Abstract
The three basic DNA-binding domain mutations of the microphthalmia associated transcription factor (Mitf), Mitfmi/mi, Mitfor/or and Mitfwh/wh, affect osteoclast differentiation with variable penetrance while completely impairing melanocyte development. Mitfor/or mice exhibit osteopetrosis that improves with age and their osteoclasts form functional multinuclear osteoclasts, raising the question as to why the Mitfor/or mutation results in osteopetrosis. Here we show that Mitfor/or osteoclasts express normal levels of acid phosphatase 5 (Acp5) mRNA and significantly lower levels of Cathepsin K (Ctsk) mRNA during receptor activator of nuclear factor kappa B (NFκB) ligand (RANKL)-mediated differentiation. Studies using chromatin immunoprecipitation (ChIP) analysis indicate that low levels of Mitfor/or protein are recruited to the Ctsk promoter. However, enrichment of Mitf-transcriptional co-activators PU.1 and Brahma-related gene 1 (Brg1) are severely impaired at the Ctsk promoter of Mitfor/or osteoclast precursors, indicating that defective recruitment of co-activators by the mutant Mitfor/or results in impaired Ctsk expression in osteoclasts. Cathepsin K may thus represent a unique class of Mitf regulated osteoclast-specific genes that are important for osteoclast function.
Keywords: Osteoclasts, Osteopetrosis, Microphthalmia-associated transcription factor, Acid phosphatase 5, Cathepsin K, Chromatin immunoprecipitation
Introduction
Regulation of osteoclast and osteoblast differentiation and function is vital to the maintenance of healthy bone homeostasis. Upon stimulation by osteoblast-derived chemokine RANKL, mononuclear myeloid precursors withdraw from cell cycle and fuse to become multinuclear bone-resorbing osteoclasts (Boyce et al., 2007; Sankar et al., 2004). Diminished osteoclast formation or function results in osteopetrosis or Albers-Schonberg disease, characterized by the accumulation of primary spongiosa in bone marrow cavities and the failure of incisors to erupt. In its severest form, the recessively inherited malignant or infantile osteopetrosis results in deafness, blindness, acute anemia and reduced immunity, and is often fatal without intervening bone marrow transplantation therapies. Benign osteopetrosis is dominantly inherited and characterized by frequent fractures of the brittle bone and poor healing.
Several irradiation and spontaneous osteopetrotic mouse mutants such as microphthalmia (mi), osteopetrosis (op/op) and grey-lethal (gl) have greatly advanced our understanding of molecular pathways that regulate osteoclast differentiation and function (Marks and Lane, 1976; Nii et al., 1995). The microphthalmia locus, located on murine chromosome 6p, encodes for a basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor called Mitf (Hallsson et al., 2000). The human MITF is mutated in families with Waardenburg syndrome type II (WS2) (Tassabehji et al., 1994). Mitf is closely related to its family members, Tfe3, TfeB and TfeC bHLH-Zip transcription factors and binds to E-box elements on promoters of target genes such as Acp5, Ctsk and E-Cadherin as homodimers or as heterodimers with other Mitf-family members, to drive target gene expression (Aksan and Goding, 1998; Luchin et al., 2000; Mansky et al., 2002b; Motyckova and Fisher, 2002). Recent studies have established that Mitf and the ets-family transcription factor PU.1 collaborate to enhance Acp5 and Ctsk transcription. RANKL relieves the association of Mitf and PU.1 with Eos, a zinc finger transcriptional repressor of the Ikaros family and other co-repressors at the promoters of Acp5 and Ctsk during osteoclast differentiation (Hu et al., 2007; Luchin et al., 2001).
The original microphthalmia mutation in mice (Mitfmi/mi) carries a 3-base pair deletion causing the loss of one of four arginines in the N-terminal DNA-binding basic domain of Mitf protein (ΔR215) (Moore, 1995). Osteoclast precursors from Mitfmi/mi mice do not fuse and the mutant mice exhibit severe osteopetrosis (Moore, 1995; Steingrimsson et al., 1994). Additionally, Mitfmi/mi mice have a white coat color and are deaf and blind due to impaired melanocyte development. The substitution of an arginine at position 216 in the basic domain with a lysine (R216K), results in the microphthalmia-oakridge (Mitfor/or) mutation. Mitfor/or mice originated from the progeny of a γ-irradiated male; exhibit white coat color, small eyes and osteopetrosis. Remarkably, the osteopetrosis in Mitfor/or mice improves (but not reversed) with age through an unknown mechanism (Nii et al., 1995). On the contrary, substitution of an isoleucine with asparagine (I212N) in the basic domain of Mitf, microphthalmia-white (Mitfwh/wh), results in a white coat color with no osteopetrosis, implying that unlike Mitfmi/mi and Mitfor/or, this basic domain mutation does not affect osteoclast differentiation (Moore, 1995). Mitfmi/mi, Mitfor/or and Mitfwh/wh, all mutations within the basic DNA-binding domain of Mitf, differently affect osteoclast differentiation while completely impairing melanocyte development. They form an allelic series that could provide crucial insights into the role of Mitf in osteoclast differentiation.
In melanocytes, Mitfmi/mi, Mitfor/or and Mitfwh/wh proteins are incapable of binding DNA as homo or heterodimers in electrophoretic mobility shift assays and act as dominant negative molecules blocking DNA binding by wild-type (WT) Mitf and Tfe3 proteins (Hemesath et al., 1994). These data beg the question of whether the differential effects of Mitfmi/mi, Mitfor/or and Mitfwh/wh proteins during osteoclast differentiation are due to differences in their DNA-binding properties or due to inabilities to recruit transcriptional co-activators to the promoters of osteoclast-specific genes upon RANKL stimulation in vivo. To address these questions, we first characterized the level of osteopetrosis in these mutants by performing careful histomorphometric analysis of Acp5-stained femur sections from newborn and 30-day old Mitfmi/mi, Mitfor/or and Mitfwh/wh mutants followed by in vitro differentiation and functional assays with osteoclast precursors derived from these mutants. Further, to correlate the level of osteopetrosis with changes in gene expression patterns, we examined the mRNA levels of Acp5 and Ctsk during in vitro differentiation of osteoclasts precursors derived from newborn and 30-day old Mitfmi/mi, Mitfor/or and Mitfwh/wh mutants using real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). Our data indicate that only Ctsk mRNA levels are significantly reduced in osteoclast precursors derived from both newborn and 30-day old Mitfor/or and Mitfmi/mi mice. We further assessed the recruitment of Mitf and its transcriptional co-activators to the Ctsk promoter in WT and Mitfor/or osteoclast precursors using ChIP assays. Data from ChIP analysis indicate recruitment of Mitfor/or to Ctsk promoter in osteoclasts. However, recruitment of the co-activators to the Ctsk promoter was significantly impaired. These data suggest that the defective recruitment of transcriptional co-activators by the mutant Mitfor/or is responsible for its inability to induce Ctsk transcription during osteoclast differentiation.
Materials and Methods
Mice and genotyping
All animals used in this study were housed and maintained under a 12-h light, 12-h dark cycle. Food and water were provided ad libitum and all care was given in compliance within NIH and Ohio State University Institutional Animal Care and Use Committee guidelines on the use of laboratory and experimental animals. Wild-type (WT), Mitfmi/mi, Mitfor/or and Mitfwh/wh were all in B6C3Fe background. Originally obtained from Jackson Laboratories (Bar Harbor, ME) these mice were genotyped based on the coat color phenotype of homozygous recessive and heterozygous mutants.
Osteoclast culture
Spleen cells from newborn (2-3 day old) mice and bone marrow cells from femurs of 30-day old or adult (3-month old) mice were cultured for three days in DMEM media with 10% heat inactivated fetal bovine serum containing 50ng/ml recombinant colony stimulating factor 1 (CSF1; a gift from David Hume, University of Queensland). The adherent cells were re-plated in differentiation media containing 50ng/ml of CSF1 and 100ng/ml of RANKL for appropriate assays. Preparation of RANKL was previously mentioned (Mansky et al., 2002a). Spleen or bone marrow cells from 4 mice were pooled (2 male and 2 female, in 30-day old or adult cohorts) in each experiment.
Acp5 staining and calcium phosphate resorption pit assays
Osteoclasts were assayed for Acp5 activity using Leukocyte Acid Phosphatase kit (Sigma, St. Louis, MO) according to manufacturer's protocol and counter-stained with 10μM bis-benzamide (Sigma). Multinuclear osteoclasts were counted using fluorescent microscopy and digital photography. An equal number of bone marrow-derived osteoclast precursors for each genotype tested was plated in differentiation medium onto BD Biocoat Osteologic 16-well multitest slides (BD Biosciences, San Jose, CA) according to manufacturer's protocol. After 12 days in differentiation media, osteoclasts were removed and the area, number and perimeter of the pits were measured using Bioquant Nova software (BIOQUANT Image Analysis Corporation, TN).
Radiography and histomorphometric analysis
The mouse skeletons were fixed in 3.7% paraformaldehyde (Polysciences, Inc, Warrington, PA) at 4°C for 48 h and transferred to 70% ethanol. Radiography was performed using a Faxitron X-ray machine (model 43855A; Hewlett Packard) at 35 kvp for 2 minutes. Glycol methacrylate infiltration and embedding of the decalcified femurs were performed using the JB-4 embedding kit (Polysciences) according to manufacturer's directions. Histomorphometric analyses on Acp5/hematoxylin stained sections (5 microns) were performed using Bioquant Nova Software as before.
Real-Time qRT-PCR
Total RNA was prepared using Trizol (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. To make the cDNA, 1 μg of RNA was incubated with 1× cDNA buffer, 1mM dNTPs, 20U of RNase inhibitor, 24U of Reverse Transcriptase (all from Roche Diagnostics, Indianapolis, IN) and 0.5μg random hexamers (Promega, Madison, WI), in a total volume of 20μl, at 50°C for 60 min and the reaction was frozen at −20°C until further use. For real-time qRT-PCR, the following primers and Taqman® probes (all from Applied Biosystems, Foster City, CA) were used: Acp5 forward 5′ GAT CTC CAA GCG CTG GAA CTT 3′; Acp5 reverse 5′ CAG TTA TGT TTG TAC GTG GAA TTT TGA 3′; Acp5 probe 6Fam-CCC AGC CCT TAC TAC CGT TTG CGC-Tamara; Cathepsin K forward 5′ ACC CAG TGG GAG CTA TGG AA 3′; Cathepsin K reverse 5′ TCC CAA ATT AAA CGC CGA GA 3′; Cathepsin K probe-6Fam-CAT CCA CCT TGC TGT TAT ACT GCT TCT GGT GA-Tamara and c-FMS forward 5′ CTG GAA TAA TCT GAC CTT TGA GCT C 3′; c-FMS reverse 5′ CGT CAC AGA ACA GGA CAT CAG AG 3′; c-FMS probe 5′ 6-Fam-CCT GCG ATA TCC CCC AGA GGT CAG TG-Tamara. GAPDH and 18S primers and probes were purchased from Applied Biosystems. The reactions were performed using Taqman Universal PCR mix or SYBR® Green PCR master mix (Applied Biosystems), according to manufacturer's directions. Real-Time q-PCR was performed in either the ABI PRISM 7700 sequence detection system (Applied Biosystems). Fold induction was calculated as the difference in cycle threshold (CT) values (ΔCT) between control and RANKL treated samples raised to the power of 2 (2ΔCT) (Livak and Schmittgen, 2001). These were further normalized against ΔCT values for GAPDH and 18S.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described earlier (Hu et al., 2007; Meadows et al., 2007; Sharma et al., 2007). Approximately 5 ×105 cell equivalent (1/6th) of the sheared soluble chromatin was pre-cleared with t-RNA blocked Protein G agarose, and 10% of the pre-cleared chromatin was set aside as input control. Immunoprecipitation was carried out with 5 μg of MITF, PU.1 or BRG1 antibodies (Sharma et al., 2007) overnight at 4°C. Immune complexes were pulled down using Protein G agarose, washed, de-cross linked and purified as described before (Hu et al., 2007; Meadows et al., 2007; Sharma et al., 2007). Samples were analyzed by real time PCR using specific probe sets from Roche universal probe library (Roche) and the Faststart TaqMan master PCR kit (Roche). The threshold for the promoter being studied was adjusted using input thresh hold values as reference values using Delta Delta Ct method of calculation (Livak and Schmittgen, 2001)and represented as relative enrichment.
Statistical analysis
We performed analysis of variance (ANOVA) for real-time qRT-PCR assays as previously described (Sankar et al., 2004). For all other experiments, the significance of differences between WT and mutant samples was tested by the Student's t-test, using Statview for Windows software, version 5.0.1 (SAS Institute). In all experiments, differences of p < 0.05 were deemed as significant.
Results
Osteopetrosis in Mitfor/or mice is milder than Mitfmi/mi and improves with age
To understand the effect of the three basic domain mutations in Mitf on osteoclast differentiation, we performed radiographic and histomorphometric analyses of Acp5 stained femur sections from newborn (3-4 day old) and 30-day old WT, Mitfwh/wh, Mitfmi/mi and Mitfor/or mice (Figure 1). At 3-4 days post-natal, the growth of the long bone is very rapid and any defect in osteoclast differentiation and function is highly discernible. Radiographic examination revealed that sclerotic lesions were present primarily in the mid-diaphysis of the newborn Mitfmi/mi and Mitfor/or and 30-day old Mitfmi/mi femurs (Figure 1A), indicating severe osteopetrosis. In contrast, the lesions presented shifted towards the distal metaphysis of the 30-day old Mitfor/or femurs (Figure 1A), indicating an improvement in the osteopetrosis that was observed in Mitfor/or newborn mice. Microscopic analysis of Acp5 and hematoxylin stained femur sections (Figure 1A) also show unresorbed endochondral trabeculae mainly in the diaphysis of newborn Mitfmi/mi and Mitfor/or and 30-day old Mitfmi/mi femurs and in the distal metaphysis of 30-day old Mitfor/or femurs.
Figure 1. Mitfor/or mice exhibit osteopetrosis that improves with age.
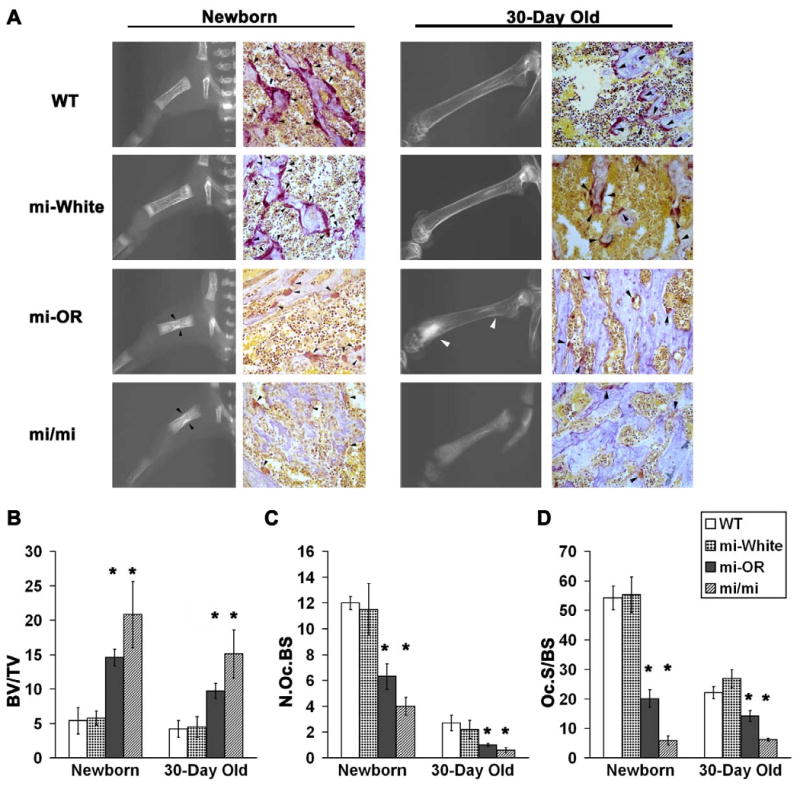
A. Representative (see below) radiographic images of long bones (left in each panel) and digital microscopic images of Acp5 and hematoxylin stained 5 μM femur sections(right in each panel) from newborn (Left panel) and 30-day old (Right panel) WT, Mitfwh/wh (mi-White), Mitfor/or (mi-OR) and Mitfmi/mi (mi/mi) mice are shown. Black arrow heads in newborn Mitfor/or and Mitfmi/mi radiographs indicate sclerotic lesions in mid-diaphysis and white arrowheads in 30-day old Mitfor/or radiographs indicate sclerotic lesions in distal metaphysis of femurs. Black arrowheads in the Acp5 stained sections indicate osteoclasts positive for Acp5 enzyme activity. B-D. Bone and osteoclast parameters measured by histomorphometric analysis of Acp5 bone sections from new born and 30-day old WT, Mitfwh/wh (mi-White), Mitfor/or (mi-OR) and Mitfmi/mi (mi/mi) mice are shown. B: BV/TV indicates the percentage of unresorbed trabecular bone volume to total bone surface; C:- N.OcS/BS indicates the number of osteoclasts per total bone surface and D:- Oc.S/BS indicates the percentage of osteoclast surface to total bone surface. For all three analyses, averages of n≥30 for newborn mice and n=20 for 30-day old mice in the respective genotypes is shown. * indicates statistical significance with p < 0.05, Student's t-test.
Histomorphometric analyses of the Acp5-stained sections were performed using three parameters: the percentage of unresorbed trabecular bone volume in the femur relative to the total bone volume (BV/TV; Figure 1B), the number of osteoclasts per total bone surface (N.Oc/BS; Figure 1C) and the percentage of osteoclast surface to total bone surface: a measure of osteoclast size and function (Oc.S/BS; Figure 1D). Overall, while both newborn and 30-day old Mitfmi/mi and Mitfor/or had significantly (p < 0.5) higher percentage of unresorbed trabeculae in their long bones compared to WT and Mitfwh/wh, the resorption in Mitfor/or was slightly better than that in Mitfmi/mi. Thus, the BV/TV in newborn Mitfor/or and Mitfmi/mi was 3-fold and 4-fold higher respectively, than that in newborn WT and Mitfwh/wh. Similarly, BV/TV was 2-fold and 3-fold higher respectively in 30-day old Mitfor/or and Mitfmi/mi than that in WT and Mitfwh/wh (Figure 1B).
Acp5-positive osteoclasts with ruffled borders were found attached to the trabecular surface in newborn and 30-day old from WT and Mitfwh/wh femurs. In contrast, femurs from newborn and 30-day old Mitfmi/mi mice had significantly fewer osteoclasts that were significantly smaller in size with weak Acp5 activity compared to WT and Mitfwh/wh (Figures 1A, 1C and 1D). As shown in Figure 1D, the percentage of Oc.S/BS was 9 fold lower in newborn and 3.5 fold lower in 30-day old Mitfmi/mi femurs compared to WT controls. Further, the number of osteoclasts and the total osteoclast surface in Mitfor/or were significantly lower than WT and Mitfwh/wh, but higher than those in age-matched Mitfmi/mi (Figures 1C and 1D). The percentage Oc.S/BS, indicative of osteoclast function, was 2.8 lower in newborn and 1.4 fold lower in 30-day old Mitfor/or than in age-matched WT femurs (Figure 1D). Together, these data present quantitative evidence to the observation that Mitfor/or mice exhibit milder osteopetrosis than that seen in Mitfmi/mi.
Osteoclast precursors from Mitfmi/mi and Mitfor/or mice form fewer functional multinuclear osteoclasts in vitro
Femurs from Mitfor/or mice contained multinuclear osteoclasts albeit in significantly fewer numbers (Figure 1A and 1C). Unlike Mitfmi/mi, Mitfor/or osteoclasts possess ruffled borders (Nii et al., 1995) and yet Mitfor/or mice exhibit mild osteopetrosis at 30-days of age, indicating that their osteoclasts are less functional in general. To investigate whether the osteoclast precursors from newborn and 30-day old mice Mitfor/or were capable of forming functional, multinuclear osteoclasts in vitro, we plated osteoclast precursors from newborn and 30-day old WT, Mitfwh/wh, Mitfor/or and Mitfmi/mi mice on gelatin-coated wells to enable the formation of multinuclear osteoclasts (Figures 2 Ai and Bi) or calcium phosphate coated wells for the formation of resorption pits by functional osteoclasts (Figures 2 Aii and Bii), in media containing CSF1 and RANKL. Further, the numbers of multinuclear osteoclasts and resorption pits were quantified and tabulated (Tables 1 and 2). Precursors from newborn and 30-day old Mitfmi/mi and Mitfor/or mice did not form any multinuclear osteoclasts with more than 5 nuclei and formed significantly fewer multinuclear osteoclasts with 3-5 nuclei than WT and Mitfwh/wh (Table 1). However, precursors from 30-day old Mitfor/or mice formed multinuclear osteoclasts with 3-5 nuclei at numbers significantly higher (3.4 fold) than Mitfmi/mi but significantly lower (5 fold) than those formed by WT or Mitfwh/wh precursors (Table 1). Thus Mitfor/or precursors formed significantly fewer multinuclear osteoclasts than WT and were unable to form multinuclear osteoclasts with more than 6 nuclei.
Figure 2. Mitfor/or precursors form fewer functional multinuclear osteoclasts than WT in vitro.
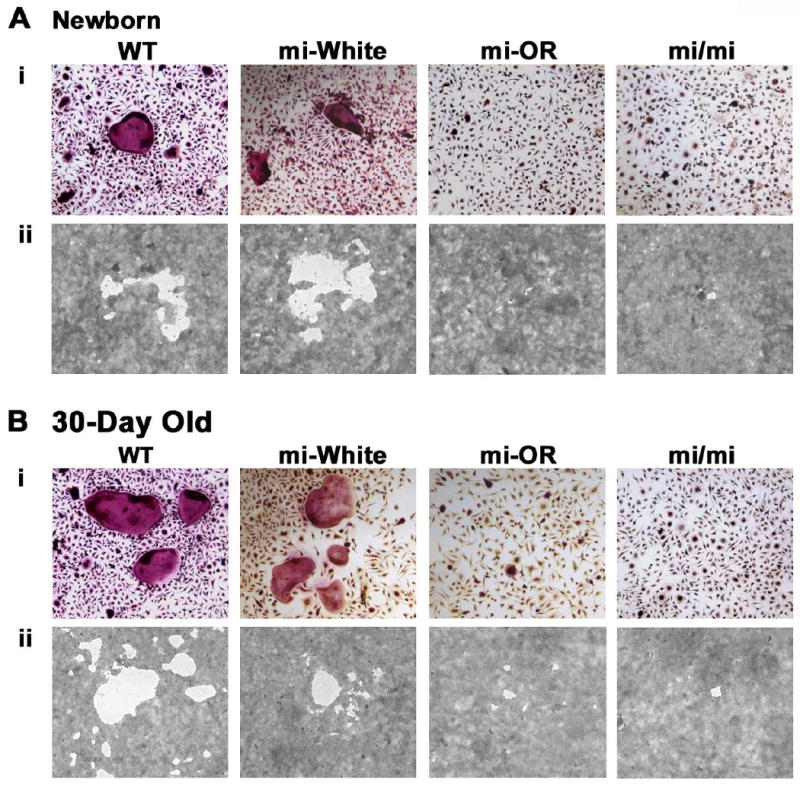
Precursors from either newborn (A) or 30-day old mice (B) of the indicated genotypes were cultured in the presence of CSF-1 and RANKL on gelatin coated wells for the formation of multinuclear osteoclasts (Ai and Bi) or on calcium phosphate coated wells for the formation of resorption pits (Aii and Bii). Representative digital microscopic images of Acp5 stained osteoclasts derived from newborn (Ai) or 30-day old (Bi) mice of the indicated genotypes. Figures Aii and Bii show representative digital microscopic images of resorption pits made by osteoclasts, from indicated genotype and age. Quantitation of the differentiation and functional assays is summarized in tables 1 and 2.
Table 1. Quantitation of Multinuclear Osteoclasts (No. of cells/mm2).
| Newboirn | 30-Day Old | |||||||
|---|---|---|---|---|---|---|---|---|
| WT | miWhite | miOR | mi/mi | WT | miWhite | miOR | mi/mi | |
| >12 nuclei | 0 | 0 | 0 | 0 | 30±11.12 | 13±1.11 | 0 | 0 |
| 6-12 nuclei | 2±1.11 | 7±1.11 | 0 | 0 | 58±6.67 | 23±1.11 | 0 | 0 |
| 3-5 nuclei | 108±12.2 | 119±3.3 | 13±4.4* | 13±1.1* | 582±18.9 | 681±26.7 | 1116±5.6* | 34±3.3* |
Average of n=3;
p<0.05, Student's t-test
Table 2. Quantitation of Osteoclast Resorption pits.
| Newborn | 30-Dav Old | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | mi-White | Mi-OR | mi/m | WT | mi-White | mi-OR | mi/mi | |||||||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | |
| Total no. of pits | 417 | 438 | 544 | 559 | 34 | 44 | 30 | 27 | 1895 | 2009 | 997 | 1323 | 94 | 129 | 58 | 55 |
| Area distribution@ | ||||||||||||||||
| <100 | 80 | 65 | 34 | 43 | 9 | 15 | 11 | 9 | 752 | 823 | 264 | 425 | 15 | 21 | 10 | 14 |
| 100-250 | 58 | 72 | 29 | 80 | 8 | 10 | 7 | 6 | 259 | 126 | 124 | 245 | 14 | 19 | 13 | 20 |
| 250-1000 | 158 | 114 | 102 | 98 | 14 | 18 | 11 | 12 | 613 | 928 | 303 | 400 | 47 | 75 | 27 | 18 |
| 1000-10,000 | 104 | 152 | 231 | 240 | 3 | 1 | 1 | 0 | 230 | 112 | 262 | 201 | 17 | 12 | 8 | 3 |
| > 10,000 | 17 | 35 | 148 | 98 | 0 | 0 | 0 | 0 | 41 | 20 | 44 | 52 | 1 | 2 | 0 | 0 |
Data from 2 individual experiments is shown
Area in square pixels
Furthermore, results from calcium phosphate resorption experiments indicate that osteoclast precursors from newborn Mitfwh/wh mice formed the highest number of pits with the largest range of surface area, while precursors from newborn Mitfmi/mi and Mitfor/or mice formed fewer pits with much smaller range of surface area, less than 10 fold of the resorption pit surface area generated by WT osteoclasts (Table 2). Further, the number and size of pits formed by newborn Mitfor/or and Mitfmi/mi osteoclasts were similar. In contrast, the number and surface area of pits formed by precursors from 30-day old Mitfor/or mice were 2-fold higher than those formed by 30-day old Mitfmi/mi mice. However, these numbers were still significantly lower than those formed by from 30-day old WT and Mitfwh/wh mice (Table 2). Collectively, these results indicate Mitfor/or osteoclast precursors are fundamentally defective in osteoclast differentiation and function.
Osteoclast precursors from newborn and 30-day old Mitfor/or mice express lower levels of Cathepsin K and normal levels of Acp5 mRNA
To determine the genetic and temporal effect of Mitf mutations on osteoclast function, we examined the expression patterns of osteoclast-specific genes regulated by Mitf such as Ctsk and Acp5 in osteoclasts derived from newborn and 30-day old mice, following stimulation with RANKL. As shown in Figures 3A and 3D, Acp5 mRNA levels are similar in differentiating osteoclasts generated from newborn and 30-day old WT, Mitfwh/wh, and Mitfor/or mice, about 7-10 fold higher than that of untreated controls at both 24 or 72 h of RANKL treatment. However, Acp5 levels are only increased by 2-4-fold in osteoclasts from newborn and 30-day old Mitfmi/mi mice (Figures 3A and 3D). These data indicate that Acp5 gene expression is not affected by the Mitfor/or mutation. In contrast to Acp5, Ctsk mRNA levels in osteoclasts from newborn Mitfor/or and Mitfmi/mi mice were always two-fold lower than those in WT and Mitfwh/wh osteoclasts, at both 24 and 72 h after RANKL treatment (Figure 3B). These differences were more pronounced in differentiating osteoclasts from 30-day old Mitfor/or and Mitfmi/mi, where the induction of Ctsk at 72 h after RANKL treatment was 3 fold and 4.5 fold lower than WT, respectively (Figure 3E). The expression level of c-fms, the gene encoding the CSF1 receptor, was included as an internal control for the changes in gene expression in osteoclast precursors treated with RANKL (Figures 3C and 3F). Further we also examined the levels of Acp5 and Ctsk in osteoclast precursors from adult (3 month old) WT and Mitfor/or mice. Our results indicate that compared to the levels in WT, Acp5 expression levels were 5.5±0.3 and 4.7±0.1 fold and Ctsk levels were 1.1±0.3 and 0.9±0.5 fold respectively, at 24 and 72 h after adding RANKL (data not shown). Taken together, these results indicate that Mitfor/or osteoclasts express Acp5 mRNA at levels similar to those expressed by WT and Mitfwh/wh osteoclasts and that the level of Acp5 expression in these cells does not change with age. However, Ctsk expression levels are significantly lower in Mitfor/or osteoclasts compared to WT and Mitfwh/wh at all time points and age groups and these levels were similar to those in Mitfmi/mi osteoclasts.
Figure 3. Mitfor/or osteoclasts express normal levels of Acp5 mRNA and significantly lower levels of Cathepsin K.
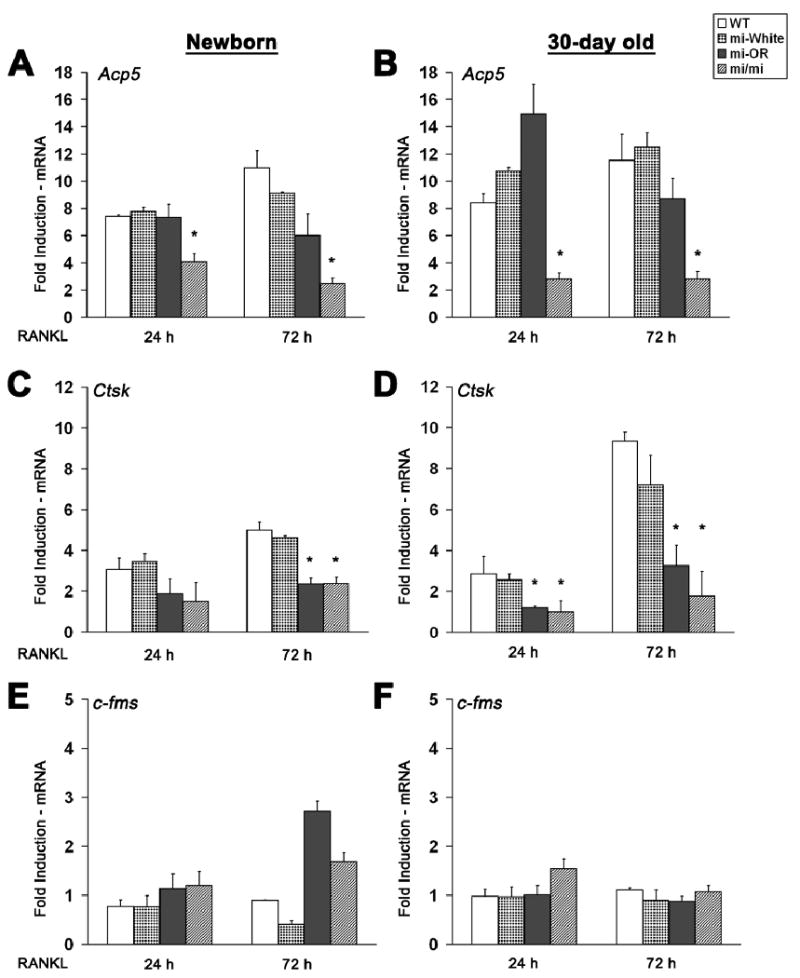
Average induction of Acp5 (A and D), Ctsk (B and E) and c-fms (C and F) mRNA levels in osteoclasts derived from newborn (left panels) or 30-day old (right panels) mice of indicated genotypes, at 24 or 72 hours in the presence of RANKL is shown. Average data (n=3), normalized to corresponding GAPDH and 18S mRNA levels, indicated as fold induction in the expression of each gene over CSF1 only treated (undifferentiated) controls, shown. Error bars indicate standard deviation and * indicates statistical significance (p<0.05, ANOVA).
Osteoclasts derived from Mitfor/or exhibit recruitment of Mitf, but not its co-activators, to Ctsk promoter during differentiation
To better understand the mechanism by which Mitfor/or mutant osteoclasts show defective activation of Ctsk gene expression, we examined the possibility of reduced Mitf recruitment to Ctsk promoter by ChIP analysis. Briefly, we immunoprecipitated the soluble chromatin from WT, Mitfwh/wh, Mitfor/or and Mitfmi/mi osteoclast precursors with Mitf antibody, at 0 and 72 h after treatment with RANKL. As expected, Mitf was highly enriched at the Ctsk promoter of WT and Mitfwh/wh cells, both prior to and at 72 h after RANKL treatment (Figure 4A), confirming our previous findings (Sharma et al., 2007). Mitfor/or and Mitfmi/mi proteins were both bound to the Ctsk promoter, both at 0 and 72 h after RANKL treatment (Figure 4A). However, the enrichment of Mitf at the Ctsk promoter of Mitfor/or and Mitfmi/mi osteoclast precursors was 2-4 fold (p < 0.05) lower than the corresponding enrichment at the Ctsk promoter of WT or Mitfwh/wh cells, both before and 72 h after treatment with RANKL (Figure 4A). These data indicate that the lower Ctsk expression in Mitfor/or osteoclasts is not due to an inability of the mutant Mitf protein to be recruited to the promoter.
Figure 4. Defective enrichment of PU.1 and Brg1 proteins at the Ctsk promoter in Mitfor/or osteoclasts.
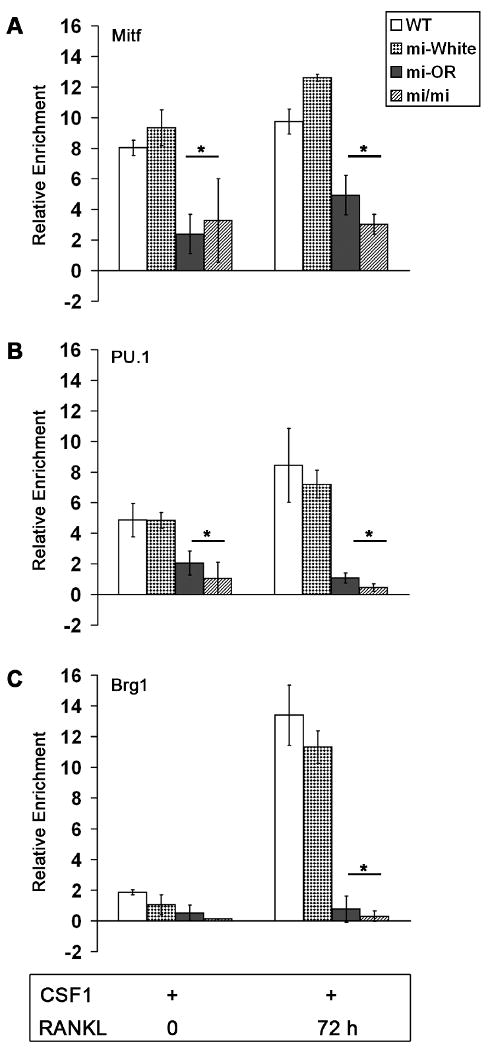
Average enrichment (n=3) of Mitf (A), PU.1 (B) and Brg1 (C) proteins at the Ctsk promoter in WT, Mitfwh/wh, Mitfor/or and Mitfmi/mi osteoclast precursors treated with CSF-1 alone or with CSF1 and RANKL for 72 h assayed by ChIP analysis. Error bars in all panels represent standard deviation and * indicates statistical significance (p<0.05, Student's t-test).
Next we investigated whether Mitfor/or is impaired in its recruitment of transcriptional co-activators of Mitf to Ctsk promoter. The myeloid transcription factor PU.1 and the ATPase-dependent chromatin remodeling subunit Brg1 conspire with Mitf to enable transcriptional activation of Ctsk in osteoclast precursors, following stimulation with RANKL (Sharma et al., 2007). As shown in Figure 4B, PU.1 was already bound at the Ctsk promoter, prior to RANKL treatment, in WT, Mitfwh/wh and Mitfor/or osteoclasts. However, the enrichment of PU.1 at the Ctsk promoter of WT and Mitfwh/wh osteoclasts was enhanced by 2-fold following treatment with RANKL. In contrast, the levels of PU.1 on Ctsk promoter of Mitfor/or osteoclasts remained unchanged following RANKL stimulation such that they were a significant 3-fold lower than the corresponding levels in WT cells (Figure 4B). Further, levels of PU.1 on Ctsk promoter of Mitfmi/mi osteoclasts were also significantly lower (8-fold) than those in WT and Mitfwh/wh osteoclasts (Figure 4B).
Enrichment of Brg1 on Ctsk promoter increased by 6-fold in WT and Mitfwh/wh osteoclasts following RANKL stimulation (Figure 4C) where as, levels of Brg1 at the Ctsk promoter of Mitfor/or osteoclast precursors was significantly lower (4-fold) than that in WT cells, following RANKL treatment (Figure 4C). In fact, results from multiple experiments indicate that the Mitf co activator Brg1 is not recruited to the Ctsk promoter of Mitfor/or or Mitfmi/mi osteoclasts following stimulation with RANKL (Figure 4C). Taken together, our data indicate that the lower Ctsk gene expression levels in Mitfor/or osteoclasts are due to defective recruitment of the co-activators PU.1 and Brg1 by the mutant Mitf protein.
Discussion
Mitfmi/mi, Mitfor/or and Mitfwh/wh, all mutations within the basic domain of the Mitf protein, form an allelic series that differentially influence osteoclast differentiation. Mitfmi/mi mutation results in severe osteopetrosis as the mutant mice harbor only mononuclear osteoclasts. In contrast, the Mitfwh/wh mice contain functional, multinuclear osteoclasts similar to WT, indicating that this Mitf mutation does not affect osteoclast differentiation. More interestingly, Mitfor/or mice exhibit a form of osteopetrosis that is milder than that in Mitfmi/mi mice. Studies from our and other (Nii et al., 1995) groups indicate that the Mitfor/or mice do possess multinuclear osteoclasts, albeit at significantly lower numbers compared to WT. Thus, precursors from Mitfor/or mice form multinuclear osteoclasts, both in vivo and in vitro, at numbers slightly higher than those formed by Mitfmi/mi precursors, but significantly lower than those formed by precursors from age-matched WT mice. Furthermore, the number, and surface area of osteoclasts and the percentage of unresorbed bone area in the long bones of 30-day old Mitfor/or mice were intermediate between those in 30-day old Mitfmi/mi and WT mice. In fact, osteopetrosis in Mitfor/or mice improves, but does not completely resolve with age.
Mitf-regulated gene expression data indicates that while Mitfor/or mice express Acp5 mRNA at levels similar to WT, they express significantly lower levels of Ctsk mRNA. Acp5 is an early response gene during osteoclast differentiation whose expression is upregulated 6-7 fold within 30 minutes of adding RANKL (Mansky et al., 2002a). Though the precise role played by Acp5 in osteoclast differentiation is unclear, Acp5 overexpression leads to mild osteoporosis and its genetic ablation results in mild, late onset osteopetrosis in transgenic mice (Angel et al., 2000; Hayman et al., 1996). The fact that Acp5 is expressed at WT levels in Mitfor/or osteoclasts derived from mice of all ages suggests that the expression of early response genes like Acp5 may not be very critical for osteoclast differentiation and function. This suggestion is supported by the milder phenotype of Acp5 knockout mice.
In contrast, Ctsk is a late response gene in osteoclasts whose expression is significantly upregulated only by 72 hours after treatment with RANKL (Figures 3B and 3E). Mice deficient in Ctsk exhibit osteopetrosis due the inability of the multinuclear osteoclasts to degrade the collagen matrix of the bone (Gowen et al., 1999; Saftig et al., 2000; Saftig et al., 1998). Mitfor/or osteoclasts express lower levels of Ctsk and form very few functional, multinuclear osteoclasts in vitro. This suggests that the late response gene Ctsk is important for osteoclast function in vitro and in vivo. Thus, Mitf-regulated genes may be categorized into two classes based on the significance of their roles during osteoclast differentiation and function: class 1 consisting of early response genes similar to Acp5 that are less critical; and class 2 consisting of late responsive genes like Ctsk that are more critical for osteoclast differentiation and function.
In melanocytes, Mitfor/or and Mitfmi/mi proteins inhibit DNA binding by WT-Mitf and Tfe3 and consequently act as dominant negative proteins (Hemesath et al., 1994). The story is more complicated in osteoclasts. The expression levels of Acp5 and Ctsk, two genes that are directly regulated by Mitf, are different in Mitfor/or osteoclasts, raising the possibility that one or more modifiers are enabling transcription from Acp5 promoter in Mitfor/or osteoclasts. It is now well established that Mitf and PU.1 are capable of activating Acp5 and Ctsk promoters in a synergistic manner and that the mutant Mitfmi/mi blocks the transactivation of Acp5 by PU.1 (Luchin et al., 2001; Sharma et al., 2007). These data suggest that PU.1 is one of the modifiers in Mitfor/or osteoclasts that enables the transcriptional activation of Acp5 promoter and that Mitfor/or does not act as a strong dominant negative with PU.1 as Mitfmi/mi on Acp5 promoter.
Our data provide a potential explanation for the lower Ctsk expression levels in Mitfor/or osteoclasts. PU.1 is a key transcriptional co-activator of Mitf for Ctsk expression, suggesting the possibility that Mitf-PU.1 interactions and the subsequent recruitment of chromatin remodeling complexes to Ctsk promoter are hampered in Mitfor/or osteoclasts,. Consistent with this, data from our ChIP analysis indicates that the recruitment of PU.1 and the chromatin remodeling protein Brg1 by Mitfor/or to Ctsk promoter are severely impaired, accounting for the inability of the mutant protein to successfully induce Cathepsin K gene expression in osteoclasts.
In conclusion, the data from this study indicates that Mitfor/or mice exhibit osteopetrosis that improves with age and that their osteoclasts form fewer functional multinuclear osteoclasts in vitro. Mitfor/or osteoclasts express Acp5 at levels similar to WT. But defective recruitment of transcriptional co-activator PU.1 and components of the SWI/SNF chromatin-remodeling complex such as Brg1 by the mutant Mitfor/or protein to the Ctsk promoter during osteoclast differentiation results in significantly reduced Ctsk gene expression. Taken together, our results indicate that Cathepsin K belongs to a class of Mitf regulated late responsive osteoclast-specific genes that are more important for osteoclast function.
Acknowledgments
This work was supported by NIAMS, National Institutes of Health Grant R01-AR-0447129 to MCO. The authors thank Krupen Patel for technical assistance and the members of the Ostrowski lab for helpful discussions.
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), National Institutes of Health Grant R01-AR-0447129 to MCO.
Literature Cited
- Aksan I, Goding CR. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol. 1998;18(12):6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel NZ, Walsh N, Forwood MR, Ostrowski MC, Cassady AI, Hume DA. Transgenic mice overexpressing tartrate-resistant acid phosphatase exhibit an increased rate of bone turnover. J Bone Miner Res. 2000;15(1):103–110. doi: 10.1359/jbmr.2000.15.1.103. [DOI] [PubMed] [Google Scholar]
- Boyce BF, Yao Z, Zhang Q, Guo R, Lu Y, Schwarz EM, Xing L. New roles for osteoclasts in bone. Annals of the New York Academy of Sciences. 2007;1116:245–254. doi: 10.1196/annals.1402.084. [DOI] [PubMed] [Google Scholar]
- Gowen M, Lazner F, Dodds R, Kapadia R, Feild J, Tavaria M, Bertoncello I, Drake F, Zavarselk S, Tellis I, Hertzog P, Debouck C, Kola I. Cathepsin K knockout mice develop osteopetrosis due to a deficit in matrix degradation but not demineralization. J Bone Miner Res. 1999;14(10):1654–1663. doi: 10.1359/jbmr.1999.14.10.1654. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Favor J, Hodgkinson C, Glaser T, Lamoreux ML, Magnusdottir R, Gunnarsson GJ, Sweet HO, Copeland NG, Jenkins NA, Steingrimsson E. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics. 2000;155(1):291–300. doi: 10.1093/genetics/155.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman AR, Jones SJ, Boyde A, Foster D, Colledge WH, Carlton MB, Evans MJ, Cox TM. Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disrupted endochondral ossification and mild osteopetrosis. Development. 1996;122(10):3151–3162. doi: 10.1242/dev.122.10.3151. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hu R, Sharma SM, Bronisz A, Srinivasan R, Sankar U, Ostrowski MC. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol Cell Biol. 2007;27(11):4018–4027. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Methods. 4. Vol. 25. San Diego, Calif: 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method; pp. 402–408. [DOI] [PubMed] [Google Scholar]
- Luchin A, Purdom G, Murphy K, Clark MY, Angel N, Cassady AI, Hume DA, Ostrowski MC. The microphthalmia transcription factor regulates expression of the tartrate-resistant acid phosphatase gene during terminal differentiation of osteoclasts. J Bone Miner Res. 2000;15(3):451–460. doi: 10.1359/jbmr.2000.15.3.451. [DOI] [PubMed] [Google Scholar]
- Luchin A, Suchting S, Merson T, Rosol TJ, Hume DA, Cassady AI, Ostrowski MC. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. The Journal of biological chemistry. 2001;276(39):36703–36710. doi: 10.1074/jbc.M106418200. [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. The Journal of biological chemistry. 2002a;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sulzbacher S, Purdom G, Nelsen L, Hume DA, Rehli M, Ostrowski MC. The microphthalmia transcription factor and the related helix-loop-helix zipper factors TFE-3 and TFE-C collaborate to activate the tartrate-resistant acid phosphatase promoter. J Leukoc Biol. 2002b;71(2):304–310. [PubMed] [Google Scholar]
- Marks SC, Jr, Lane PW. Osteopetrosis, a new recessive skeletal mutation on chromosome 12 of the mouse. J Hered. 1976;67(1):11–18. doi: 10.1093/oxfordjournals.jhered.a108657. [DOI] [PubMed] [Google Scholar]
- Meadows NA, Sharma SM, Faulkner GJ, Ostrowski MC, Hume DA, Cassady AI. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. J Biol Chem. 2007;282(3):1891–1904. doi: 10.1074/jbc.M608572200. [DOI] [PubMed] [Google Scholar]
- Moore KJ. Insight into the microphthalmia gene. Trends Genet. 1995;11(11):442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- Motyckova G, Fisher DE. Pycnodysostosis: role and regulation of cathepsin K in osteoclast function and human disease. Curr Mol Med. 2002;2(5):407–421. doi: 10.2174/1566524023362401. [DOI] [PubMed] [Google Scholar]
- Nii A, Steingrimsson E, Copeland NG, Jenkins NA, Ward JM. Mild osteopetrosis in the microphthalmia-oak ridge mouse. A model for intermediate autosomal recessive osteopetrosis in humans. Am J Pathol. 1995;147(6):1871–1882. [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Hunziker E, Everts V, Jones S, Boyde A, Wehmeyer O, Suter A, von Figura K. Functions of cathepsin K in bone resorption. Lessons from cathepsin K deficient mice. Adv Exp Med Biol. 2000;477:293–303. doi: 10.1007/0-306-46826-3_32. [DOI] [PubMed] [Google Scholar]
- Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A. 1998;95(23):13453–13458. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankar U, Patel K, Rosol TJ, Ostrowski MC. RANKL coordinates cell cycle withdrawal and differentiation in osteoclasts through the cyclin-dependent kinase inhibitors p27KIP1 and p21CIP1. J Bone Miner Res. 2004;19(8):1339–1348. doi: 10.1359/JBMR.040321. [DOI] [PubMed] [Google Scholar]
- Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S, Ostrowski MC. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J Biol Chem. 2007;282(21):15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D'Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8(3):256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8(3):251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]


