Abstract
Recently discovered bacterial homologues of eukaryotic pentameric ligand-gated ion channels, such as the Gloeobacter violaceus receptor (GLIC), are increasingly used as structural and functional models of signal transduction in the nervous system. Here we present a one-microsecond-long molecular dynamics simulation of the GLIC channel pH stimulated gating mechanism. The crystal structure of GLIC obtained at acidic pH in an open-channel form is equilibrated in a membrane environment and then instantly set to neutral pH. The simulation shows a channel closure that rapidly takes place at the level of the hydrophobic furrow and a progressively increasing quaternary twist. Two major events are captured during the simulation. They are initiated by local but large fluctuations in the pore, taking place at the top of the M2 helix, followed by a global tertiary relaxation. The two-step transition of the first subunit starts within the first 50 ns of the simulation and is followed at 450 ns by its immediate neighbor in the pentamer, which proceeds with a similar scenario. This observation suggests a possible two-step domino-like tertiary mechanism that takes place between adjacent subunits. In addition, the dynamical properties of GLIC described here offer an interpretation of the paradoxical properties of a permeable A13′F mutant whose crystal structure determined at 3.15 Å shows a pore too narrow to conduct ions.
Keywords: allosteric transition, hydrophobic gate, cys-loop receptor family
Neurotransmitter receptors mediate chemical communication in the nervous systems. In the case of ligand-gated ion channels typified by the nAChR (1, 2), the receptor is a transmembrane pentamer carrying the neurotransmitter binding sites and the ion channel on distinct extracellular (ECD) and transmembrane (TMD) domains, respectively, and acetylcholine binding elicits channel opening through a global conformational transition, creating a flux of cations along their electrochemical gradient.
nAChRs belong to the cys-loop receptors family (CLRs) that includes serotonin, glycine, and GABA receptors (3), and their gating plausibly involves an allosteric pathway that links the neurotransmitter sites to the channel (4), the interface between TMD and ECD playing a critical role in this process (4). Furthermore, intermediate priming states in the gating process have recently been suggested (5, 6).
The structural data of eukaryotic CLRs is limited to x-ray structures of the ECD (7, 8) and to a model of Torpedo nAChR derived from cryo-EM data at 4-Å resolution (9). The discovery of CLR sequences in bacteria (10) and the demonstration that a Gloeobacter violaceus homologue (called GLIC), functions as a proton-gated cationic channel (11) were incentive to the resolution at 3.3 Å of the crystallographic structure of a full CLR from Erwinia chrysanthemi (12) in a closed conformation (called ELIC) and of GLIC at 2.9 Å resolution after crystallization at pH 4.6 in an open conformation (13, 14). ELIC and GLIC structures resemble that of nAChRs but lack the unconserved intracellular domain.
Comparison of the GLIC and ELIC structures suggests a plausible mechanism of CLR gating that notably involves a quaternary twist movement previously identified by normal mode analysis (15). A recent mixed elastic network study of GLIC and ELIC TMDs corroborates these findings (16) but without inferring the sequence of events elicited by agonist binding, because the ECD was absent from the simulation.
Ultrafast conformational changes of CLRs have been investigated computationally on a time scale of up to 60 ns. A recent 30-ns molecular dynamics (MD) simulation of ELIC (17) shows a remarkable stability of the channel, which remains dehydrated and thus closed throughout the simulation. Because the pharmacology of ELIC is unknown, its gating could not be studied. Other MD simulations used the cryo-EM structure of the nAChR or various homology models derived from it (18, 19).
The present MD study was carried out with the atomic structure of GLIC, the only bacterial CLR with a known agonist. Recently, microsecond-scale MD simulations on large membrane proteins have been described for rhodopsin (43,222 atoms) (20), the β2-adrenergic receptor (∼99,000 atoms) (21), and the integral Kv1.2 ion channel (∼120,000 atoms) (22). Here, a 200,000-atoms model of the fully hydrated membrane-inserted GLIC channel was simulated for 1 μs. We start from the open form of the channel at acidic pH after 20 ns of equilibration as described previously (13). In order to describe GLIC’s pH-elicited gating (here closing), and because little is known about the precise coupling between protonation state and conformational change (see refs. 23 and 24), we artificially split the process into two distinct steps. We first perform an instantaneous pH change, by setting the charges of the ionizable residues of the channel to their standard state at neutral pH. Then we observe the conformational transition elicited by this change (see Fig. 1). The simulation reveals an early closure of the channel followed by a concerted—yet still partial—transition, with two adjacent subunits following the same scenario, in a progressive manner, leading to a fully closed channel.
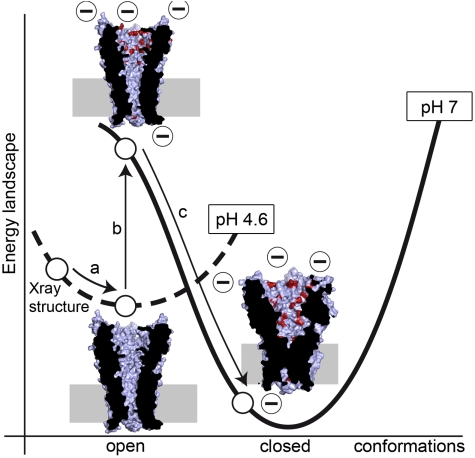
Fig. 1.
Schematic representation of the simulation protocol. Starting from the crystal structure at pH 4.6, a brief equilibration is carried out (step a, 20 ns). Then, an instantaneous change in pH (step b) is made (Fig. S1a and Table S1), followed by the relaxation towards a closed conformation (step c, 1.06 μs). Only results from step c are discussed in this work. The curves with broken and plain lines represent energy landscapes for pH 4.6 and 7.0, respectively. Cross-sections of the channel illustrate each state involved in this scheme: an equilibrated open state at pH 4.6, an open state at pH 7 (simulation at 0 μs), and a state at pH 7 (simulation at 1.06 μs) on the way to the closed conformation. The protein’s surface is represented in light blue with residues changing charge during the pH jump in red. The protein’s cross-section is shown in black. The approximate position of the membrane is indicated by a gray rectangle.
Results
A Rapid Asymmetric Closure of the Pore in less than 100 ns.
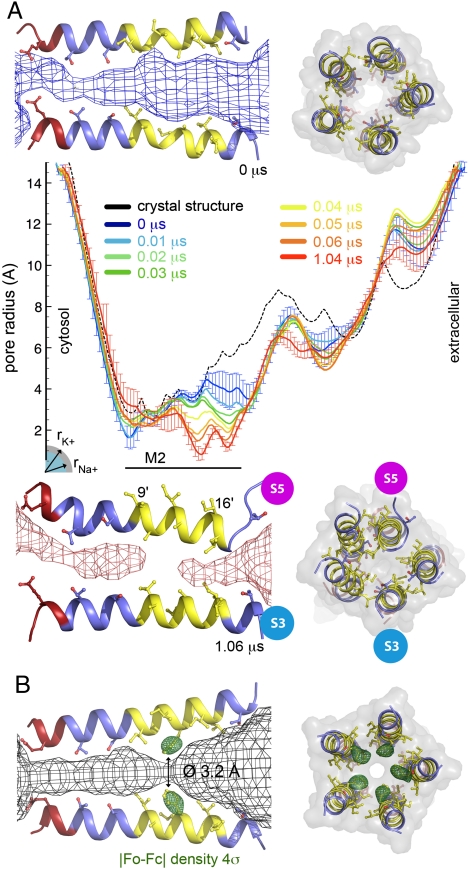
The state of the channel was monitored by using the “Hole” software (25). Fig. 2A shows that, in the initial open conformation of the receptor, the ion permeation pathway consists of a wide extracellular vestibule (more than 6 Å diameter) and of a narrower transmembrane pore (less than 4 Å diameter; labeled M2). By taking into account conformational fluctuations (represented as error bars), the overall shape of the outer vestibule and of M2’s cytoplasmic intracellular mouth region can be considered constant. In contrast, the extracellular mouth of M2 undergoes a major reorganization during the first 100 ns; especially, the I9′ to I16′ region shows a radius drop from 4 Å to less than 1 Å, thereby fully closing the pore. Thereafter, this initially highly flexible region becomes rigid. An exhaustive plot of the whole channel pore profile is given by Fig. S2a. Fig. 2A further provides side views of two M2 helices with the pore volume shown as a mesh, confirming its obstruction in the upper part of M2 during the simulation.
Fig. 2.
Pore radius. (A) Side and top views of GLIC M2 helices are shown for the start (Top) and end (Bottom) of the simulation; side chains facing the pore are depicted. Hydrophobic, polar, and negative residues are colored yellow, blue, and red, respectively. In the side view, only two subunits are shown, including the S5 subunit with a particular motion towards the channel axis resulting in partial unwinding at its top. The channel pathway is represented as a mesh. The central panel shows the pore radius along the channel axis for the GLIC crystal structure (Black, Broken Line) and for several stretches of the molecular dynamics simulation. The simulation results are averages over 20-ns windows, starting at 0.0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06 (blue to orange) and at 1.04 μs (red). Standard deviations are shown as error bars for the 0.0- and 1.04-μs windows. The radii of Na+ and K+ ions liganded in a protein environment are indicated. (B) Pore structure of the A13′F mutant, whose crystal structure was solved at 3.15 Å resolution. Same views as above with the density from initial Fourier difference maps contoured at 4σ represented as green mesh around the mutated A13′F position. The primed residue numbers are a common numbering scheme for all cys-loop receptors in the M2 helices, starting at its cytosolic end. GLIC’s V225 is V1′.
A Global Quaternary Twist Movement During the 1 μs Simulation.
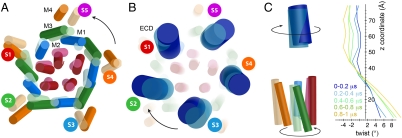
The overall conformational transition of the channel is illustrated by superposing the initial and final inertial axes of each helix of the TMD (Fig. 3A) and of the whole ECD (Fig. 3B). The behavior of each subunit is unique, and the evolution of the inertial axes displays asymmetry. However, the closing transition features an overall twist movement where ECD and TMD move in opposite directions (see arrows in Fig. 3A and B and ref. 15). This twist can be further quantified by analyzing cumulated rotations of z slices of the protein as presented in Fig. 3C, reaching +8° counterclockwise rotation in the TMD and -6° clockwise rotation in the ECD. The twist follows the initial tilt direction of the axes of inertia and is the same as in the comparison of the two known crystal conformations of GLIC and ELIC (13). As previously inferred from this comparison, the ECDs rather move as rigid bodies (Fig. S2b).
Fig. 3.
Quaternary changes. (A) Top view of the principal axes of inertia for TMD helices M1–M4 of each subunit (colored in blue, red, green, and orange, respectively) at the end of the simulation. The principal axes at the start of the simulation are superposed in lighter colors. The number of each subunit is indicated with colors red, green, blue, orange, and magenta for S1–S5, respectively. (B) Same view as in A for the ECDs shown in dark blue. (C) Theta twist angle in successive z slabs normal to the membrane. The snapshot on the left is a side view of subunit S1. The indicated twist direction corresponds to positive (Bottom) and negative (Top) theta angles, respectively. On the right, time evolution is provided by superposition of averages on successive 0.2-μs windows.
Two Asymmetric but Concerted Transitions Occur Within the Channel During the 1 μs Simulation.
The transition of M2 helices can be described by following their axis orientation characterized by two angles: δ (inclination with respect to the protein symmetry axis) and θ (azimuthal orientation of the helix in the plane of the membrane). The stereographic projection plots shown in Fig. 4A display the trajectories defined by following δ and θ angles of each M2 helix, as seen from the extracellular region. Time-independent clusters of helix orientation can be defined. For each helix, these clusters are arbitrarily named A, B, C, and D according to their sequential apparition during the simulation. Fig. 4B displays a timeline of cluster occupation showing that subunits switch between clusters both in concert and independently. This representation highlights two major events that result in a concerted switch of all M2 helices: First, all helices leave their initial A cluster after about 0.05 μs of simulation and twist clockwise around the pore axis; second, between 0.4 and 0.6 μs of simulation, M2 helices S2–S5 tilt their upper part toward the pore, whereas S1 moves away from it (see Fig. S2c for more details).
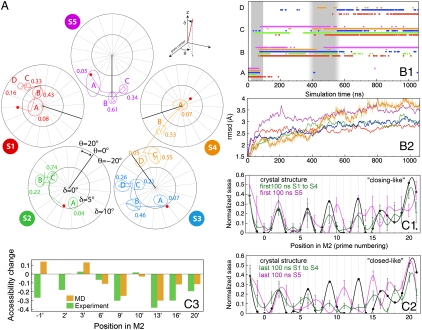
Fig. 4.
M2 helix movement analysis. (A) Stereographic projections of polar δ and azimuthal θ angles of the M2 helix principal axis of inertia in each subunit (same subunit color code as in Fig. 3). Red dots represent initial positions. Lines depict the trajectory between average orientations at 0.1-μs intervals. The sampling of the angle space of each subunit is represented by covariance ellipsoids centered on mean positions (see Full Methods). The fraction of time spent in each cluster (normalized to 1) is indicated. (B1) Exploration of A, B, C, and D clusters along the simulation, for each subunit, using the same color code as in A. (B2) Subunit rmsd is calculated as compared to the beginning of the simulation. Curves are smoothed for clarity, with an example of the raw curve given for subunit 4. (C1) Normalized solvent accessible surface areas for the M2 helices determined for the crystal structure (Black Line), averaged over subunits S1–S4 (Green Line), and for subunit S5 (Purple Line). The top C1 panel displays the average accessible areas for the start of the simulation (0–0.1 μs); the bottom C2 panel shows the averages over 0.96–1.06 μs. (C3) Comparison of solvent accessibility changes between MD simulation data and substituted cysteine accessibility method (SCAM) measurements. The MD result for each residue was obtained by calculating the difference between the mean normalized SASA of S1–S5 over the last 100 ns and the crystal structure. The displayed SCAM results are -0.05 ln(k + /k - ), where k+ and k- are second-order rate constants in the presence and absence of acetylcholine, respectively (26).
The conformational changes of the M2 helices can be further characterized by measuring their solvent accessible surface area (SASA) as shown in Fig. 4C. Normalized SASA was averaged over the first 100 ns of the simulation, reflecting both the open-channel state and the early closing motion, and for the last 100 ns, reflecting the end state. The initial SASA patterns for subunits S1–S4 are almost identical to that of the crystal structure. S5 shows changes resulting from its high mobility (indicated by larger error bars). The S5 helix rotates around its own axis, thereby exposing residues at 10′ and 14′ positions to the lumen. This rotation is associated with a secondary deformation with loss of the upper helix turn, starting at 16′. At the end of the simulation, all subunit patterns clearly differ from the crystal pattern. S5 still markedly differs from the other subunits with a unique motion involving, in addition to the deformation, a rotation, as illustrated by the difference at positions 7′, 11′, and 15′.
The qualitative agreement between the experimental results of Pascual and Karlin on nAChRs (26) and the data calculated from the simulation is good (Fig. 4C) except at the bottom of the channel (intracellular end) at positions 1′ and 2′. Rearrangements of salt links are described in Fig. S3.
Rearrangements of the M2 Helices Drive Global Reorganization of the Protein.
The unique behavior of subunit S5, particularly at the beginning of the simulation, is a striking feature of GLIC’s transition. At 25 ns after the pH switch, the M2 helix of this subunit swings into the pore (Fig. 4A) and substantially closes the channel (Fig. 2A, green curve). In addition, in the first 200 ns, this subunit experiences a strong pivot in its ECD (Fig. S4b, Top) and undergoes, in advance of other subunits, a global tertiary deformation as can be seen from its structural drift (Fig. 4B). The second ensemble of events, which starts at around 400 ns, primarily affects subunit S4, in a manner similar to subunit S5: M2 movement occurs between 400 and 500 ns (Fig. 4A), followed by an important pivot (of the TMD this time) and tertiary deformation (Fig. S4b, Bottom and Fig. 4B). Meanwhile, M2 helices of S2 and S3 visit positions similar to ELIC (δ = 9.3 and θ = 113), especially in their cluster B.
Experimental Evidence for Flexibility in the Upper Part of the M2 Helix.
On the basis of the GLIC structure, we hypothesized that mutation of a pore-facing side chain within the gate into a bigger hydrophobic one would stabilize the closed state by increasing hydrophobic interactions. We therefore constructed the A13′F mutant. Its 3.15 Å structure was solved by using crystals grown at acidic pH, in the same conditions as for the wild-type open structure. Both structures superimpose almost perfectly, except for the bulkier 13′F whose side chains point inside the channel and should thus hamper conduction (Fig. 2B). However, functional data show that the channel does open at acidic pH (see Fig. S5). This apparent contradiction might be resolved by an unusually high flexibility in solution in the vicinity of the mutation, allowing the 13′F side chain to open a passage for the flux of ions through the channel.
Water and Ions in the Channel.
Channel hydration analysis initially shows a continuous water column that is disrupted early on in the upper half of M2, underlining the presence of a hydrophobic gate (Fig. S6a). Furthermore, we observe that cations are attracted by the channel and accumulate within three distinct reservoirs, with an entirely cation-free gating region (see details in SI Text and Fig. S6b).
Discussion
This simulation provides an attempt to describe the gating dynamics in the μs time range of a CLR, namely, the GLIC channel closure at the atomic level. It extends and complements recent approaches aimed at describing nAChR gating (4, 27, 28). Even if the particular driving forces at the origin of the transition remain difficult to identify because (i) the conformational transition was not completed at the end of the 1 μs simulation, (ii) the simulation starts with a significant perturbation, namely, the instantaneous pH switch, and (iii) we have only limited statistics from a single trajectory, the trajectory exhibits a number of critical features that characterize channel gating. In particular, we observe that the hydrophobic gate undergoes large conformational changes disrupting channel hydration. A first subunit undergoes a two-step tertiary transition very early in the simulation, followed at 450 ns by a similar transition of its immediate neighbor in the pentamer. A mechanistic hypothesis for channel gating might be derived from these results, referred to as the “domino” mechanism, where a sequence of localized events propagating between adjacent subunits is nested within a global quaternary twist conformational change of the molecule.
GLIC’s Hydrophobic Gate is Located Between I9′ and I16′.
Locating the channel gate(s) in CLRs remains a challenging task. Experiments on substituted cysteine or histidine accessibility suggest a location at either the intracellular (29, 30) or the extracellular side of M2 (27, 28, 31). Our analysis, combined with the recent report on ELIC (17), supports a location of the gate of GLIC and ELIC within two rings of hydrophobic residues in the upper part of the channel, as assessed independently from the pore radius analysis (Fig. 2), from channel dehydration (Fig. S6a), and from the existence of cation-excluded regions (Fig. S6b). Whether our results are transferable to other CLRs remains to be established, but previous work already pointed to such a hydrophobic girdle as contributing to the gate in nAChRs (19, 28, 32, 33). The fast ∼0.1 μs gating time scale observed herein is consistent with studies based on shorter calculations (19) but still out of reach of current electrophysiological approaches limited to a ∼10 μs resolution.
Water and Ions as Indicators for Gating.
A hydrophobically gated channel is expected to show at least partial dehydration, thus disrupting the connection between intra- and extracellular media (34). This disruption is precisely what happens here (Fig. S6a). Such a change in hydration, related to conformational changes of the M2 helices, has recently been suggested on the basis of experimental data and Φ-value analysis (35). These observations are corroborated by the absence of ions in this region, whereas nearby areas are cation reservoirs (Fig. S6b). The most potent reservoir, R3, is located in the lower part of the channel where selectivity is thought to be operating. Electron density has been measured in this region of GLIC and was attributed to Cs+, Rb+, and Zn2+ cations (14). These results on hydration and ion propensity are also in good qualitative agreement with an 11-ns MD simulation on nAChR (33).
Highly Flexible M2 Helices Initiate Gating.
Conformational changes of the M2 helices are a hallmark of the gating process. Several types of motions were previously proposed for M2 helices: translation, rotation(s) of and around the helix axis, and backbone deformation. Here, all three types are observed (Fig. 4). In particular, the subunit 5 M2 helix (S5, at the top) deviates from the others and swings early to close the pore. The propagation of this and subsequent events from one subunit to the other and from the M2 helices to the rest of the protein is still an ongoing process at the end of the 1-μs run.
Even though the transition is not complete, the channel is fully closed and M2 motions agree with several observations. First, measurement of the rates of 2-aminoethyl methanethiosulfonate modification of cysteine mutants in M2 showed that positions 1′, 2′, 6′, 9′, 13′, 16′, and 20′ display faster accessibility for the open than for the closed channel (26). Our simulation fits these findings, as demonstrated by the difference between the mean accessible surfaces in the closed channel (0.96–1.06 μs) and the ones in the open-channel crystal structure (Fig. 4C). Second, in the initial period, important fluctuations of S5 SASAs are observed. In the final period, the upper part of the helix loses the pattern typical of a tightly packed pore-lining helix. These independent facts denote flexibility and/or loose packing, a feature experimentally observed by using cysteine accessibility scanning of the GABA receptors (36) or looking at the ability to form inter-M2 disulfide bridges (37). Third, the S5 subunit exhibits a unique motion involving a rotation (see the difference at positions 7′, 11′, and 15′ in Fig. 4C) in addition to its deformation. Such a rotation was predicted on the basis of the nAChR EM structure (9). Finally, we note that the M2 helix of S5 loses its upper C-terminal turn. Backbone deformations of the upper part of the helix were inferred by studying amide-to-ester mutations that prevent main chain hydrogen bond formation (38).
Additional indirect evidence for the mobility of the M2 helices comes from our A13′F mutant crystal structure combined with electrophysiological data. The structure (Fig. 2B) shows a channel constriction in the gating region. This mutant channel is, however, functional, which implies that it can open a passage for ion permeation. Hence, the vicinity of the 13′F side chain has to be flexible. The same conclusion can be drawn from the large amplitude movements that we observed in this region of the protein during our simulation. Both observations lead to the interpretation that the mutant crystal structure is trapped in a closed state, where M2 flexibility is blocked, but will assume a conformational equilibrium in a membrane environment because of the enhanced flexibility at the top of the M2 helices.
A Possible Previously Undescribed Gating Mechanism in the Cys-Loop Receptor Family.
There is too little experimental data at present to guide and validate the extrapolation of the described process to a complete transition, by using, for instance, the conformation of the S4 and S5 subunits and the fivefold symmetry to generate the 3 other ones, thereby producing a quantitative structural model for the closed state. Nevertheless, we wish to propose an hypothetical, qualitative model of the full conformational transition that, accordingly, would progress from one subunit to the neighboring one, through a “domino-like” process, and involve at least two steps: large fluctuations at the top of the M2 helix, followed by a more global tertiary rearrangement of the whole subunit.
In the first phase, large fluctuations would occur in one subunit Sn in the form of fast, local, asymmetric conformational changes at the top of the M2 helix. The second phase would consist of slower, long-range, concerted reorganizations implying tertiary deformations of the 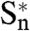 subunit and propagation to the whole TMD, driven by the M2 helices. Propagation would occur between adjacent subunits, from
subunit and propagation to the whole TMD, driven by the M2 helices. Propagation would occur between adjacent subunits, from 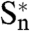 to Sn-1, similar to a domino chain. Simultaneously, the TMD changes elicit a global twist motion, whereas the ECDs twist in the opposite direction. These changes would lead to quaternary reorganization, initiated in the same subunit and then affecting the whole pentamer. This “domino model” might plausibly apply to the entire CLR family.
to Sn-1, similar to a domino chain. Simultaneously, the TMD changes elicit a global twist motion, whereas the ECDs twist in the opposite direction. These changes would lead to quaternary reorganization, initiated in the same subunit and then affecting the whole pentamer. This “domino model” might plausibly apply to the entire CLR family.
It is remarkable that the first events occur in the ECD–TMD interface of a single subunit as large amplitude fluctuations, later stabilized by the rest of the subunit, rather than the opposite scenario involving first a conformational change at the interface between subunits in the ECD, at the level of the known agonist binding site, that would subsequently propagate to the TMD to close the gate. In our simulation, the M2 fluctuations occur spontaneously, perhaps because agonists (protons) have been removed for all subunits simultaneously after the abrupt global pH change. It is not known for certain if these large fluctuations are directly caused by a protonation change of the whole subunit or if they are independent of the presence of the agonist. However, the observed sequence of events, namely, the tertiary relaxation after the large M2 movements, suggests that the latter are transiently occurring events, either in the presence or in the absence of agonist, whose binding then merely stabilizes and selects this new conformation by a “population shift” mechanism (see refs. 39 and 40).
More simulations with different starting points are needed to assess the universality of this mechanism but are beyond the scope of the present study.
Comparison with Experimental Data for the Cys-Loop Receptor Family.
It is difficult to confront the present simulation to electrophysiological data because they concern different time windows: nano- to microseconds for molecular dynamics simulations and tens of micro- to milliseconds for single channel recordings. Still, the model has several implications that appear to fit recent electrophysiological data.
First, even with the symmetric homopentameric GLIC channel, the transition starts with a specific motion of just one subunit initiating channel closing. This observation suggests that, in the case of heteromeric nAChRs such as the muscle type, gating may be initiated by any one particular subunit. Mutational analyses indicate that alpha subunit(s) mutations have stronger allosteric effects than at homologous positions in nonalpha subunits. This difference suggests that alpha subunits might be the initiating ones in this case (4).
Second, a major prediction of the simulation is that closure is initiated in the upper part of the channel. It is noteworthy that the three rings of hydrophobic residues that drive the process were shown to play a role in receptor gating, and their mutation into hydrophilic residues produces pleiotropic gain-of-function phenotypes in virtually all CLR subtypes (41). For GLIC, mutation I9′A strongly slows deactivation (11), a feature consistent with the idea that this process is facilitated through its interaction with hydrophobic side chains. In addition, by measuring the single channel opening and closing rate constants of numerous mutations of the muscle-type nAChR, Auerbach and colleagues proposed that structure of the transition state in the activation process resembles the open conformation in the ECD and that of the resting (closed) conformation in the TMD (27). This scheme suggests that activation is initiated in the ECD but conversely that deactivation is initiated in the TMD, because their mutation alters preferentially the closing rate constants; this is consistent with the present simulation.
Conclusion
The present work provides a possible atomistic model of the gating process for the bacterial GLIC receptor that may be valid for the whole CLR family. Gating occurs very rapidly with an initial closure event on the sub-0.1 μs time scale. A hydrophobic gate forms between the 9′ and 16′ pore-lining residues involving a global twist motion. An early local intrusion of the top of the M2 helix into the pore is observed, followed by a tertiary rearrangement of the whole subunit and propagation of these changes to the neighboring subunit. Extrapolation of this scenario might provide a model for the transition that can altogether be described as a two-step domino mechanism for channel gating, where closing is initiated by large fluctuations inside the pore. Further studies aiming at sampling the initial state at the time of the pH jump are necessary to consolidate these findings. In particular, the inverse experiment of opening a closed channel should be possible, provided that the issue of the precise protonation state of all aspartate and glutamate residues at pH 4.0–4.5 is settled. We are currently working on this problem that requires refined methods to predict side-chain pKas for membrane proteins in their native environment.
Methods
Molecular Dynamics Simulations.
Full methods are in SI Text including details about additional control simulations (Figs. S1 and S7). Molecular dynamics simulations were performed with GLIC inserted into a fully hydrated palmitoyloleoyl-phosphatidylcholine bilayer [content of the 108 Å × 106 Å × 168 Å simulation box: 5 protein chains, 307 lipids, 43,992 water molecules, and 54 Na+, 89(pH 4.6)/34(pH 7.0)Cl- ions]. The crystal structure was equilibrated at pH 4.6 during a 20-ns production run (13). Acidic side chains were then assigned partial charges according to standard deprotonated states at pH 7.0 and the production simulation of 1.06 μs was run.
Crystal Structure of the A13′F GLIC Mutant.
The A13′F mutant was produced as described (13). Datasets at a maximum resolution of 3.15 Å of single frozen crystals were recorded at Source Optimisée de Lumière d’Energie Intermédiaire du Laboratoire d’utilisation du rayonnement électromagnétique and European Synchrotron Radiation Facility. After integration of the spot intensities with XDS, scaling of batches with CCP4, and refinement with BUSTER, the final crystallographic factors are R = 21.5% and Rfree = 22.4%. The structure falls in the 93rd percentile among structures of comparable resolution according to the MolProbity validation server (see Table S2 for details).
Supplementary Material
Acknowledgments.
This work was performed by using high-performance computing resources from Grand Equipement National de Calcul Intensif Institut du développement et des resources en informatique scientifique (Grant 2009-072292 to M.D.). M.B. thanks the French Agency for Research for funding (Grant ANR-07-CIS7-003-01). H.N. is funded by the European Neurocypres project.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001832107/DCSupplemental.
Data deposition: Coordinates of the A13′F mutant have been deposited in the Protein Data Bank under accession number 3LSV.
References
- 1.Lester RA. Activation and desensitization of heteromeric neuronal nicotinic receptors: implications for non-synaptic transmission. Bioorg Med Chem Lett. 2004;14(8):1897–1900. doi: 10.1016/j.bmcl.2004.02.081. [DOI] [PubMed] [Google Scholar]
- 2.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440(7083):448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 3.Enna SJ, Moehler H. The GABA Receptors. Totowa, NJ: Humana Press; 2007. [Google Scholar]
- 4.Lee WY, Sine SM. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 2005;438(7065):243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- 5.Mukhtasimova N, Lee WY, Wang HL, Sine SM. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature. 2009;459(7245):451–454. doi: 10.1038/nature07923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lape R, Colquhoun D, Sivilotti LG. On the nature of partial agonism in the nicotinic receptor superfamily. Nature. 2008;454(7205):722–727. doi: 10.1038/nature07139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411(6835):269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 8.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat Neurosci. 2007;10(8):953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- 9.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346(4):967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 10.Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6(1):R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bocquet N, et al. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445(7123):116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 12.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452(7185):375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 13.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457(7225):111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 14.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457(7225):115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 15.Taly A, et al. Normal mode analysis suggests a quaternary twist model for the nicotinic receptor gating mechanism. Biophys J. 2005;88(6):3954–3965. doi: 10.1529/biophysj.104.050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu F, Hummer G. Gating transition of pentameric ligand-gated ion channels. Biophys J. 2009;97(9):2456–2463. doi: 10.1016/j.bpj.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X, Ivanov I, Wang H, Sine SM, McCammon JA. Molecular-dynamics simulations of ELIC—A prokaryotic homologue of the nicotinic acetylcholine receptor. Biophys J. 2009;96(11):4502–4513. doi: 10.1016/j.bpj.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Wang H, Grant B, Sine SM, McCammon JA. Targeted molecular dynamics study of C-loop closure and channel gating in nicotinic receptors. PLoS Comput Biol. 2006;2(9):e134. doi: 10.1371/journal.pcbi.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, et al. Mechanics of channel gating of the nicotinic acetylcholine receptor. PLoS Comput Biol. 2008;4(1):e19. doi: 10.1371/journal.pcbi.0040019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khelashvili G, Grossfield A, Feller SE, Pitman MC, Weinstein H. Structural and dynamic effects of cholesterol at preferred sites of interaction with rhodopsin identified from microsecond length molecular dynamics simulations. Proteins. 2009;76(2):403–417. doi: 10.1002/prot.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dror RO, et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci USA. 2009;106(12):4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjelkmar P, Niemelä PS, Vattulainen I, Lindahl E. Conformational changes and slow dynamics through microsecond polarized atomistic molecular simulation of an integral Kv1.2 ion channel. PLoS Comput Biol. 2009;5(2):e1000289. doi: 10.1371/journal.pcbi.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitch CA, Whitten ST, Hilser VJ, García-Moreno EB. Molecular mechanisms of pH-driven conformational transitions of proteins: insights from continuum electrostatics calculations of acid unfolding. Proteins. 2006;63(1):113–126. doi: 10.1002/prot.20797. [DOI] [PubMed] [Google Scholar]
- 24.Whitten ST, García-Moreno EB, Hilser VJ. Local conformational fluctuations can modulate the coupling between proton binding and global structural transitions in proteins. Proc Natl Acad Sci USA. 2005;102(12):4282–4287. doi: 10.1073/pnas.0407499102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graphics. 1996;14(6):354–360. doi: 10.1016/s0263-7855(97)00009-x. 376. [DOI] [PubMed] [Google Scholar]
- 26.Pascual JM, Karlin A. State-dependent accessibility and electrostatic potential in the channel of the acetylcholine receptor. Inferences from rates of reaction of thiosulfonates with substituted cysteines in the M2 segment of the alpha subunit. J Gen Physiol. 1998;111(6):717–739. doi: 10.1085/jgp.111.6.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purohit P, Mitra A, Auerbach A. A stepwise mechanism for acetylcholine receptor channel gating. Nature. 2007;446(7138):930–933. doi: 10.1038/nature05721. [DOI] [PubMed] [Google Scholar]
- 28.Beckstein O, Sansom MS. A hydrophobic gate in an ion channel: The closed state of the nicotinic acetylcholine receptor. Phys Biol. 2006;3(2):147–159. doi: 10.1088/1478-3975/3/2/007. [DOI] [PubMed] [Google Scholar]
- 29.Wilson GG, Karlin A. The location of the gate in the acetylcholine receptor channel. Neuron. 1998;20(6):1269–1281. doi: 10.1016/s0896-6273(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 30.Paas Y, et al. Pore conformations and gating mechanism of a Cys-loop receptor. Proc Natl Acad Sci USA. 2005;102(44):15877–15882. doi: 10.1073/pnas.0507599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panicker S, Cruz H, Arrabit C, Slesinger PA. Evidence for a centrally located gate in the pore of a serotonin-gated ion channel. J Neurosci. 2002;22(5):1629–1639. doi: 10.1523/JNEUROSCI.22-05-01629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corry B. An energy-efficient gating mechanism in the acetylcholine receptor channel suggested by molecular and Brownian dynamics. Biophys J. 2006;90(3):799–810. doi: 10.1529/biophysj.105.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haddadian EJ, Cheng MH, Coalson RD, Xu Y, Tang P. In silico models for the human alpha4beta2 nicotinic acetylcholine receptor. J Phys Chem B. 2008;112(44):13981–13990. doi: 10.1021/jp804868s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckstein O, Sansom MS. The influence of geometry, surface character, and flexibility on the permeation of ions and water through biological pores. Phys Biol. 2004;1(1–2):42–52. doi: 10.1088/1478-3967/1/1/005. [DOI] [PubMed] [Google Scholar]
- 35.Jha A, Purohit P, Auerbach A. Energy and structure of the M2 helix in acetylcholine receptor-channel gating. Biophys J. 2009;96(10):4075–4084. doi: 10.1016/j.bpj.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goren EN, Reeves DC, Akabas MH. Loose protein packing around the extracellular half of the GABA(A) receptor beta1 subunit M2 channel-lining segment. J Biol Chem. 2004;279(12):11198–11205. doi: 10.1074/jbc.M314050200. [DOI] [PubMed] [Google Scholar]
- 37.Bera AK, Akabas MH. Spontaneous thermal motion of the GABA(A) receptor M2 channel-lining segments. J Biol Chem. 2005;280(42):35506–35512. doi: 10.1074/jbc.M504645200. [DOI] [PubMed] [Google Scholar]
- 38.England PM, Zhang Y, Dougherty DA, Lester HA. Backbone mutations in transmembrane domains of a ligand-gated ion channel: Implications for the mechanism of gating. Cell. 1999;96(1):89–98. doi: 10.1016/s0092-8674(00)80962-9. [DOI] [PubMed] [Google Scholar]
- 39.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17(8):1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weikl TR, von Deuster C. Selected-fit versus induced-fit protein binding: Kinetic differences and mutational analysis. Proteins. 2009;75(1):104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 41.Revah F, et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature. 1991;353(6347):846–849. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.