Abstract
Recent work has demonstrated that the diversity of skin-associated bacterial communities is far higher than previously recognized, with a high degree of interindividual variability in the composition of bacterial communities. Given that skin bacterial communities are personalized, we hypothesized that we could use the residual skin bacteria left on objects for forensic identification, matching the bacteria on the object to the skin-associated bacteria of the individual who touched the object. Here we describe a series of studies de-monstrating the validity of this approach. We show that skin-associated bacteria can be readily recovered from surfaces (including single computer keys and computer mice) and that the structure of these communities can be used to differentiate objects handled by different individuals, even if those objects have been left untouched for up to 2 weeks at room temperature. Furthermore, we demonstrate that we can use a high-throughput pyrosequencing-based ap-proach to quantitatively compare the bacterial communities on objects and skin to match the object to the individual with a high degree of certainty. Although additional work is needed to further establish the utility of this approach, this series of studies introduces a forensics approach that could eventually be used to independently evaluate results obtained using more traditional forensic practices.
Keywords: bacterial forensics, human microbiome, pyrosequencing, skin microbiology, microbial ecology
The human skin surface harbors large numbers of bacteria that can be readily dislodged and transferred to surfaces upon touching, hence the importance of proper hand hygiene by health care practitioners (1, 2). These skin bacteria may persist on touched surfaces for prolonged periods because many are highly resistant to environmental stresses, including moisture, temperature, and UV radiation (3, 4). Therefore, we likely leave a persistent “trail” of skin-associated bacteria on the surfaces and objects that we touch during our daily activities.
Recent work has demonstrated that our skin-associated bacterial communities are surprisingly diverse, with a high degree of interindividual variability in the composition of bacterial communities at a particular skin location (5–9). For example, only 13% of the bacterial phylotypes on the palm surface are shared between any two individuals (8), and a similar level of interpersonal differentiation is observed at other skin locations (5, 9). In addition, skin bacterial communities are relatively stable over time: palm surface bacterial communities recover within hours after hand washing (8); and, on average, interpersonal variation in community composition exceeds temporal variation within people, even when individuals are sampled many months apart (5, 9). Given that individuals appear to harbor personally unique, temporally stable, and transferable skin-associated bacterial communities, we hypothesized that we could use these bacteria as “fingerprints” for forensic identification.
To demonstrate that we can use skin bacteria to link touched surfaces to specific individuals, the following criteria must be met: (i) bacterial DNA recovered from touched surfaces allows for adequate characterization and comparison of bacterial communities; (ii) skin bacterial communities persist on surfaces for days to weeks; and (iii) surfaces that are touched can be effectively linked to individuals by assessing the degree of similarity between the bacterial communities on the object and the skin of the individual who touched the object. To establish these criteria and to demonstrate the potential utility of the approach for forensic identification, we carried out three interrelated studies that combine recent developments in phylogenetic community analyses (10) with high-throughput pyrosequencing methods (11). First, we compared bacterial communities on individual keys of three computer keyboards to the communities found on the fingers of the keyboard owners. Second, we examined the similarity between skin-associated bacterial communities on objects stored at −20 °C (a standard method for storing samples before DNA extraction) versus those objects stored under typical indoor environmental conditions for up to 14 days. Finally, we linked objects to specific individuals by comparing the bacteria on their computer mice against a database containing bacterial community information for more than 250 hand surfaces, including the hand of the owner.
Results and Discussion
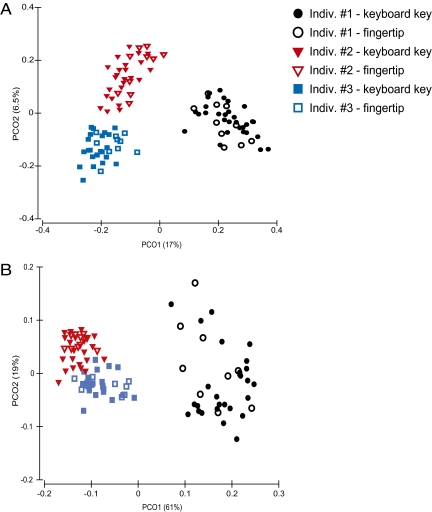
To establish criteria i and iii, we swabbed individual keys from three personal computer keyboards and compared the communities on those keys to the bacterial communities on the fingertips of the key-board owners. We also sampled individual keys from other private and public computer keyboards so that we could quantify the degree of correspondence between the bacterial communities on the owner’s fingers and keyboard versus other keyboards never touched by that person. Bacterial DNA was extracted from the swabs, and bacterial community composition was determined using the barcoded pyrosequencing procedure described previously (8), obtaining an average of over 1,400 bacterial 16S rRNA gene sequences per sample. We found that bacterial communities on the fingertips or keyboard of a given individual are far more similar to each other than to fingertips or keyboards from other individuals (Fig. 1 and Fig. 2). Likewise, the bacterial communities on the fingers of the owner of each keyboard resembled the communities on the owner's keyboard (Fig. 1 and Fig. 2), which suggests that differences in keyboard-associated communities are likely caused by direct transfer of fingertip bacteria. The discrimination between individuals is stronger with the unweighted UniFrac metric than with the weighted metric, suggesting that differences in community membership (rather than community structure) discriminate best among individuals. The patterns evident in Fig. 1 are confirmed by ANOSIM analyses, which demonstrate that each keyboard harbors a distinct bacterial community, the finger-associated bacterial communities are unique to each of the three individuals, and that the interindividual differences in fingertip and keyboard communities exceed the differences between bacterial communities on the fingers and keyboards belonging to a given individual (Table S1). Together these results demonstrate that bacterial DNA can be recovered from relatively small surfaces, that the composition of the keyboard-associated communities are distinct across the three keyboards, and that individuals leave unique bacterial ‘fingerprints’ on their keyboards.
Fig. 1.
Match between bacterial communities on individual keyboards and the fingers of the owners of the keyboards. Principal coordinates plots showing the degree of similarity between bacterial communities on fingertips of the three individuals sampled as part of this study and their respective keyboards. Plots were generated using the pairwise unweighted (A) and weighted (B) UniFrac distances (22, 23), respectively. The UniFrac algorithm uses the degree of phylogenetic overlap between any pair of communities with points that are close together representing samples with similar bacterial communities.
Fig. 2.
Bacterial community distances between keyboard keys and fingertips. Mean pairwise distances between keys from the same keyboard (black bar), between individual's fingertips and their own keyboard keys (hatched bar), and between individual's fingertips and keys from keyboards not belonging to them (gray bar). Average unweighted and weighted UniFrac distances for each individual are shown (A and B, respectively). Lower UniFrac values indicate that the communities are more similar on average. Bars show 95% confidence intervals for the means. Graph demonstrates that the fingertips of an individual harbor bacterial communities more similar to those found on the keys of that individual's keyboard than to those communities found on keyboard keys not touched by the individual.
For the ‘keyboard’ study described above, the keyboards were swabbed 1–2 h after having last been touched. To demonstrate the longer-term temporal stability of skin-associated communities on nonskin surfaces, we conducted a smaller-scale study to assess how bacterial communities may shift in composition after exposure to typical indoor environmental conditions. The skin surface from two individuals was swabbed and the swabs were either frozen immediately at -20 °C or left in open containers on a bench in the laboratory at ≈20 °C. Storage under typical indoor conditions had little to no influence on bacterial community composition, or the ability to resolve differences between the bacterial communities on the skin of the two individuals, even after two weeks (Fig. 3 and Table S2). These results demonstrate the potential utility of this approach for forensic identification given that, under standard indoor conditions, skin-associated bacteria persist on objects with the overall structure and composition of these communities remaining essentially unchanged for days after the object was last handled.
Fig. 3.
Effect of storage conditions on skin-associated bacterial communities collected on dry cotton swabs. (A and B) Principal coordinates plots generated using the unweighted and weighted UniFrac distance matrices, respectively. Samples were stored at either −20 °C or +20 °C with DNA extracted from the swabs after 3 days and 14 days, but storage temperature had minimal effects on bacterial community composition. (C) Relative abundances of the most abundant bacterial taxa after 14 days at either −20 °C or at +20 °C. Classifications are to the genus (gen.), family (fam.), or order level. For each taxon, the phylum or subphylum is also indicated: Actino., Actinobacteria; Bacter., Bacteroidetes; Firmic., Firmicutes. Taxa are classified to the highest taxonomic level to which they could be confidently assigned.
Since the keyboard results summarized in Figs. 1 and 2 indicate that we can use skin-associated bacteria to link an object to its owner, we designed a more targeted study to determine the efficacy of this approach for forensic identification. We wanted to determine whether the bacteria on a personal object are more similar to the bacteria found on the owner's skin than to the general population. We sampled bacteria from nine computer mice (from personal computers) that had not been touched for more than 12 h and from the palms of the mouse owners. We then calculated the phylogenetic distance between the bacterial communities on each mouse and mouse owner's hand, comparing this distance to the distances between the mouse bacterial communities and the communities on 270 hands that had never touched the mouse. These 270 hand bacterial communities came from a database of individuals sampled for various studies conducted over the past 2 years using the same sampling and community analysis tech-nique described above. If the approach were to hold promise as a tool for forensic identification, we would expect the communities on the mice to be more similar to the communities on their owner's hands than to all of the other hands in the database.
In all nine cases, the bacterial community on each mouse was significantly more similar to the community on the owner’s hand than to other hands in the database, regardless of the distance metric used (Fig. 4), indicating that the technique has potential to serve as a robust means of forensic identification. However, just as other forensics techniques have required considerable testing and refinement long after they were initially conceived, further research is required to assess how the accuracy of this technique might compare with more standard, and widely accepted, forensic tools. In particular, it will be important to assess how the accuracy of the approach might be improved by compiling a larger database of hand-associated bacterial communities, obtaining more sequences per sample, collecting multiple specimens per object or hand, developing new distance metrics to improve our ability to resolve differences between communities, or using only a subset of the bacterial community in the analyses (i.e., that portion of the hand-associated bacterial communities that is most personally identifying). Likewise, to further establish the utility of this technique, additional studies will be needed to assess how well it works with objects of different surface materials, objects touched less frequently, or objects that come into contact with multiple skin locations on a given individual.
Fig. 4.
Accuracy of forensic identification using bacterial communities. Phylogenetic distance between the bacterial communities found on the computer mouse (with the nine mice identified with the x axis labels) and the hand swab from the individual that used the mouse (the unfilled symbols) versus the average phylogenetic distance between the bacterial communities on the computer mouse and the 270 other hand swab samples in the database (filled symbols). Error bars represent 95% confidence intervals. Phylogenetic distance measured using either the unweighted or weighted UniFrac algorithm (red squares and blue circles, respectively); the more similar the communities the lower the distance. Note that in nearly all cases the bacterial community on a given mouse is significantly more similar to those on the owner's hand than to the other hands in the database.
Conclusions
The approach described here could provide independent confirmation of forensic results obtained using other methods (e.g., human DNA analysis or fingerprint analysis) and the approach might represent a valuable alternative to these more standard techniques under certain conditions and scenarios. For example, unless there is blood, tissue, semen, or saliva on an object, it is often difficult to obtain sufficient human DNA for forensic identification. However, given the abundance of bacterial cells on the skin surface and on shed epidermal cells (12), it may be easier to recover bacterial DNA than human DNA from touched surfaces (although additional studies are needed to confirm that this is actually true). Furthermore, the technique might be useful for identifying objects from which clear fingerprints cannot be obtained (e.g., fabrics, smudged surfaces, highly textured surfaces).
Together, these studies demonstrate that research on human-associated microbial communities, such as the Human Microbiome Project (13), will not only yield valuable contributions in the fields of microbiology and medicine, but also unexpected and novel applications to other fields and disciplines. Specifically, we have leveraged the recent and surprising discovery that our microbes our highly personalized to initiate the development of a unique forensic approach. The further development of this approach warrants careful consideration by bioethicists seeking to understand the ethical, legal, and social implications of the Human Microbiome Project; even identical twins harbor substantially different microbial communities (14), suggesting that the collective genomes of our microbial symbionts may be more personally identifying than our own human genomes.
Methods
Sample Collection.
For the keyboard study, we swabbed individual keys of three personal computer keyboards (25–30 keys per keyboard) and the skin on the ventral surface of the distal joint of each fingertip of the owner and nearly exclusive user of each keyboard. All three individuals were healthy at the time of sampling, had not taken antibiotics for at least 6 months, and were between 20 and 35 years of age. Two of these individuals shared the same office space. Keyboards and fingertips were swabbed within 10 min of one another, but the keyboards had not been touched for more than 30 min before sampling. To compare the bacterial communities on these keyboards to other miscellaneous keyboards, we swabbed space bar keys from 15 other private and public computer keyboards located on the University of Colorado campus. Skin surfaces and keyboard keys were sampled using autoclaved cotton-tipped swabs premoistened with a sterile solution (8, 15). Swabbing has been shown to be a suitable method for skin sample collection for microbial community analysis (7). The entire exposed surface of each keyboard key was swabbed lightly for 10 s. All swabs were stored at −80 °C for less than 1 week before DNA extraction.
For the “storage” study, we used the swabbing technique described above to sample the right axillary (armpit) skin surface of two healthy adult individuals. This skin surface was chosen because it harbors taxa similar to those found in other skin habitats (9), yet the biomass levels are likely high enough to allow us to get sufficient amounts of bacterial biomass onto all of the replicate swab samples that were collected. The entire skin surface was simultaneously swabbed with 16 moistened swabs per individual, rotating the swabs to ensure homogeneity in the skin area contacted by each swab. Half of these swabs were immediately frozen at −20 °C with the other half left in uncapped 15-mL conical tubes on the laboratory bench. Conditions in the laboratory were typical of indoor environments: the temperature was held at ≈20 °C for the duration of the experiment with fluorescent lighting on for ≈8 h per day. Bacterial DNA was extracted from four replicate swabs per storage condition after either 3 days or 14 days, with the DNA stored at −80 °C before analysis.
For the computer mouse study, we recruited nine healthy adults (four female and five male, all 20–35 years of age) who worked in the same building on the University of Colorado campus. Using the swabbing technique described above, the entire exposed surface of each computer mouse and the palm surface of the individual's dominant hand (the hand typically used to operate the mouse) was swabbed. Care was taken to ensure that the mouse had last been touched by the owner 12 h before the swabbing (the mice remained at room temperature during this period). Palm surfaces were sampled midday and the volunteers were told to follow their typical hand hygiene practices before the sampling. All swabs were stored at −80 °C before DNA extraction. We estimated the accuracy of matching the mouse to the owner of the mouse by measuring the degree of similarity between bacterial communities on each computer mouse to the hands of the mouse's owner and to the hands that had never touched the mouse. We compiled a database of bacterial communities from 270 other hands sampled for other projects (8, 9). The 270 hands bacterial communities included in this database came from both left and right palm surfaces belonging to male and female volunteers in equal proportions that were healthy and between the ages of 18 and 40 years. The palms were sampled and the bacterial communities analyzed using procedures identical to those described here.
For all three studies described above, the individuals were made aware of the nature of the study and gave written informed consent in accordance with the sampling protocol approved by the University of Colorado Human Research Committee (protocol 0109.23).
DNA Extraction and Pyrosequencing.
Genomic DNA was extracted from the swabs using the MO BIO PowerSoil DNA Isolation kit. The cotton tips of frozen swabs were broken off directly into bead tubes to which 60 μL of Solution C1 had been added. Tubes were incubated at 65 °C for 10 min and then shaken horizontally at maximum speed for 2 min using the MO BIO vortex adapter. The remaining steps were performed as directed by the manufacturer.
For each sample, we amplified 16S rRNA genes using the primer set described in Fierer et al. (8) that had been optimized for the phylogenetic analysis of pyrosequencing reads (16). PCR reactions were carried out in triplicate 25-μL reactions with 0.6 μM forward and reverse primers, 3 μL template DNA, and 1× of HotMasterMix (5 PRIME). Thermal cycling consisted of initial denaturation at 94 °C for 3 min followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 50 °C for 30 s, and extension at 72 °C for 90 s, with a final extension of 10 min at 72 °C. Replicate amplicons were pooled and visualized on 0.1% agarose gels using SYBR Safe DNA gel stain in 0.5× TBE (Invitrogen). Amplicons were cleaned using the UltraClean-htp 96-well PCR Clean-up kit (MO BIO).
Amplicon DNA concentrations were measured using the Quant-iT PicoGreen dsDNA reagent and kit (Invitrogen). Following quantitation, cleaned amplicons were combined in equimolar ratios into a single tube. The final pool of DNA was precipitated on ice for 45 min after the addition of 5 M NaCl (0.2 M final concentration) and 2 volumes of ice-cold 100% ethanol. The precipitated DNA was centrifuged at 7,800 × g for 40 min at 4 °C, and the resulting pellet was washed with an equal volume of ice-cold 70% ethanol and centrifuged again at 7,800 × g for 20 min at 4 °C. The supernatant was removed and the pellet was air dried for 10 min at room temperature and then resuspended in nuclease-free water (MO BIO). Pyrosequencing was carried out on a 454 Life Sciences Genome Sequencer FLX instrument (Roche) by the Environmental Genomics Core Facility at the University of South Carolina (Columbia).
Sequence Analyses and Community Comparisons.
Sequences were processed and analyzed following the procedures described previously (8, 11). Sequences were removed from the analysis if they were less than 200 or more than 300 bp in length, had a quality score less than 25, contained ambiguous characters, contained an uncorrectable barcode, or did not contain the primer sequence. Remaining sequences were assigned to samples by examining the 12-nt barcode. Similar sequences were clustered into operational taxonomic units (OTUs) using cd-hit (17) with a minimum coverage of 97% and a minimum identity of 97%. A representative sequence was chosen from each OTU by selecting the longest sequence that had the largest number of hits to other sequences in the OTU. Representative sequences were aligned using NAST (18) and the Greengenes database (19) with a minimum alignment length of 150 and a minimum identity of 75%. The PH Lane mask was used to screen out hypervariable regions after alignment. A phylogenetic tree was inferred using Clearcut (20) with Kimura's two-parameter model. Taxonomy was assigned using the RDP classifier with a minimum support threshold of 60% and the RDP taxonomic nomenclature (21).
For each of the samples included in the three studies described above (including those in the database of 270 palm surfaces used to estimate the accuracy of the computer mouse assignments) we obtained a minimum of 800 quality sequences (range 800–1,500 sequences per sample) with sequences averaging 240 bp in length.
To determine the amount of dissimilarity (distance) between any pair of bacterial communities, we used the UniFrac metric (10, 22, 23). UniFrac distances are based on the fraction of branch length shared between two communities within a phylogenetic tree constructed from the 16S rRNA gene sequences from all communities being compared. A relatively small UniFrac distance implies that two communities are compositionally similar, harboring lineages sharing a common evolutionary history. In unweighted UniFrac, only the presence or absence of lineages is considered. In weighted UniFrac, branch lengths are weighted based on the relative abundances of lineages within communities. We used the analysis of similarities (ANOSIM) (24) function in the program PRIMER (25) to test for differences in community composition among groups of samples.
Supplementary Material
Acknowledgments
This work was funded in part by grants from the National Science Foundation (to N.F.) and grants from the National Institutes of Health, the Crohn's and Colitis Foundation of America, and the Howard Hughes Medical Institute (to R.K.). We thank the volunteers who participated in these studies and members of the Fierer and Knight laboratories for their help with sample collection, data analysis, and manuscript editing. Micah Hamady provided assistance with the analyses of the sequence data.
Footnotes
The authors declare no conflict of interest.
Data deposition: Data have been deposited in the GenBank Short Read Archive (SRA0102034.1).
This article is a PNAS Direct Submission.
See Commentary article on page 6125.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000162107/DCSupplemental.
References
- 1.Jarvis WR. Handwashing—the Semmelweis lesson forgotten? Lancet. 1994;344:1311–1312. doi: 10.1016/s0140-6736(94)90687-4. [DOI] [PubMed] [Google Scholar]
- 2.Pittet D, Allegranzi B, Boyce J, World Health Organization World Alliance for Patient Safety First Global Patient Safety Challenge Core Group of Experts The World Health Organization guidelines on hand hygiene in health care and their consensus recommendations. Infect Control Hosp Epidemiol. 2009;30:611–622. doi: 10.1086/600379. [DOI] [PubMed] [Google Scholar]
- 3.Smith SM, Eng RHK, Padberg FT., Jr Survival of nosocomial pathogenic bacteria at ambient temperature. J Med. 1996;27:293–302. [PubMed] [Google Scholar]
- 4.Brooke JS, Annand JW, Hammer A, Dembkowski K, Shulman ST. Investigation of bacterial pathogens on 70 frequently used environmental surfaces in a large urban U.S. university. J Environ Health. 2009;71:17–22. [PubMed] [Google Scholar]
- 5.Grice EA, et al. NISC Comparative Sequencing Program Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Z, Tseng CH, Pei ZH, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, et al. NISC Comparative Sequencing Program A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamady M, Walker J, Harris J, Gold N, Knight R. Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredricks DN. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 18.DeSantis T, et al. NAST: A multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheneman L, Evans J, Foster JA. Clearcut: A fast implementation of relaxed neighbor joining. Bioinformatics. 2006;22:2823–2824. doi: 10.1093/bioinformatics/btl478. [DOI] [PubMed] [Google Scholar]
- 21.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke K. Non-parametric multivariate analysis of changes in community structure. J Aus Ecol. 1993;18:117–143. [Google Scholar]
- 25.Clarke K, Gorley R. Plymouth, UK: PRIMER-E Ltd.; 2006. PRIMER. ver. 6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.