Abstract
Chemical biology aims for a perfect control of protein complexes in time and space by their site-specific labeling, manipulation, and structured organization. Here we developed a self-inactivated, lock-and-key recognition element whose binding to His-tagged proteins can be triggered by light from zero to nanomolar affinity. Activation is achieved by photocleavage of a tethered intramolecular ligand arming a multivalent chelator head for high-affinity protein interaction. We demonstrate site-specific, stable, and reversible binding in solution as well as at interfaces controlled by light with high temporal and spatial resolution. Multiplexed organization of protein complexes is realized by an iterative in situ writing and binding process via laser scanning microscopy. This light-triggered molecular recognition should allow for a spatiotemporal control of protein-protein interactions and cellular processes by light-triggered protein clustering.
Keywords: biofunctionalized interface, light-triggerd chemical biology, molecular recognition, protein interaction, self-assembly by light
A key challenge in life science is the qualitative and quantitative analysis of protein function and protein-protein interactions up to dynamic protein networks involved in cellular logistics. This way of looking at protein functions in cells has motivated significant efforts in the field of protein labeling combined with high-resolution optical microscopy (1, 2). Genetically encoded in vivo labeling of biological complexes via autofluorescent proteins (AFPs), including photoactivatable and photoswitchable AFPs, which change their spectral properties upon irradiation, quickly became a popular and very powerful tool (3–5). A complementary strategy is the labeling of recombinant proteins with chemical probes. Various small sequence tags have been developed, allowing for covalent as well as noncovalent protein modifications (6, 7). Major advantages of synthetic fluorophors over AFPs are their high quantum yield, small size, high photostability, and the wide spectrum of existing colors.
One of the missing key features is the possibility to instantaneously trigger protein interactions via small molecules by external stimuli. As examples, changes in temperature, pH, small molecule concentration, and light intensity could allow for temporal control of peptide, protein, or DNA modifications in solution as well as at interfaces (8–14). Among these, light is a versatile trigger, as it induces photoreactions not only instantaneously but also with high spatial resolution. Light is compatible with many applications since most cells and even model animals, such as zebrafish or Caenorhabditis elegans, are virtually transparent for wavelengths above 350 nm. From the usage of light, strategies for dose-regulated and orthogonal activation immediately come to one’s mind. Triggering the activity of biochemical compounds by light has been previously demonstrated (15–17). Exciting developments have been reported exploiting channel rhodopsins or phytochromes for the perturbation or neuronal and signaling networks (18, 19). However, it is still a challenge to change the affinity between two biomolecules from low to high by a light pulse. Caged biotin has been used for the light induced interaction with (strept)avidin (20–22). Another recent example is the synthesis of caged O6-benzylguanine substrates for the labeling, dimerization, and immobilization of SNAP®(O-6-alkylguanine DNA alkyltransferase)-tagged proteins (23). However, both approaches rely on large protein domains (> 15 kDa) and do not allow for a noncovalent, reversible protein modification. Reversibility is desirable, for example, if in a single-molecule analysis the fluorescent label needs to be “refreshed” after photobleaching (24).
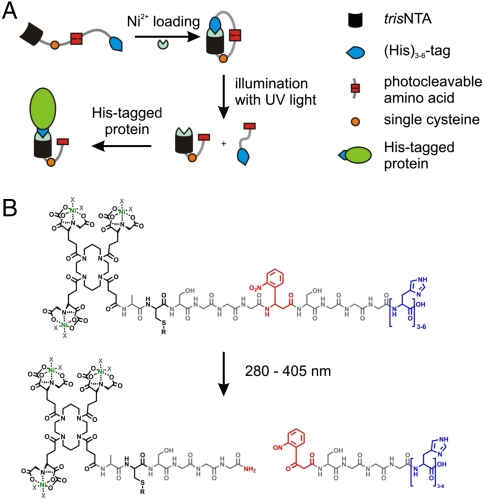
Here we present a unique, generic approach changing the binding affinity by light from almost zero to high (nanomolar) over 6–7 orders of magnitude. The concept is implemented with a small-molecule, lock-and-key element based on a multivalent chelator head (MCH), which is self-inactivated by a tethered intramolecular ligand. Activation is achieved by a photoreaction separating the intramolecular ligand from the MCH. We chose the well-characterized and widely used interaction between His-tagged proteins and a trivalent N-nitrilotriacetic acid chelator head (tris-NTA). Nickel-loaded tris-NTA is known to bind His-tagged proteins with nanomolar affinities, leading to a site-specific, stable, but reversible interaction (25, 26). Part of this concept includes inactivation of the tris-NTA by complexation of intramolecular exposed histidines (Fig. 1A). The number of inactivating (intramolecular) histidines control the stringency of self-inactivation. Activation is achieved by a light-induced separation of the intramolecular histidines, providing a free-to-interact high-affinity tris-NTA group. The versatility of these photoactivatable (PA) tris-NTA compounds for the site-specific interaction with proteins is demonstrated in solution and at interfaces. In situ activation of the biofunctional interfaces by light allows for multiplexed binding of different His-tagged proteins or protein complexes. This newly designed strategy allows in situ, instant, and multiplexed protein assembly.
Fig. 1.
Design of the photoactivatable tris-NTAs (PA tris-NTAs). (A) Schematic illustration of the PA tris-NTA concept relying on a tris-NTA moiety, a single cysteine for further modifications, the UV-sensitive amino acid (Anp), and cumulated histidines (blue). Addition of Ni2+ ions forces PA tris-NTA in a closed self-inactivating conformation. Photocleavage disrupts the intramolecular self-inactivation, allowing subsequent binding to His-tagged proteins. (B) Chemical structure of the PA tris-NTA compounds. Upon illumination the peptide backbone is photocleaved at the Anp (red).
Results and Discussion
Design and Concept of Photoactivatable Multivalent Chelator Heads.
The design of the photoactivatable tris-NTA has to comply with the following three demands: (i) self-inactivation of the high-affinity tris-NTA by tethered intramolecular histidines, (ii) activation by a photocleavage reaction, (iii) modification to follow photon-triggered binding events and to couple proteins or ligands (Fig. 1A). The tris-NTA compounds are based on a flexible, photocleavable peptide backbone, which contain a tris-NTA group at its N-terminus and a variable number of intramolecular histidines at its C-terminus. A flexible glycine-serine linker harboring a photo-sensitive 3-amino-3-(2-nitrophenyl)-propionic acid (Anp) separates these two functional groups. The photocleavable linker is optimized to afford intramolecular complex formation (self-inactivation of the tris-NTA). A single cysteine in proximity to the tris-NTA allows for a variable modification of the PA tris-NTA, such as for attaching fluorophors, ligands, or proteins, and for immobilization. This leads to the following sequence: tris-NTA-ACSGGGAnpSGGGHn with n = 3–6 (PA tris-NTA-Hisn, Fig. 1B). The photocleavable peptide was synthesized by solid-phase Fmoc chemistry. Carboxy-functionalized, OtBu-protected tris-NTA was coupled manually to the free N-terminus of the peptides bound to the resin. Our concept is based on the photocleavable amino acid Anp, separating the tris-NTA from the inactivating histidines upon illumination (Fig. 1B). Using conventional solid-phase chemistry offers the advantage that the number of histidines, the linker length, the position of the photocleavable element, as well as the amino acid composition and therewith the characteristics of the PA tris-NTAs can be easily varied and optimized. Synthesis and characterization of the PA tris-NTAs are detailed in SI Text.
Complex Formation and Protein Labeling Triggered by Photons.
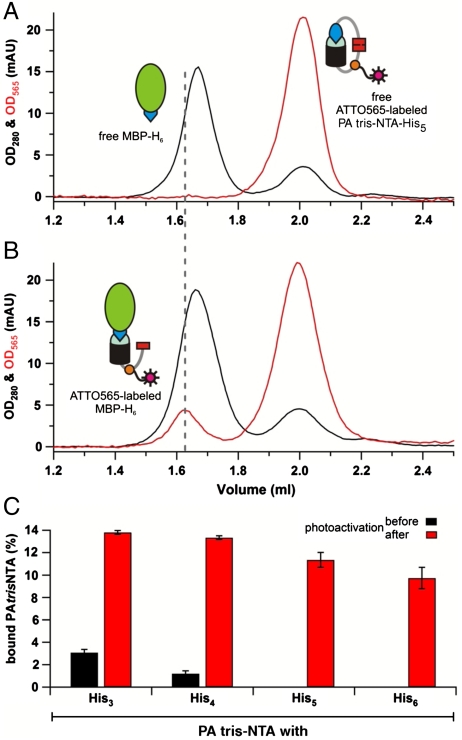
To follow site-specific protein modification triggered by light, ATTO565-labeled PA tris-NTA-His5 was incubated with His6-tagged proteins at an equimolar ratio. The maltose-binding protein (MBP-H6) was chosen as a model. Complex formation was analyzed via size exclusion chromatography by monitoring the absorption at 280 nm and 565 nm (Fig. 2A, black and red, respectively). Before photoactivation, the MBP-H6 and PA tris-NTA-His5 did not interact and elute as separate peaks based on their different hydrodynamic radii. Upon photoactivation, however, a stable MBP-H6-tris-NTA(ATTO565) complex was formed as demonstrated by a coinciding absorption of both wavelengths. In addition, the protein peak is shifted to a slightly larger apparent size (Fig. 2B). Based on peak integration, ∼12% of the protein was labeled which is explained by competition between His-tagged proteins and the photocleaved His5-tag. Notably, no interaction was observed in the presence of imidazole or EDTA. Furthermore, proteins lacking a His-tag did not bind to tris-NTA, demonstrating that the light-triggered interaction is site-specific. Since low power UV illumination was used, we did not observe a significant photobleaching. Exposure to increasing light doses showed saturation of binding after 20 min of illumination (Fig. S1). This is in good agreement with the investigation of the photoreaction of the Anp by reversed-phase HPLC. After 20 min illumination with light at 366 nm, no further changes in the product peaks were detectable.
Fig. 2.
Complex formation triggered by light. Size exclusion chromatography of equimolar mixtures PA tris-NTA-His5 and MBP-H6 before (A) and after photoactivation (B) demonstrates a stable complex formation by coinciding absorption at 280 nm (black) and 565 nm (red) within the MBP peak. (C) Using the absorption at 565 nm the percentage of labeled MBP-H6 before (gray) and after (red) can be calculated. Data are derived from triplicate measurements. The error bars show the standard deviations of the mean.
A systematic analysis of the series of PA tris-NTA compounds revealed that full self-inactivation was achieved by five and six intramolecular histidines, whereas three and four intramolecular histidines are not sufficient to block the intermolecular binding (Fig. 2C). After photoactivation, the amount of labeled proteins decreases slightly with an increasing number of inactivating histidines within the PA tris-NTA. These observations can be rationalized by considering the fact that the photoreaction generates a free His-peptide, which can be displaced by the His-tagged protein for the intermolecular complex formation. Importantly, the PA tris-NTA can be conjugated not only to a fluorophor, but to any molecule of interest, including other probes, activating or inhibiting ligands, DNA, RNA, and proteins, thus allowing for specific and light-triggered assembly of protein complexes.
Photons Trigger the Molecular Organization of Proteins.
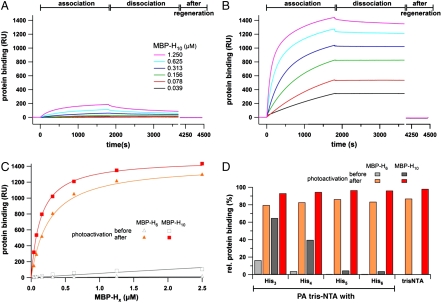
We next investigated the light-triggered interaction by surface plasmon resonance (SPR) using gradually self-inactivated PA tris-NTAs in parallel microfluidics. A dextran surface was functionalized to allow thiol-specific coupling of the different photoactivatable and nonactivatable tris-NTAs. Association and dissociation kinetics of His10-tagged MBP (MBP-H10) interacting with PA tris-NTA-His5 were monitored before (Fig. 3A) and after photoactivation (Fig. 3B). Before photoactivation, a very low and transient protein binding to the self-inactivated state was observed. These findings can be explained by the high effective concentration of intramolecular histidines competing for binding of His-tagged proteins. This behavior changed radically after photoactivation: A drastic increase in the amount of bound protein and a stable, specific binding of His-tagged proteins was observed (compare Fig. 3A vs. 3B). Similar results have been obtained for His6-tagged MBP (Fig. S2). Because of the reduced number of histidines in the case of MBP-H6, not even a transient binding signal was detected before photoactivation. The binding characteristics of MBP-H6 and MBP-H10 before and after photoactivation are summarized in Fig. 3C. While hardly any interaction was detected in the inactivated state, a high-affinity binding, similar to the reported, nonactivatable tris-NTA (25), was observed after photoactivation.
Fig. 3.
Light-induced interaction of His-tagged proteins followed by SPR. The association and dissociation kinetics of MBP-H10 before (A) and after photoactivation (B) reveal a drastic change in affinity. Before photoactivation almost no interaction is observed, while after illumination His-tagged proteins are stably bound at high density. (C) Protein binding to the PA tris-NTA-His5 surface (30 min after dissociation) are plotted against the concentration of MBP-H6 and MBP-H10 before (open) and after (closed symbols) photoactivation. (D) Comparison of the relative binding signals obtained for MBP-H6 and MBP-H10 (625 nM each) interacting with different PA tris-NTAs. The relative amounts of bound proteins (30 min after dissociation) are shown before (gray) and after photoactivation (red) in comparison to the binding observed with nonactivatable tris-NTA. Maximum binding of MBP-H6/10 after photoactivation for each interface is normalized to 100%.
Relative binding signals for all four PA tris-NTA interfaces at a fixed protein concentration are summarized in Fig. 3D. The maximal binding signal at the end of the association phase was normalized to 100%. Comparison of the relative amount of protein transiently bound to the unexposed PA tris-NTA surfaces reveals an increasing stringency of self-inactivation with an increasing number of intramolecular histidines. After photoactivation, PA tris-NTA share the same high-affinity characteristics and reversibility for His-tagged proteins as the nonactivatable tris-NTA (25).
Two-Dimensional Protein Assembly by Light.
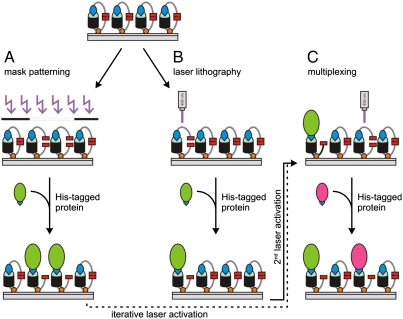
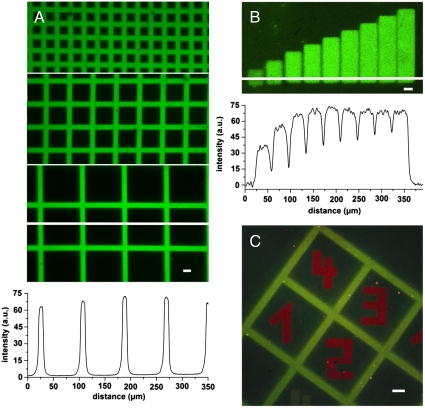
Guided by this quantitative analysis, we explored the PA tris-NTA-His5 interface for the two-dimensional organization of His-tagged proteins. Two different strategies have been explored to produce structured protein microarrays: (A) conventional mask lithography and (B) in situ laser lithography (Scheme 1). In the first approach, a quartz mask with grid patterns of different sizes was placed on top of the PA tris-NTA surface during illumination. After photoactivation, fluorescently labeled MBP-H10 was specifically captured at photoactivated areas. Sharp and well-defined edges of fluorescent patterns demonstrate stable protein organization with high spatial control (Fig. 4A). The very high contrast reflects the differences in protein binding between photoactivated and self-inactivated areas. Regeneration of the photostructured tris-NTA surfaces was readily afforded by washing with imidazole and EDTA. Mask lithography allows patterning large areas in a single operation.
Scheme 1.
Assembly of protein complexes by light. (A) Patterning with UV light using a chrome mask during illumination. (B) In situ photocleavage of the PA tris-NTAs using laser scanning microscopy (405 nm). (C) Multiplexing of different His-tagged proteins by successive activation of different chip areas using in situ laser scanning lithography is possible.
Fig. 4.
In situ assembly of proteins by light. (A) High contrast fluorescence images of Alexa488-MBP-H10 interacting with photo-patterned PA tris-NTA-His5 interfaces revealing sharp, diffraction limited edges. For photo patterning three distinct grid masks of different feature sizes were used. The white line indicates the position for which the fluorescence intensity profile is presented below the images. (B) Freely chosen regions of a PA tris-NTA-His5 interface were in situ exposed by laser scanning microscopy. Increasing time-dependent UV doses (exposure times 0–5 min) generate a protein gradient of Alexa488-MBP-H10 molecules. The corresponding fluorescence intensity profile emphasizes the stepwise increase in protein binding. (C) Orthogonal multiplexing of protein binding at a PA tris-NTA-His5 interface is demonstrated by using mask and in situ laser lithography in combination. The green pattern of Alexa488-MBP-H10 originates from the mask photo-patterned PA tris-NTA interface. Subsequently, in situ laser lithography was employed to write red numbers of ATTO565-MBP-H10 molecules into areas initially shadowed by the photo mask. The scale bars have a length of 20 μm.
The second approach utilizes in situ laser scanning lithography, offering an appealing advance over conventional mask lithography. Any region of interest can be gradually photoactivated by different, time-dependent UV doses, demonstrating the spatiotemporal organization of proteins at interfaces (Fig. 4B). In contrast to mask lithography, areas composed of different protein densities, e.g. receptor gradients, can be implemented.
In situ laser lithography is also a powerful method to realize multiplexed binding of different His-tagged proteins by iterative photoactivation of user-defined areas (Scheme 1C). Multiplexing was confirmed by a combination of both mask and in situ laser lithography as shown in Fig. 4C. Alexa488-MBP-H10 was first patterned by light activation of PA tris-NTA via a grid mask, followed by in situ laser lithography to write new patterns of a red fluorescent ATTO565-MBP-H10. Patterns with well-defined edges and high contrast were obtained for the orthogonal pair, demonstrating the high spatial resolution of the technique. Notably, only a very small amount of ATTO565-MBP-H10 was detectable on the gridlines, which could be either due to free tris-NTA binding sites or due to a slow, almost negligible protein exchange. In summary, our results show that multiplexing can be achieved comfortably using the PA tris-NTA-His5 interface. As a special benefit, size and shape of the in situ laser activated areas can be chosen on demand as needed. The resulting target protein densities within activated areas can be adjusted by controlling the dose during illumination. Therefore, the proposed methodology enables interaction studies on biofunctionalized interfaces with engineered gradients of protein density.
Functional Receptor Patterns Sensing Human Rhinovirus Particles.
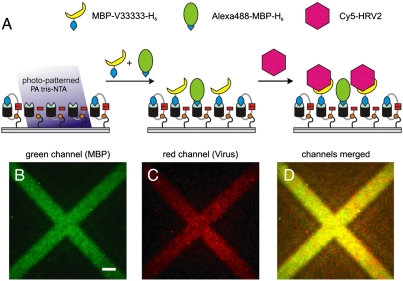
Advanced protein chip technology does not only demand site-specific and multiplexed binding of different proteins but also has to be practically compatible with complex physiologically or medically relevant samples. An important prerequisite for the highly selective binding of physiological targets is the functional integrity of immobilized proteins under physiological conditions. As an example, we chose the very low-density lipoprotein (VLDL) receptor, which specifically recognizes minor group human rhinoviruses such as HRV2 particles. HRVs are one of the main causes of the common cold and have a strong impact on human health (27). Here we photo-patterned the engineered MBP-VLDL-receptor fusion protein MBP-V33333-H6, which consists of a pentameric V3 repeat and binds HRV2 with subnanomolar affinity (28). To visualize the receptor arrays, we doped the VLDL receptor with Alexa488-MBP-H6 as direct labeling can disable receptor function. Interaction between the His-tagged VLDL receptor and Cy5-labeled HRV2 was investigated by total-internal reflection fluorescence (TIRF) microscopy. The receptor (Fig. 5B, green) and the virus particle layer (Fig. 5C, red) were imaged subsequently. Merged images confirmed that the virus particles were selectively bound to receptors patterned by light (Fig. 5D). No binding of viral particles and receptors was observed in the absence of nickel ions. In conclusion, PA tris-NTA interfaces represent a highly flexible platform for detection and analysis of clinically relevant virus particles.
Fig. 5.
Two-dimensional organization of receptor-virus particle by light. (A) Schematic representation of capturing viral particles onto PA tris-NTA interfaces. The virus specific VLDL receptor (MBP-V33333-H6) doped with Alexa488-MBP-H6 was preimmobilized onto photo-patterned PA tris-NTA-His5 interfaces. Subsequently, Cy5-labeled HRV2 was bound by its receptor. Highly sensitive total-internal reflection fluorescence microscopy was used to detect the two-dimensional organization of receptor proteins (MBP-V33333-H6 was detected indirectly via cocapturing of fluorescent Alexa488-MBP-H6) (B) and Cy5-labled HRV2 particles (C). An overlay image of (B) and (C) is presented in (D). The scale bar has a length of 10 μm.
Conclusion
In summary, we have developed small photoactivatable recognition elements with striking characteristics in terms of site-specific protein labeling, stable complex formation, and two-dimensional assembly of proteins triggered by light. The self-inactivation by intramolecular ligands was found to be very efficient. In contrast to previous attempts, which involved drastic, stepwise, protein incompatible surface modifications (29, 30), this strategy allows in situ, instant, and multiplexed protein organization. As an example, different proteins were multiplexed by iterative in situ writing and binding with temporal and spatial control. Importantly, we confirmed the applicability of this generic concept to affinity capture virus particles. In situ photopatterning creates arbitrarily shaped interaction and reference spots as well as continuous or discontinuous protein gradients. Despite the many advantages, there is still room and need for further improvements within the self-inactivation concept. The photoactivatable multivalent chelator heads will be further improved by placing the photocleavable group within the self-inactivating histidines. Further developments will aim at investigating cellular processes by photon-triggered protein-protein interactions (e.g. receptor clustering), one of the major objectives in chemical biology.
Materials and Methods
Details of the synthesis and analytic characterization of the photoactivatable tris-NTAs (Table S1) as well as protein purification, labeling, and surface chemistry are provided in SI Text.
Complex Formation in Solution.
Five μM of MBP-H6 was incubated with equimolar ATTO565-labeled PA tris-NTA in HBS buffer (20 mM Hepes, 150 mM NaCl, pH 7.5). Photoactivation was performed in a 96-well plate on ice. The distance between sample and lamp was 1.5 cm. After illumination for 30 min at 366 nm (2 × 8 W, Benda), the sample was incubated in the dark for 30 min at room temperature. As negative control, the samples were treated identically except from exposure to UV light. Complex formation was analyzed using a Superdex™ 200 PC 3.2/30 column (GE Healthcare) in an ETTAN™ system (GE Healthcare). The experiments were performed at 10 °C using HBS buffer as running buffer with a flow rate of 70 μL/min. For analysis, 10 μL of the PA tris-NTA/MBP-H6 mixtures were taken before and after photoactivation.
Protein Binding at Interfaces.
The binding of His-tagged MBP to PA tris-NTA functionalized surfaces was followed in real-time by surface plasmon resonance (Biacore® T100, GE Healthcare). A carboxymethylated dextran surface (CM5 sensor chip, GE Healthcare) was functionalized via the 2-(2-pyridinyldithio)ethaneamine linker, which allowed coupling of the PA tris-NTAs via its single cysteine. For direct comparison, each PA tris-NTA was coupled separately to each of the four flow cells. To this end 200 μM of PA tris-NTA in citric acid buffer (0.1 M, pH 2.5) were injected into the respective flow cell for 15 min. All kinetic experiments were carried out in HBS buffer supplemented with 50 μM EDTA (HBSE buffer) and 0.05% Tween20 at a flow rate of 10 μL/min. In the beginning of each binding cycle, the surfaces were washed successively with imidazole (1 M, 2 min) and EDTA (100 mM, 2 min), followed by loading of the tris-NTA groups with 10 mM of Ni2+ ions for 2 min. Association of MBP-H6 (39 nM to 10 μM) and MBP-H10 (39 nM to 2.5 μM) was measured at a flow rate of 5 μL/min for 30 min. The dissociation was monitored for 30 min under constant buffer flow (10 μL/ min). Finally, affinity captured protein was removed by injection of imidazole (1 M, 2 min) and EDTA (100 mM, 2 min). Omitting the nickel-loading completely abolished MBP binding, underlining the highly specific interaction between tris-NTA and His-tagged MBP. For photoactivation, a UV lamp (366 nm; 2 × 8 W; Benda) was placed 2 cm above the chip and exposure was carried out for different periods of time on ice. Using progressive illumination doses, a maximum protein binding was observed after 20 min of illumination (Fig. S3). Protein binding was analyzed as described before. The binding kinetics was corrected for bulk refractive index contribution by subtraction of binding curves obtained from measurements without nickel ions.
Mask Patterning.
Quartz masks with chrome grids of different sizes were used to fabricate laterally structured PA tris-NTA surfaces on glass slides (cover glass 24 mm diameter, thickness 160–190 μm, Menzel). The mask was placed on top of the PA tris-NTA chip during exposure to UV light of a 200 W Hg/Xe lamp equipped with a dichroic mirror (280–400 nm, Newport Spectra-Physics). Irradiation was carried out for 12 min with a small amount of HBSE buffer between mask and surface. After removal of the mask, the surface was successively washed with 1 M of imidazole and 100 mM of EDTA.
Virus-Receptor Assembly.
Tris-NTA groups were loaded with 10 mM of Ni2+ ions for 10 min, followed by a specific immobilization of the respective His-tagged protein. Alexa488-MBP-H10 was used at a concentration of 200 nM for 20–30 min. The viral receptor MBP-V33333-His6 (500 nM) was doped with Alexa488-MBP-H6 (20∶1 molar ratio) to allow fluorescence imaging. After receptor patterning, the surface was rinsed thoroughly with HBS buffer containing 2 mM of CaCl2 (HBSCa buffer). Subsequently, the surface was incubated with 10 nM of Cy5-labeled HRV2 particles in HBSCa buffer for 2 h. Imaging was performed after removal of unbound HRV2 in HBSCa buffer.
In Situ Assembly of Protein Complexes by Light.
Protein binding was monitored by confocal laser scanning microscopy (CLSM) (LSM 510, Carl Zeiss) with an inverted microscope (Axiovert 200 M, Carl Zeiss). The protein chips were mounted in a home-built liquid cell and imaged under HBSE buffer using a Plan-Apochromat 20x objective (NA 0.75, Carl Zeiss). Alexa488-MBP-H10 was excited at a wavelength of 488 nm using an argon laser (17.5% maximal power output, 30 mW). A He/Ne laser (1 mW) with a wavelength of 543 nm and 84% maximal power output was used to excite the ATTO565-MBP-H10. For in situ patterning the 405 nm laser (diode laser, 25 mW) of the CLSM setup was used. The surface was incubated with ATTO565-MBP-H10 (200 nM). The generated structures were imaged in HBSE buffer using the CLSM. Alternatively, photopatterned interfaces were analyzed using a TIRF microscope (DMI 6000 B, Leica Microsystems) with a home-built liquid cell. Images were acquired with a 100x oil-immersion objective (HCX Planapo, NA 1.46, Leica Microsystems). The Alexa488-MBP-H6 within the receptor layer was visualized using a 488 nm laser. The Cy5-HRV2 was excited with a 633 nm laser (laser source: AM TIRF MC, Leica Microsystems). Fluorescence images were processed with the Leica LAS AF software (Leica Microsystems).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Gerhard Spatz-Kümbel for technical assistance and Helge Großmann for helpful discussions. The work was supported by the Bundesministerium für Bildung und Forschung (BMBF, Nanobiotechnology 0312031 and 0312034), the Deutsche Forschungsgemeinschaft (DFG, Ta157/6), and the Austrian Science Foundation (FWF).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
See Commentary on page 6123.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912617107/DCSupplemental.
References
- 1.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 3.Nienhaus GU, Wiedenmann J. Structure, dynamics and optical properties of fluorescent proteins. ChemPhysChem. 2009;10:1369–1379. doi: 10.1002/cphc.200800839. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan KF. Fluorescent Proteins. 2nd Ed. San Diego: Academic; 2008. [Google Scholar]
- 5.Andresen M, et al. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat Biotechnol. 2008;26:1035–1040. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- 6.Johnsson N, Johnsson K. A fusion of disciplines: Chemical approaches to exploit fusion proteins for functional genomics. ChemBioChem. 2003;4:803–810. doi: 10.1002/cbic.200200603. [DOI] [PubMed] [Google Scholar]
- 7.O’Hare HM, Johnsson K, Gautier A. Chemical probes shed light on protein function. Curr Opin Struct Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 9.Stevens MM, et al. Molecular level investigations of the inter- and intramolecular interactions of pH-responsive artificial triblock proteins. Biomacromolecules. 2005;6:1266–1271. doi: 10.1021/bm049369x. [DOI] [PubMed] [Google Scholar]
- 10.Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 11.Heckel A, Mayer G. Light regulation of aptamer activity: An anti-thrombin aptamer with caged thymidine nucleobases. J Am Chem Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- 12.Chen CT, Wagner H, Still WC. Fluorescent, sequence-selective peptide detection by synthetic small molecules. Science. 1998;279:851–853. doi: 10.1126/science.279.5352.851. [DOI] [PubMed] [Google Scholar]
- 13.Chockalingam K, Blenner M, Banta S. Design and application of stimulus-responsive peptide systems. Protein Eng Des Sel. 2007;20:155–161. doi: 10.1093/protein/gzm008. [DOI] [PubMed] [Google Scholar]
- 14.Mendes PM. Stimuli-responsive surfaces for bio-applications. Chem Soc Rev. 2008;37:2512–2529. doi: 10.1039/b714635n. [DOI] [PubMed] [Google Scholar]
- 15.Dorman G, Prestwich GD. Using photolabile ligands in drug discovery and development. Trends Biotechnol. 2000;18:64–77. doi: 10.1016/s0167-7799(99)01402-x. [DOI] [PubMed] [Google Scholar]
- 16.Pelliccioli AP, Wirz J. Photoremovable protecting groups: Reaction mechanisms and applications. Photochem Photobiol. 2002;1:441–458. doi: 10.1039/b200777k. [DOI] [PubMed] [Google Scholar]
- 17.Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew Chem Int Ed Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 19.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundberg SA, et al. Spatially-addressable immobilization of macromolecules on solid supports. J Am Chem Soc. 1995;117:12050–12057. [Google Scholar]
- 21.Pirrung MC, Huang CY. A general method for the spatially defined immobilization of biomolecules on glass surfaces using “caged” biotin. Bioconjugate Chem. 1996;7:317–321. doi: 10.1021/bc960013v. [DOI] [PubMed] [Google Scholar]
- 22.Blawas AS, Oliver TF, Pirrung MC, Reichert WM. Step-and-repeat photopatterning of protein features using caged-biotin-BSA: Characterization and resolution. Langmuir. 1998;14:4243–4250. [Google Scholar]
- 23.Banala S, Arnold A, Johnsson K. Caged substrates for protein labeling and immobilization. ChemBioChem. 2008;9:38–41. doi: 10.1002/cbic.200700472. [DOI] [PubMed] [Google Scholar]
- 24.Guignet EG, Segura JM, Hovius R, Vogel H. Repetitive reversible labeling of proteins at polyhistidine sequences for single-molecule imaging in live cells. ChemPhysChem. 2007;8:1221–1227. doi: 10.1002/cphc.200700065. [DOI] [PubMed] [Google Scholar]
- 25.Lata S, Reichel A, Brock R, Tampé R, Piehler J. High-affinity adaptors for switchable recognition of histidine-tagged proteins. J Am Chem Soc. 2005;127:10205–10215. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 26.Tinazli A, Piehler J, Beuttler M, Guckenberger R, Tampé R. Native protein nanolithography that can write, read and erase. Nat Nanotechnol. 2007;2:220–225. doi: 10.1038/nnano.2007.63. [DOI] [PubMed] [Google Scholar]
- 27.Couch RB. Rhinoviruses. In: Knipe DM, et al., editors. Fields Virology. 5th Ed. Vol 1. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 893–910. [Google Scholar]
- 28.Wruss J, et al. Attachment of VLDL receptors to an icosahedral virus along the 5-fold symmetry axis: Multiple binding modes evidenced by fluorescence correlation spectroscopy. Biochemistry. 2007;46:6331–6339. doi: 10.1021/bi700262w. [DOI] [PubMed] [Google Scholar]
- 29.Alonso JM, Reichel A, Piehler J, del Campo A. Photopatterned surfaces for site-specific and functional immobilization of proteins. Langmuir. 2008;24:448–457. doi: 10.1021/la702696b. [DOI] [PubMed] [Google Scholar]
- 30.Bhagawati M, et al. Organization of motor proteins into functional micropatterns fabricated by a photoinduced Fenton reaction. Angew Chem Int Ed Engl. 2009;48:9188–9191. doi: 10.1002/anie.200904576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.