 4B). PSIPRED software (19) predicted the assignment of 94% of the amino acids to the same secondary structural elements. The correspondence between the cryoEM findings and the prediction of so many α-helices in VP5 suggested that mapping of the amino acid sequence onto the secondary structural elements in the cryoEM density map might be possible, as attempted and confirmed previously (20, 21). The density map also permits the identification of the connectivity between many of the α-helices (SI Text).
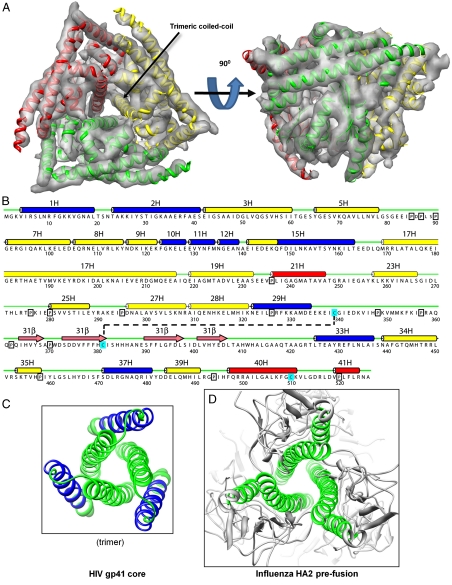
Fig. 4.
Fold of VP5 and prediction of secondary structure from its amino acid sequence. (A) Sixfold averaged density map of the VP5 trimer, viewed from top and side, with an embedded ribbon model. See also Movie S3. (B) Secondary structure prediction. Predicted helices are marked by cylinders, blue (amphipathic), red (hydrophobic), and yellow (other). Predicted β-strands are marked by pink arrows, loops by thin green lines, prolines by black boxes, and cysteines by cyan boxes. The black, dashed line represents a putative disulfide bond. (C, D) Ribbon diagrams of the central coiled-coil helix bundle and a triplet of peripheral helices in fusion proteins gp41 of HIV (PDB ID code 1SZT) (24) and HA2 of influenza (PDB ID code 1HGD) (26). Blue ribbons represent amphipathic regions of α-helices; green ribbons represent nonamphipathic regions of α-helices.
The deduced three-dimensional (3D) model (Fig. 5A, Fig. S3A, and Movie S4) of the prepenetration state of VP5 has many supporting features. First, the amphipathic α-helical region #1H-2H at the N-terminal end (22, 23) and four more amphipathic α-helical regions (#10H-12H, #15H, #28-29H, and #37H) (blue amino acid strings in Fig. 4B; helix wheels in Fig. S4) are on the exterior surface of the protein (blue cylinders in Fig. 5A; blue helices in Fig. 5B and Fig. S3). Second, three copies of one of these, amphipathic helix #37H, are on the top surface (Fig. 5A and Fig. S3A), well positioned to roll and expose their hydrophobic undersides to the endosomal membrane. Third, as noted, the amphipathic helical region #1H-2H is positioned on the exposed (side) surface of VP5 (Fig. 5A and Fig. S3). This region has already been identified as capable of membrane penetration (11, 23). Moreover, amphipathic helices #10H-12H, #15H, and #28-29H are similarly positioned on the exposed surface. Correspondingly, the peripheral triplet of α-helices of the HIV trimeric fusion protein gp41 (24), amino acids 40–68, contains a long amphipathic string of amino acids, amino acids 42–60 (blue turns in Fig. 4C and Fig. S5 A–B). Fourth, the trimer of α-helices #33H/#34H in the VP5 trimer forms a coiled-coil helix bundle that runs up the center of the trimer (Fig. 4A; central triplet of helices in Fig. 5B), similar to what is found among the fusion proteins of the enveloped HIV (25) (Fig. 4C and Fig. S5 A–B), influenza virus (26) (Fig. 4D and Fig. S5C), vesicular stomatitis virus (VSV) (27) (Fig. S5D), herpesvirus (28) (Fig. S5E), and in the penetration protein of the nonenveloped rotavirus (29). Fifth, as expected, in the 3D model, prolines (boxes in Fig. 4B) are consistently located at kinks within helices, between helices and loops, and between loops. Finally, one of the three cysteine residues (amino acid 339, cyan in Figs. 4B and 5A) is located just after the end of α-helix #29H, close in three dimensions to another cysteine 381 at the end of the second (hence “returning”) β-strand in the β-sheet. The density map shows at that point a dense region that bridges the two secondary structures, a region that might correspond to a disulfide bond (indicated by the cyan S-S cylinder in Fig. 5A). This observation is consistent with a thiol/disulfide rearrangement often being necessary during membrane penetration and uncoating processes, as shown in a number of enveloped viruses (see review in ref. 30) and also more recently in human polyoma viruses (31). This observation indicates the presence of such a thiol/disulphide bond and its possible role in virus entry in any complex nonenveloped virus.
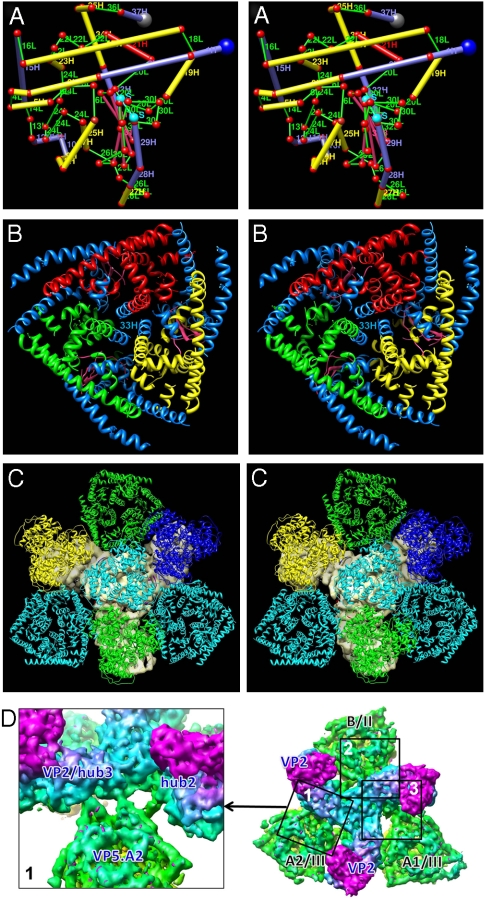
Fig. 5.
Mapping of the secondary structures predicted from the amino acid sequence of VP5 to the secondary structures revealed by the cryoEM density map. (A) In this stereo, side view of a cylinder model of one monomer of VP5, α-helices are represented by thick cylinders (blue = amphipathic, red = hydrophobic, and yellow = other), β-strands by intermediate thickness cylinders (pink), loops by thin cylinders (green), and the putative disulfide bond (S-S) by a thin cylinder (cyan). (B) In this stereo, top view of a VP5 trimer, amphipathic helices are colored blue, but each of the three monomers’ nonamphipathic α-helices has its own color (red, green, and yellow). The central triplet of helices includes amphipathic helix #33H. See also Movie S4 and Fig. S3. (C) Viewed in stereo from the interior of the virus toward the outer surface of the virus, the two (cyan) VP5.A and the one (green) VP5.B conformer in the inner coat fit into the three gaps between the three legs of the solid gray VP2 triskelion in the outer coat. (The hub of the VP2 triskelion is largely obscured by the VP7.Q trimer (cyan); its three tip domains are largely obscured by the VP7.P (green), VP7.R (blue), and VP7.S (yellow) trimers.) (D) Fitting into the three gaps of the VP2 triskelion, the three VP5 penetration trimers interact with at most two hub domains. Box 1 shows three thin fingers of density connecting to two hub domains. Boxes 2 and 3 are shown in Fig. S6. See also Tables S1.
Interactions Among VP2, VP5, and Underlying Core Protein VP7.
The VP2 triskelion in the outer coat has three legs, thereby creating three gaps, filled by three quasi-equivalent VP5 trimers (Figs. 1C Top and 5C). The density map shows that just one, three, and three thin fingers of density (hence weak interactions) connect the VP2 hub domains—not tip domains—to the three VP5 trimers (Fig. 5D, Fig. S6, Table S1, and details in SI Text). Similarly, the VP5 trimer makes few, weak interactions with its subjacent core VP7 trimers (Fig. S7, Tables S2 and S3, and details in SI Text). Just 9 and 10 thin fingers of density connect the A and B conformers of the VP5 trimer to five of their six subjacent VP7 trimers (Fig. S7 and details in SI Text). These weak interactions have the virtue that they would permit conformational changes of VP5 during the penetration process and shedding of the outer coat. By contrast, the outer-coat VP2 tip domains connect to their underlying VP7 trimers (Fig. 2B and Fig. S8A) by moderately thick fingers of density, and the hub domain connects to its underlying VP7 trimer by a wall of density (Fig. S8A and details in SI Text), implicating a large number of amino acid residues (e.g., ∼13 to 69) in the VP7 trimers involved in the interaction with the hub (Fig. S8B). Nonetheless, the VP2 protein detaches readily from the underlying core at high salt concentration (32).
Discussion
In order to infect host cells, viruses must transport their genomic material across a lipid bilayer via special structural proteins. For BTV, access of the transcriptionally active core particles to the cytoplasm involves receptor-mediated endocytosis and low pH-triggered activation of a membrane-permeabilizing protein. As a nonenveloped virus, BTV lacks lipid-envelope proteins, so these functions must be performed by the two outer-coat proteins VP2 and VP5. Previous biochemical studies have suggested that VP2 is the host attachment protein, whereas VP5 is directly involved in the membrane penetration process (11). However, to date it has not been possible to determine the mechanism of either. The 7-Å structures of VP2 and VP5 that we present here give us some clues of how these coat proteins accomplish their tasks.
Our VP2 structure suggests the presence of two binding sites, one on the tip domains of the protein, the other—which appears to bind sialic acid—on the hub domains. Indeed, our biochemical data demonstrate the importance of the putative SA-binding site in VP2 (Fig. 2). This SA-binding domain is ∼40 Å inward from the outer end of the tip domain of VP2, a distance well within the range of lengths of chains of sugars on glycoproteins (Fig. 3C). Conceivably, subsequent to the attachment of the tip domain to the host cell surface, the SA binding of VP2 might further stabilize the interaction with the tip domain (Fig. 3C), thus facilitating infection of host cells. In addition, SA binding may promote adsorption of the virus onto the surface of erythrocytes, thereby increasing the probability of ingestion by the blood-feeding Culicoides vector.
However, it should be noted that in the presence of wheat germ agglutinin, which binds sialic acid, BTV infectivity (Fig. 3B) is not completely abolished, indicating that beyond simple recognition of SA residues there must be further cellular factors involved in the interaction of BTV with receptors. Interestingly, similar data for canine parvovirus (CPV) have demonstrated that although CPV haemagglutinates RBCs via SA interaction, SA is not essential for infection in cultured cells (33, 34). These findings therefore suggest that the interaction of BTV with SA might play its most important role during in vivo infection or in transmission of virus between vertebrate host and the Culicoides vector.
Following attachment mediated by VP2, how does VP5 accomplish membrane penetration? For influenza virus, from prefusion to postfusion, a terminal string of amino acids anchored at the top of the HA2 protein is launched toward the membrane (35). Rotavirus is also thought to employ such an umbrella-opening-like mechanism, except that its penetration protein is predominantly composed of β-sheets (29). For BTV, in analogy with influenza and rotavirus, we speculate that each VP5 trimer is anchored to the membrane by its triplets of amphipathic helix #37H and C terminal, hydrophobic helices #40H and #41H (Fig. 4B). The external surface of VP5 with its 12 additional amphipathic helical regions could swing up to the membrane, where the amphipathic helices could roll to make extensive hydrophobic contact with the membrane and perforate it. This unfurling of VP5 would also detach it and the rest of the coat (VP2) from the core.
Prevention of the unfurling of VP5 and thus penetration and uncoating might be a strategy for development of stable vaccines. Like the putative disulfide bond, Cys339 to Cys381, additional cysteines could be engineered to internally cross-link VP5 and hold it in its prepenetration conformation. Alternatively, additional cysteines could be placed in both VP5 and VP7 near their normal interaction sites to prevent unfurling.
Materials and Methods
Sample Preparation.
BTV-1 (South African strain) was recovered from 10 single-stranded RNA segments synthesized in vitro from cDNA clones with the reverse genetics approach previously described (36). Baby hamster kidney cells were infected with BTV-1 (36) at approximately 0.03 infectious units per cell and cultured at 35 °C for 67 h. Infected cells were lysed in 3-mL 100-mM Tris-HCl buffer (pH8.8) containing 50 mM NaCl, 10 mM EDTA, and 0.1% v/v NP-40 per 107 cells at 4 °C for 10 min. The lysate was centrifuged at 2,000 g for 10 min to pellet nuclei. The supernatant was loaded onto a double cushion [1.8 mL 66% w/w sucrose and 6 mL 50% w/v sucrose in 20-mM Tris-HCl (pH8.8) containing 0.3% v/v NP-40] for centrifugation at 100,000 g for 1 h at 4 °C. Virus and cellular material were collected at the interface of the two sucrose cushions, diluted 10-fold in 20-mM Tris-HCl (pH8.8), and clarified further from aggregates by centrifugation at 16,000 g for 10 min. The supernatant was centrifuged at 37,000 g for 90 min at 4 °C to pellet the virions, and then the pellet was resuspended in 20 μL of 20 mM Tris-HCl (pH8.8) prior to cryoEM sample preparation.
CryoEM, Data Processing, and Visualization.
To prepare cryoEM grids, 3-μL aliquots of sample were placed on thin continuous carbon films for 5 min, blotted with filter paper, and plunged into liquid ethane. Images were recorded at a magnification of 80,000× on a 4 k × 4 k CCD camera (TVIPS) in an FEI TF20 cryo-electron microscope operated at 200 kV. The defocus was determined to be from -1.1 μm to -5.2 μm with the CTFind program (37).
The Boxer program in the EMAN package (38) automatically selected 3,123 particle images from 224 CCD frames. We first reconstructed a low-resolution structure from about 100 particle images with Image Management and Icosahedral Reconstruction System (IMIRS) (39) using orientation and center parameters determined by the cross-common-line method, and this structure was used as the initial model. Further orientation determination, 3D reconstruction, and structural refinement were carried out with Frealign (40) and IMIRS (39). Briefly, the orientation and center parameters of each of the 3,123 particle images were first determined by a global search by maximizing the cross-correlation coefficient of the particle image and projections computed from the current best model. These parameters were iteratively refined by maximizing the cross-correlation coefficient of the particle image and projections around the current particle orientation by including image data truncated to the current resolution of the model. We discarded 23% of the particles with poor correlation coefficients during the refinement. The iterative process was terminated until no further improvement in the resolution of the 3D reconstruction could be obtained. The final map was reconstructed from 2,406 particle images and has an effective resolution of 7 Å based on the Fourier shell correlation coefficient criterion between two independent maps (41) (Fig. S1A).
Local averages from the three monomers in each of the trimers of VP2, the A conformer of VP5, and the B conformer of VP5 were obtained as previously described (42). The secondary structures of VP2 and of VP5 were modeled by manually docking standard polyalanine helix models into the cryoEM density maps with UCSF Chimera (43).
Supplementary Material
Acknowledgments.
This research was supported in part by grants from the National Institutes of Health (GM071940 and AI069015 to Z.H.Z. and AI045000 to P.R.) and BBSRC (to P.R.). We acknowledge the use of the cryoEM imaging facility at the UCLA Electron Imaging Center for NanoMachines supported in part by NIH (1S10RR23057).
Footnotes
The authors declare no conflict of interest .
*This Direct Submission article had a prearranged editor.
Data deposition: The electron density maps and associated models reported here have been deposited in the European Bioformatics Institute (EBI), www.ebi.ac.uk (EBI ID code EMD-5147), and Protein Data Bank, www.pdb.org (PDB ID code PDB-3IYK).
This article contains supporting information online at www.pnas.org/cgi/content/full/0913403107/DCSupplemental.
References
- 1.Gibbs EP, Tabachnick WJ, Holt TJ, Stallknecht DE. U.S. concerns over bluetongue. Science. 2008;320:872. doi: 10.1126/science.320.5878.872a. [DOI] [PubMed] [Google Scholar]
- 2.Wilson AJ, Mellor PS. Bluetongue in Europe: Past, present and future. Philos Trans R Soc Lond B Biol Sci. 2009;364:2669–2681. doi: 10.1098/rstb.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy P, Boyce M, Noad R. Prospects for improved bluetongue vaccines. Nat Rev Microbiol. 2009;7:120–128. doi: 10.1038/nrmicro2052. [DOI] [PubMed] [Google Scholar]
- 4.Urakawa T, Ritter DG, Roy P. Expression of largest RNA segment and synthesis of VP1 protein of bluetongue virus in insect cells by recombinant baculovirus: Association of VP1 protein with RNA polymerase activity. Nucleic Acids Res. 1989;17:7395–7401. doi: 10.1093/nar/17.18.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauber N, et al. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J Virol. 1997;71:7220–7226. doi: 10.1128/jvi.71.10.7220-7226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 7.Roy P. Bluetongue virus: Dissection of the polymerase complex. J Gen Virol. 2008;89:1789–1804. doi: 10.1099/vir.0.2008/002089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 9.Nason EL, et al. Interactions between the inner and outer capsids of bluetongue virus. J Virol. 2004;78:8059–8067. doi: 10.1128/JVI.78.15.8059-8067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya B, Roy P. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J Virol. 2008;82:10600–10612. doi: 10.1128/JVI.01274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forzan M, Wirblich C, Roy P. A capsid protein of nonenveloped Bluetongue virus exhibits membrane fusion activity. Proc Natl Acad Sci USA. 2004;101:2100–2105. doi: 10.1073/pnas.0306448101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 13.Hassan SS, Roy P. Expression and functional characterization of bluetongue virus VP2 protein: Role in cell entry. J Virol. 1999;73:9832–9842. doi: 10.1128/jvi.73.12.9832-9842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormitzer PR, Sun ZY, Wagner G, Harrison SC. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002;21:885–897. doi: 10.1093/emboj/21.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton BT, Crameri GS. The site of bluetongue virus attachment to glycophorins from a number of animal erythrocytes. J Gen Virol. 1989;70(12):3347–3353. doi: 10.1099/0022-1317-70-12-3347. [DOI] [PubMed] [Google Scholar]
- 16.Allen AK, Neuberger A, Sharon N. The purification, composition and specificity of wheat-germ agglutinin. Biochem J. 1973;131:155–162. doi: 10.1042/bj1310155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liemann S, et al. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou ZH, et al. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat Struct Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa A, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure. 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Roy P. Functional mapping of bluetongue virus proteins and their interactions with host proteins during virus replication. Cell Biochem Biophys. 2008;50:143–157. doi: 10.1007/s12013-008-9009-4. [DOI] [PubMed] [Google Scholar]
- 23.Hassan SH, Wirblich C, Forzan M, Roy P. Expression and functional characterization of bluetongue virus VP5 protein: Role in cellular permeabilization. J Virol. 2001;75:8356–8367. doi: 10.1128/JVI.75.18.8356-8367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan K, et al. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weissenhorn W, et al. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 26.Sauter NK, et al. Binding of influenza virus hemagglutinin to analogs of its cell-surface receptor, sialic acid: Analysis by proton nuclear magnetic resonance spectroscopy and x-ray crystallography. Biochemistry. 1992;31:9609–9621. doi: 10.1021/bi00155a013. [DOI] [PubMed] [Google Scholar]
- 27.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 28.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 29.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenouillet E, Barbouche R, Jones IM. Cell entry by enveloped viruses: Redox considerations for HIV and SARS-coronavirus. Antioxid Redox Signal. 2007;9:1009–1034. doi: 10.1089/ars.2007.1639. [DOI] [PubMed] [Google Scholar]
- 31.Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009;83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huismans H, van der Walt NT, Cloete M, Erasmus BJ. Isolation of a capsid protein of bluetongue virus that induces a protective immune response in sheep. Virology. 1987;157:172–179. doi: 10.1016/0042-6822(87)90326-6. [DOI] [PubMed] [Google Scholar]
- 33.Tresnan DB, et al. Analysis of the cell and erythrocyte binding activities of the dimple and canyon regions of the canine parvovirus capsid. Virology. 1995;211:123–132. doi: 10.1006/viro.1995.1385. [DOI] [PubMed] [Google Scholar]
- 34.Basak S, Turner H, Parr S. Identification of a 40- to 42-kDa attachment polypeptide for canine parvovirus in A72 cells. Virology. 1994;205:207–16. doi: 10.1006/viro.1994.1614. [DOI] [PubMed] [Google Scholar]
- 35.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 36.Boyce M, Celma CC, Roy P. Development of reverse genetics systems for bluetongue virus: Recovery of infectious virus from synthetic RNA transcripts. J Virol. 2008;82:8339–8348. doi: 10.1128/JVI.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 38.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semi-automated software for high resolution single particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 39.Liang Y, Ke EY, Zhou ZH. IMIRS: A high-resolution 3D reconstruction package integrated with a relational image database. J Struct Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 40.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci USA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.