Abstract
The lamellipodium of migrating animal cells protrudes by directed polymerization of a branched actin network. The underlying mechanisms of filament growth, branching, and capping can be studied in in vitro assays. However, conflicting results have been reported for the force–velocity relation of such actin networks, namely both convex and concave shapes as well as history dependencies. Here we model branching as a reaction that is independent of the number of existing filaments, in contrast to capping, which is assumed to be proportional to the number of existing filaments. Using both stochastic network simulations and deterministic rate equations, we show that such a description naturally leads to the stability of two qualitatively different stationary states of the system, namely a ± 35° and a +70/0/-70° orientation pattern. Changes in network growth velocity induce a transition between these two patterns. For sufficiently different protrusion efficiency of the two network architectures, this leads to hysteresis in the growth velocity of actin networks under force. Dependent on the history of the system, convex and concave regimes are obtained for the force–velocity relation. Thus a simple generic model can explain the experimentally observed anomalies, with far reaching consequences for cell migration.
Keywords: cytoskeleton, cell motility, stochastic simulations, rate equations
Cell migration of animal cells relies on the polymerization of an actin network, the so-called lamellipodium, pushing the advancing edge forward (1). Studies with many different cell types, including fibroblasts, epithelial cells, growth cones, and keratocytes, have shown that the underlying molecular mechanisms are well preserved (2). Indeed these mechanisms are sufficiently universal that they can be used to propel pathogens like Listeria in different host cells and in in vitro assays (3). For the same reason, it has been possible to harness the power of growing actin networks for the propulsion of plastic beads (4) and lipid vesicles (5, 6). A quantitative understanding of the generic aspects of growing actin networks thus appears possible and highly desirable (7).
By letting the actin network grow against a deformable object like a micropipette (8) or the cantilever of an atomic force microscope (AFM) (9), it is possible to measure force–velocity relations for growing actin networks. However, results from different experiments are conflicting. On the one hand, a convex decline was measured (8, 10), while on the other hand, a load-independent phase (11) often followed by a concave velocity decrease near the stall force was observed (9, 12). A particularly remarkable feature that was found in the context of one of these experiments was that the network growth velocity at a given force can be altered by the loading history of the network (9). Although in principle these discrepancies might be attributed to technical differences in the experiments (13), here we show theoretically that a simple generic mechanism exists which can explain most of the observed anomalies.
Motivated by the universal aspect of the underlying mechanisms, growing actin networks have been subject to a large body of modeling work (reviewed in refs. 13–15). The standard model for growing actin networks is the dendritic nucleation/array treadmilling model describing the balance between polymerization at the advancing side and depolymerization at the retracting side (16). The advancing and retracting sides correspond to the barbed and pointed ends of the polar actin filaments, respectively. Single filaments grow with a typical velocity of up to 1 μm/s. Branching occurs only in a very narrow region close to the leading edge, which is defined by the presence of the activated branching agent Arp2/3. This protein complex is able to nucleate a new filament on an existing one with the emerging barbed end in a direction around 70° from the original one. After a typical time in the subsecond range, polymerization at the barbed ends is terminated by a capping protein. Because the lamellipodium of migrating cells is very flat, the interplay of growth, branching, and capping can theoretically be studied in two dimensions.
A first theoretical model based on these rules already suggested that two different orientation patterns might exist (17), namely, a ± 35° pattern and a +70/0/-70° pattern. However, guided by an apparent dominance of the ± 35° pattern, as well as by corresponding experimental observations with electron microscopy (18), it was concluded that only the ± 35° pattern is physiologically relevant. Also the exact mechanism for the establishment of a stationary state was left open. This problem was solved by the autocatalytic model by Carlsson (19). Here filament creation and annihilation close to the leading edge are balanced and maintain a stationary system at a constant filament number proportional to the applied force on the network. Later stochastic network simulations confirmed that a first-order rate for branching leads to a unique steady state of the network dominated by a ± 35° pattern (20). However, these models cannot explain the anomalies observed for the force–velocity relation. Moreover, it has been recently pointed out that the orientation patterns observed in electron microscopy are more variable than formerly appreciated (21). In addition, biochemical evidence suggests the availability of activated branching agents rather than the availability of actin filaments is the rate-limiting factor for branching (22). This implies that branching should be modeled as a zero-order rather than as a first-order process in the number of actin filaments.
Extending earlier work by Maly and Borisy (17) and Carlsson (19), here we view this assumption as a plausible hypothesis and explore the consequences of modeling branching as a zero-order process. Complementing stochastic simulations with an analytical mean field theory, we show that in this case, both ± 35° and +70/0/-70° patterns are stable and that transitions between the two can be triggered by changes in the protrusion velocity of the bulk network. In combination with a network protrusion efficiency that is sufficiently distinct between the two patterns, this allows us to explain the experimentally observed anomalies regarding force–velocity dependencies and hysteresis effects as a consequence of the competition between the two different orientation patterns.
Model
Stochastic Network Simulations.
We have implemented two-dimensional stochastic network simulations for growing actin networks according to a simplified version of the dendritic nucleation model (16, 23). We are using periodic boundaries in the horizontal direction and let the network grow in the vertical direction. To speed up the simulations, filament growth is treated deterministically, i.e., each filament barbed end grows with velocity vfil. That means each barbed end elongates by one unit length increment δfil per unit time. Close to optimal conditions, single filaments grow approximately every 3 ms by 3 nm (22) (the SI Text has details about the physical values of the model parameters). Stochastic capping and branching events are simulated within a branching region of constant width dbr = 2δfil close to the leading edge of the network, using a Gillespie-like procedure (24). New filaments are able to branch from barbed ends of existing filaments. Adopting earlier used values, the branching angle relative to the mother filament is chosen randomly from a Gaussian distribution with mean 70° and standard deviation 5°. The side of the mother filament from which the new filament grows is selected randomly. As the activation of Arp2/3 by WASp/Scar proteins has been suggested to be the limiting step in generating new filament branches (22), the branching reaction is assumed to be of zero order. This assumption has been used before in analytical treatments (19) and network simulations (25), but its consequences have not yet been fully explored. As usual, capping is assumed to be of first order. As we disregard uncapping reactions, capped filaments will eventually be outgrown by the bulk network and thus leave the simulation box. We do not model their further fate, which in practice might be remodeling into a lamellum or direct depolymerization by actin depolymerization factors/cofilin proteins.
Because capping is assumed to be a first-order process, in our simulations the capping probability per unit time is adjusted by the new filament number in the branching region after each reaction. In contrast, branching is assumed to be a zero-order process and therefore the branching probability remains constant during the simulations. As an important consequence of this treatment, the steady-state growth velocity of the network vnw is not uniquely defined. However, this velocity is a crucial aspect of the system as it determines up to which orientation filaments are able to keep up with the bulk network. Without any additional constraints, only filaments pointing in the forward direction would persist. In our simulations, we therefore discriminated between two populations of filaments at the leading edge. The first population is defined by the assumption that there exists a fixed number Nfront of topmost filaments that are stalled by the obstacle. Those filaments neither grow nor form branches nor get capped. The physical mechanisms underlying stalling might be very different in different systems of interest, being appropriately modeled, e.g., by a ratchet or end-tracking motor model (13, 26). Here they are represented in an effective manner by the model parameter Nfront. The identification of the Nfront leading filaments is updated after each simulation step. The second population are the filaments in the branching region which can grow, branch, or get capped as described above. Our simulations revealed that, for a constant number Nfront of stalled filaments, the networks grow into a steady state with constant network growth velocity vnw and constant total number of filaments. By adjusting Nfront, we therefore can simulate growing networks with the protrusion velocity vnw varying over a large range.
Deterministic Rate Equation Approach.
We also developed a deterministic rate equation approach to network growth. Following earlier approaches of this kind (17, 19), we introduce a distribution function N(θ,t) for the number of uncapped barbed ends in the branching region pointing in direction θ at time t. In this region, the filaments are able to branch and get capped. In addition, they might leave the branching region if outgrown by the bulk network. The rate equation therefore reads
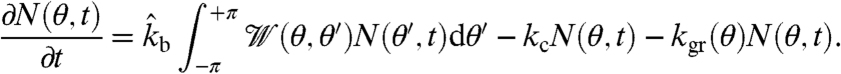 |
[1] |
The three terms on the right-hand side represent branching, capping, and outgrowth. As in earlier work, capping is assumed to be a first-order reaction with a constant rate kc. In the branching term, 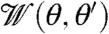 is a branching angle dependent weighting factor distribution, which we assume to be a Gaussian around an absolute angle difference of 70° with standard deviation 5° between the branching mother and daughter filament barbed ends at θ′ and θ, respectively. Because here we assume branching to be a zero-order reaction, the branching rate
is a branching angle dependent weighting factor distribution, which we assume to be a Gaussian around an absolute angle difference of 70° with standard deviation 5° between the branching mother and daughter filament barbed ends at θ′ and θ, respectively. Because here we assume branching to be a zero-order reaction, the branching rate 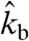 is normalized with regard to the total number of new filament ends:
is normalized with regard to the total number of new filament ends:
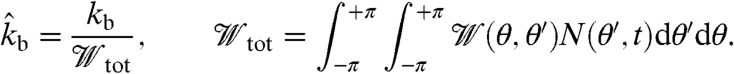 |
[2] |
This definition makes the central model Eq. 1 very different from similar equations studied before in refs. 17 and 19.
Like in the network simulations, we again assume that network growth velocity is fixed at a certain value. In general, this value has to be between zero and vfil. Due to the competition between single filament growth and network growth, a critical orientation angle θc = arccos(vnw/vfil) exists above which the single filament cannot follow the network anymore and therefore is outgrown by the bulk. Hence, for a given filament orientation θ, we write the rate of outgrowth as a function of the network growth velocity:
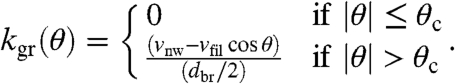 |
[3] |
The factor of 2 arises because we assume that new filaments are generated on average in the middle of the growing region. Because we use dbr = 2δfil, new filaments are assumed to appear at a distance δfil from the leading edge. Taken together, Eq. 3 states that the rate of outgrowth is zero up to the critical angle and then increases up to a maximal value at |θ| = 180°.
In order to analyze Eq. 1, we convert the continuous distribution into a discrete one by binning the angles around specific values 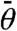 . In principle, this can be done numerically to high precision by choosing a small bin width and solving the self-consistent equations by an iterative procedure. As an instructive alternative, we also used the binning approach to arrive at an analytical solution of our model. We first assume that branching events from mother filaments with an orientation angle |θ| ≳ 90° can be neglected as these filaments grow out of the branching region sufficiently quickly. We next choose the size of the angle bins equal to half the branching angle and the branching angle itself to be 70°. Then we arrive at a system of five coupled ordinary differential equations, namely the reaction equations for the
. In principle, this can be done numerically to high precision by choosing a small bin width and solving the self-consistent equations by an iterative procedure. As an instructive alternative, we also used the binning approach to arrive at an analytical solution of our model. We first assume that branching events from mother filaments with an orientation angle |θ| ≳ 90° can be neglected as these filaments grow out of the branching region sufficiently quickly. We next choose the size of the angle bins equal to half the branching angle and the branching angle itself to be 70°. Then we arrive at a system of five coupled ordinary differential equations, namely the reaction equations for the 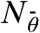 with
with 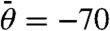 , -35, 0, +35, +70°. This system of equations is given in the SI Text and is termed the reduced rate equation approach below.
, -35, 0, +35, +70°. This system of equations is given in the SI Text and is termed the reduced rate equation approach below.
Results
Two Competing Network Architectures.
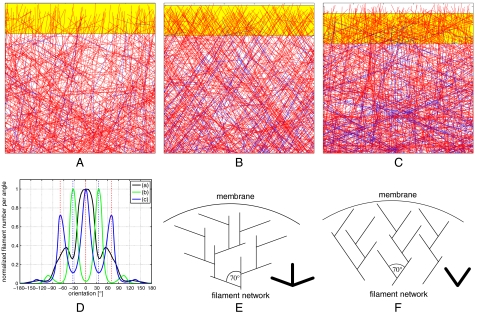
We first analyzed the results from the stochastic network simulations. Fig. 1 A–C shows representative snapshots of the network organization in steady state with decreasing protrusion velocity vnw, i.e., increasing Nfront. The total number of filaments in the simulation box was held approximately constant by adjusting the branching probability for each of the three runs. As shown in Fig. 1D, our simulations revealed that the filament orientation distribution changes strongly as a function of network growth velocity. For fast and slow growing networks, a dominant +70/0/-70° distribution was found, whereas for a network of intermediate velocity, a ± 35° pattern emerged. In Fig. 1 E and F, we show schematic representations of the two different orientation patterns. In the following, we will symbolize the two competing network orientation patterns by the symbols shown at the lower right of these subfigures. A movie is provided in SI Text which shows network organization changes in the computer simulations as a function of protrusion velocity.
Figure 1.
Stochastic network simulations. (A, B, C) Snapshots of the steady-state network organization. Growing and capped filaments are shown in red and blue, respectively. The yellow region at the top corresponds to the branching region, where capping and branching reactions occur. The network velocity vnw is adjusted by assuming that a constant number of filaments at the top Nfront is obstructed by the obstacle and not able to grow, branch, or get capped. The larger Nfront, the lower the yellow region and the smaller vnw. In these simulations, the capping probability was chosen constant at 0.05 per filament, whereas the integral branching rate pb was adjusted for an approximately constant filament density within the simulation box. All uncapped barbed ends deterministically grow with velocity vfil. (A) pb = 30, Nfront = 2, vnw/vfil ≃ 0.95. (B) pb = 22, Nfront = 20, vnw/vfil ≃ 0.76. (V) pb = 9, Nfront = 140, vnw/vfil ≃ 0.30. (D) Normalized filament orientation distributions for the three situations. (E) Schematic representation of the dominant +70/0/-70 distribution corresponding to the networks in A and C. (F) Competing distribution with peaks at ± 35 corresponding to the network in B.
We next performed a linear stability analysis of the reduced rate equation approach and found that this model has two physically meaningful steady-state solutions which are given explicitly in the SI Text and which correspond exactly to the two different network architectures revealed in our stochastic simulations. The conclusion that exactly two competing orientation patterns exist is also confirmed by a numerical analysis of Eq. 1 based on 360 angle bins. Therefore the emergence of two competing orientation patterns is a robust property of both the stochastic network simulations and the rate equation approach.
Phase Diagram for the Competing Orientation Patterns.
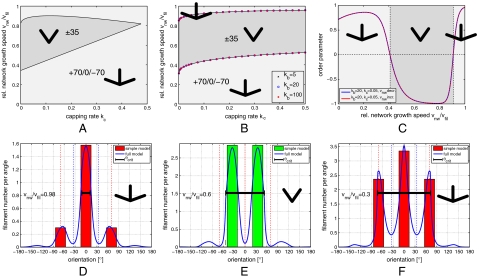
Our rate equation approach allows us to investigate network growth in a systematic manner for all parameter values. Again we use the linear stability analysis of the reduced rate equation approach (details are given in SI Text). Our equations show that the stability of the two steady states is independent of the branching rate kb. In the stationary state, this rate only determines the filament density in the network as the filament number in each angle bin is proportional to the branching rate. Thus there are only two relevant parameters left, namely, network velocity vnw and capping rate kc. For each possible parameter combination, we find that the two fixed points always correspond to one attracting node and one repelling saddle point. Thus we can divide the two-dimensional phase space into two regions, one in which the ± 35 steady state is stable and one in which the +70/0/-70 steady state is stable. The resulting phase diagram is shown in Fig. 2A and predicts that the ± 35 pattern only occurs at intermediate network velocities and small capping rates.
Figure 2.
Phase diagrams resulting from the rate equation approach. The dark and bright gray regions correspond to stability of the ± 35 and +70/0/-70 orientation patterns, respectively. (A) Analytical results from a linear stability analysis of the reduced model. (B) Numerical solution of the full model with three different branching rates kb = {5,20,100}. (C) Evolution of the order parameter with changing network velocity vnw from the numerical solution of the full model. The network behavior for increasing (red) and decreasing (blue) velocity are the same, thus the system is always in a stationary regime. (D, E, F) Comparison of the steady states of the analytically solved reduced model (bars) and the numerically solved full model (curves) for kb = 20 and kc = 0.05. Network bulk velocity vnw is decreasing from D to F: (D) vnw/vfil = 0.98, (E) vnw/vfil = 0.6, and (F) vnw/vfil = 0.3. The vertical red and blue dotted lines mark the prominent angles +70/0/-70 and ± 35, respectively. The horizontal solid black line marks the orientation range |θ| ≤ θc in which single filaments are not outgrown by the bulk network.
In order to compare the analytical results of the reduced rate equation model to the full model Eq. 1, we used numerical iteration with 360 angle bins. In addition, we then used three different values for the branching rate kb. The result is shown in Fig. 2B and proves that indeed, in regard to the kind of resulting orientation pattern, the exact value of the branching rate is irrelevant. A comparison with Fig. 2A shows that, in the reduced rate equation model, the ± 35° region of stability is eliminated for large capping rate kc. This results from the assumption that branching does not occur from mother filaments with an orientation angle larger than 87.5°. This restricting assumption is required to arrive at an analytical treatment, but is not shared by the full rate equation model or the stochastic network growth model, which predict that the stability region for the ± 35° pattern depends only weakly on the capping rate (Fig. 2B and SI Text).
In order to compare the two versions of the rate equation approach in more detail, in Fig. 2D–F, we compare the steady-state filament orientation distribution for the reduced and the full rate equation approach (for a reasonable small capping rate of 0.05 per unit time). From Fig. 2D–F, the network bulk velocity is reduced and the stationary distribution switches from the +70/0/-70 state in the fast growth phase (Fig. 2D) through the ± 35 orientation pattern in the medium growth phase (Fig. 2E) back to another +70/0/-70 distribution in the slow growth phase (Fig. 2F). The agreement between the two approaches is excellent. In each case shown, the orientation range in which growth is possible (that is up to the critical angle θc) is indicated by a horizontal black line. For very fast network growth, the critical angle θc is close to zero and filaments with larger orientations are suppressed. So the dominant orientation will be close to zero with branching angles around ± 70°. Once the network growth velocity reduces to a point where the critical angle is sufficiently large to prevent filaments at ± 35° from growing out of the network, the ± 35 solution dominates and the system changes to the medium growth phase. As the growth velocity is further reduced, the critical angle increases above 70° and the +70/0/-70 pattern dominates again.
In order to simplify the identification of the two different orientation patterns, we define as an appropriate order parameter the relative difference of filaments in the angle bin around 0° to the one around 35°,
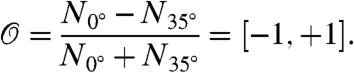 |
[4] |
For a perfect +70/0/-70 distribution, this parameter will approach +1, whereas for the competing ± 35 pattern, it will approach -1. Fig. 2C shows the evolution of the order parameter as a function of the bulk network velocity. The order parameter is shown both for stepwise decreasing and stepwise increasing velocities, showing that the network can adapt sufficiently fast to attain the steady-state values.
Force-Velocity Relation for Growing Actin Networks.
Until now, we have been concerned with the structural properties of growing actin networks as a function of network growth velocity. In order to address the relation between force and velocity, we now ask which network growth velocity vnw results for a given external force Fext on the network. For this purpose, we have to extend our growth model by a mechanical component. For a given force and a given network orientation distribution, we can determine the angle θmax up to which load is carried by the network by summing over the contributions from the individual filaments:
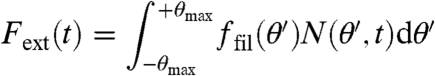 |
[5] |
where ffil(θ) is the force carried by a single filament with orientation θ. The growth of differently oriented single filaments under load depends on the details of the system of interest and different models have been suggested in this context, including a variety of ratchet models and the end-tracking motor model (13, 26). Focusing on mechanical aspects, here we assume that each single filament behaves like a Hookean spring, thus it is able to carry a load
| [6] |
with an indentation length x orthogonal to the leading edge, which is assumed to be the same for all filaments. This treatment had to be replaced by more detailed mechanical models for specific situations of interest and also for obstacles with curvature. The orientation-dependent effective spring constant κ(θ) used here represents bending of an elastic beam for sufficiently large angles (27). Due to the fast decay with increasing orientation angle, it leads to a strong difference in the ability of the two competing network architectures to grow forward while carrying a certain level of load. At small angles, the expression in Eq. 6 is not valid and compression is expected to dominate, thus we assume filaments with an orientation angle smaller than 10° to carry a constant maximal force 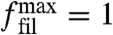 .
.
In steady state, the angle θmax has to correspond to the critical angle θc of the growing network, which in turn defines network growth velocity. In this way, network growth velocity is determined by the given external force
| [7] |
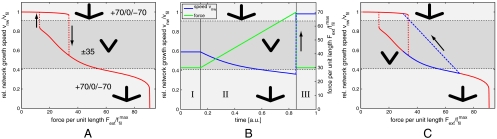
For a given external force, we now can solve the coupled Eqs. 1, 5, and 7 numerically by iterating the orientation distribution with 360 angle bins until a steady state is achieved. Fig. 3A shows a force–velocity relation obtained in this way for typical parameter values. The external force acting on the network Fext is given relative to 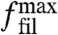 . For zero force, the network is prepared in its fast growth state with the +70/0/-70 orientation pattern. As force increases, the velocity only slowly decreases, because there are sufficiently many filaments almost perpendicular to the leading edge to carry the load. Yet the critical angle continuously grows because increasing load has to be accommodated. If the force passes a certain threshold at around
. For zero force, the network is prepared in its fast growth state with the +70/0/-70 orientation pattern. As force increases, the velocity only slowly decreases, because there are sufficiently many filaments almost perpendicular to the leading edge to carry the load. Yet the critical angle continuously grows because increasing load has to be accommodated. If the force passes a certain threshold at around 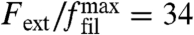 , the +70/0/-70 network is not able to maintain the high velocity anymore and an abrupt change occurs to the ± 35 network, which, due to the single filament force relation Eq. 6, can carry this amount of load only at a relatively low velocity with high filament density. As load is increased further, the critical angle grows and the network gradually reorganizes into the +70/0/-70 pattern. In contrast to the first transition, this one goes into a phase which is especially suited to carry load according to the assumed single filament force relation Eq. 6, thus an instability does not occur and the force–velocity relation is relatively flat. As load increases even further, the orientation distribution starts to broaden, compare Fig. 2C, leading to a decreasing number of filaments at small orientation angles. At this point, the velocity of the network rapidly drops with a concave shape.
, the +70/0/-70 network is not able to maintain the high velocity anymore and an abrupt change occurs to the ± 35 network, which, due to the single filament force relation Eq. 6, can carry this amount of load only at a relatively low velocity with high filament density. As load is increased further, the critical angle grows and the network gradually reorganizes into the +70/0/-70 pattern. In contrast to the first transition, this one goes into a phase which is especially suited to carry load according to the assumed single filament force relation Eq. 6, thus an instability does not occur and the force–velocity relation is relatively flat. As load increases even further, the orientation distribution starts to broaden, compare Fig. 2C, leading to a decreasing number of filaments at small orientation angles. At this point, the velocity of the network rapidly drops with a concave shape.
Figure 3.
(A) Predicted force-velocity relation of an actin network protruding against a changing external force for kc = 0.05 and kb = 20. The most prominent feature is the hysteresis cycle involving the fast and medium growth phases. Abrupt transitions are shown as dashed lines and their directions are indicated by arrows. The full force–velocity curve contains regions with convex, force insensitive, and concave regimes. (B and C) Explanation of the hysteresis experiment (9). (B) The green line is a prescribed change in external force. The blue line is the resulting network velocity. In I, the initial velocity as well as the force are constant. In II, the force is linearly increased to prepare the system in the slow growth phase and the +70/0/-70 orientation pattern. After a rapid decrease in force to its original value in III, the system does not reorganize in the medium growth phase, but rather attains the steady-state velocity in the fast growth phase, which is much higher than the initial one. (C) The same results shown in force–velocity space (blue). The dashed blue line corresponds to the jump in the external force. For comparison, the hysteresis curve of A is shown in red.
Fig. 3A also shows that, if we initially prepare the network in the intermediate growth phase and then decrease force, the transition back to the fast growth phase occurs at a smaller value for forces, thus resulting in a hysteresis loop. The size of the predicted hysteresis loop changes with model parameters, but in general it exists over a large range of model parameters, as revealed by a sensitivity analysis described in the SI Text. In particular, the standard deviation of the branching angle can be increased to a value around 15° before the hysteresis loop vanishes. The existence of a hysteresis loop is an immediate consequence of the competition between the two different orientation patterns and can be used to explain the experimentally observed history dependance (9) as demonstrated in Fig. 3 B and C. We start by initiating the network in the medium growth phase within the hysteresis cycle. For a constant force of 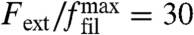 , the network protrudes with a constant velocity vnw/vfil ≃ 0.6 in this phase. Next the force is linearly increased (green line in Fig. 3B) and so the transition to the slow growth phase is initiated (blue line in Fig. 3B). At some point, the force has grown large enough and the system will eventually develop the final +70/0/-70 orientation pattern characteristic for this phase. Now the force is instantaneously reduced to its initial value
, the network protrudes with a constant velocity vnw/vfil ≃ 0.6 in this phase. Next the force is linearly increased (green line in Fig. 3B) and so the transition to the slow growth phase is initiated (blue line in Fig. 3B). At some point, the force has grown large enough and the system will eventually develop the final +70/0/-70 orientation pattern characteristic for this phase. Now the force is instantaneously reduced to its initial value 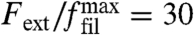 . Instead of going back to the medium growth phase, the system now jumps right into the fast growth phase, because this one is also stable at this force level and in addition is structurally similar to the existing slow growth phase. In Fig. 3C, this scenario is plotted in force–velocity space.
. Instead of going back to the medium growth phase, the system now jumps right into the fast growth phase, because this one is also stable at this force level and in addition is structurally similar to the existing slow growth phase. In Fig. 3C, this scenario is plotted in force–velocity space.
The effects demonstrated here are generic and can be obtained for different realizations of the force–velocity relation of individual filaments, given that the advantage in performance of the two +70/0/-70 dominated phases compared to the competing ± 35 regime is sufficiently large. However, for filaments elongating as a Brownian ratchet (27), this is not the case and the difference in performance of the two competing orientation patterns is too weak to show significant hysteresis effects (shown in SI Text).
Discussion
We have shown that in growing actin networks a competition between two self-organized filament orientation patterns occurs if branching is assumed to be a zero-order reaction. This means that only a certain number of branching points can be established in a given region at the leading edge, independent of the number of actin filaments. If Arp2/3 is activated before filament binding, this would imply that its activation time is larger than all other time scales involved in actin filament branching (22). Recent evidence suggests that Arp2/3 might be activated after filament binding (28). In this case, successful branching might be limited by the availability of nucleation promoting factors localized to the leading edge. If this process takes longer than the nascent branching points take to grow away from the leading edge, then branching is effectively also a zero-order process.
Our theoretical analysis shows that, for large and small network growth velocities, the stable steady state is a +70/0/-70 pattern, whereas for intermediate network growth velocity, the stable steady state is a ± 35 pattern. We then showed that, when combined with a simple generic model for the ability to carry mechanical work, the competition between the two orientation patterns leads to unusual force–velocity relations and history-dependent behavior. In our model, these results follow from the fact that the two patterns have different efficiency in generating protrusion under the constraint of carrying a certain level of load.
With the proposed model, different puzzling experimental observations find a simple explanation. The orientation analysis of electron micrographs from lamellipodia of migrating cells yielded differently orientated patterns in the actin network dependent on the phase of cell migration, with more filaments being orientated with a larger angle after reduction of protrusion velocity during phases of pausing or retraction (21). In our model, this corresponds to the transition from ± 35 to +70/0/-70 as velocity is decreased, (see Fig. 2 E and F). For the force–velocity relation, regions of load-independent network protrusion (11) and rapid concave growth velocity decay near the stall force have been measured (9, 12). These observations might correspond to the high force part of the force–velocity curve of Fig. 3A. One experiment reported a sharp drop in velocity at very small loading force (12). This observation might correspond to the abrupt transition in the low force part of the force–velocity curve of Fig. 3A. Other experiments reported a convex dependence of network growth velocity with force (8, 10), which might correspond to the intermediate force part of the force-velocity curve of Fig. 3A when initially prepared in the medium growth phase. Most importantly, the predicted competition between two different orientation patterns provides a generic mechanism to explain the experimentally observed hysteresis loop (9) as demonstrated in Fig. 3 B and C.
In our model, velocity is not determined by the growing actin network, but appears as a boundary condition. This agrees with the experimental observation that in migrating cells most relevant processes are localized to the leading edge, including Arp2/3-activation by WASp/Scar proteins and assembly of adhesion complexes. In general, the exact details that determine velocity might be very different in different systems, depending on how single barbed filament ends interact with the leading edge. For example, different ratchet mechanisms have been suggested to allow incorporation of new monomers into the growing filament (27). At the leading edge of migrating cells, end-tracking motors might also play an essential role (26, 29). Our model suggests that, irrespective of the details of these processes, the main determinant for the structure of the growing actin network is the resulting velocity.
In order to test our predictions in more detail, different lines of experiments seem promising. First, one should systematically correlate the orientation distribution of actin networks from electron micrographs with the network growth velocity, both for reconstituted assays and for experiments with migrating cells. Second, the full hysteresis cycle should be measured experimentally, for example in an AFM setup. Third, the role of branching and capping rates can be investigated by biochemical interference with reconstitution assays and live cells. For example, a reduction of the activation time of Arp2/3 might change the zero-order process into a first-order process and thus stabilize the ± 35 pattern. The insight gained in this way would not only strongly increase our understanding of a fundamental biological process, it also would allow us to develop new protocols to control the growth of actin networks outside the cell, thus opening up exciting new perspectives for applications of this fascinating biomaterial.
Supplementary Material
Acknowledgments.
This work was supported by the research unit for systems biology Viroquant and the cluster of excellence CellNetworks at the University of Heidelberg.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913730107/DCSupplemental.
References
- 1.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84:359–370. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 4.Cameron LA, Footer MJ, Van Oudenaarden A, Theriot JA. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci USA. 1999;96:4908–4913. doi: 10.1073/pnas.96.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Upadhyaya A, Chabot JR, Andreeva A, Samadani A, van Oudenaarden A. Probing polymerization forces by using actin-propelled lipid vesicles. Proc Natl Acad Sci USA. 2003;100:4521–4526. doi: 10.1073/pnas.0837027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giardini PA, Fletcher DA, Theriot JA. Compression forces generated by actin comet tails on lipid vesicles. Proc Natl Acad Sci USA. 2003;100:6493–6498. doi: 10.1073/pnas.1031670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcy Y, Prost J, Carlier MF, Sykes C. Forces generated during actin-based propulsion: A direct measurement by micromanipulation. Proc Natl Acad Sci USA. 2004;101:5992–5997. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nat Cell Biol. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 10.McGrath JL, et al. The force-velocity relationship for the actin-based motility of Listeria monocytogenes. Curr Biol. 2003;13:329–332. doi: 10.1016/s0960-9822(03)00051-4. [DOI] [PubMed] [Google Scholar]
- 11.Wiesner S, et al. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol. 2003;160:387–398. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mogilner A. On the edge: Modeling protrusion. Curr Opin Cell Biol. 2006;18:32–39. doi: 10.1016/j.ceb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Mogilner A. Mathematics of cell motility: Have we got its number? J Math Biol. 2009;58:105–134. doi: 10.1007/s00285-008-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard TD, Berro J. Mathematical models and simulations of cellular processes based on actin filaments. J Biol Chem. 2009;284:5433–5437. doi: 10.1074/jbc.R800043200. [DOI] [PubMed] [Google Scholar]
- 16.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 17.Maly IV, Borisy GG. Self-organization of a propulsive actin network as an evolutionary process. Proc Natl Acad Sci USA. 2001;98:11324–11329. doi: 10.1073/pnas.181338798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verkhovsky AB, et al. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol Biol Cell. 2003;14:4667–4675. doi: 10.1091/mbc.E02-10-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlsson AE. Growth velocities of branched actin networks. Biophys J. 2003;84:2907–2918. doi: 10.1016/S0006-3495(03)70018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaus TE, Taylor EW, Borisy GG. Self-organization of actin filament orientation in the dendritic-nucleation/array-treadmilling model. Proc Natl Acad Sci USA. 2007;104:7086–7091. doi: 10.1073/pnas.0701943104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koestler SA, Auinger S, Vinzenz M, Rottner K, Small JV. Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat Cell Biol. 2008;10:306–313. doi: 10.1038/ncb1692. [DOI] [PubMed] [Google Scholar]
- 22.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanism controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 23.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. 1977;81:2340–2361. [Google Scholar]
- 25.Schaub S, Meister JJ, Verkhovsky AB. Analysis of actin filament network organization in lamellipodia by comparing experimental and simulated images. J Cell Sci. 2007;120:1491–1500. doi: 10.1242/jcs.03379. [DOI] [PubMed] [Google Scholar]
- 26.Dickinson RB. Models for actin polymerization motors. J Math Biol. 2009;58:81–103. doi: 10.1007/s00285-008-0200-4. [DOI] [PubMed] [Google Scholar]
- 27.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283:7135–7144. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson RB, Purich DL. Clamped-filament elongation model for actin-based motors. Biophys J. 2002;82:605–617. doi: 10.1016/S0006-3495(02)75425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.