Abstract
Mutations in the genes encoding the RNaseH2 and TREX1 nucleases have been identified in patients with Aicardi-Goutieres syndrome (AGS). To determine if the AGS RNaseH2 mutations result in the loss of nuclease activity, the human wild-type RNaseH2 and four mutant complexes that constitute the majority of mutations identified in AGS patients have been prepared and tested for ribonuclease H activity. The heterotrimeric structures of the mutant RNaseH2 complexes are intact. Furthermore, the ribonuclease H activities of the mutant complexes are indistinguishable from the wild-type enzyme with the exception of the RNaseH2 subunit A (Gly37Ser) mutant, which exhibits some evidence of altered nuclease specificity. These data indicate that the mechanism of RNaseH2 dysfunction in AGS cannot be simply explained by loss of ribonuclease H activity and points to a more complex mechanism perhaps mediated through altered interactions with as yet identified nucleic acids or protein partners.
Keywords: Aicardi-Goutieres syndrome, RNaseH2, Nuclease, Immune activation
Introduction
Aicardi-Goutieres syndrome (AGS) is a rare genetic encephalopathy that phenotypically mimics congenital viral infection characterized by elevated interferon-α levels in cerebrospinal fluid [1–3]. Specific genetic defects that cause AGS have now been identified in four genes that encode two nucleic acid metabolizing enzymes [4, 5]. The three genes RNASEH2A, RNASEH2B, and RNASEH2C encode the heterotrimeric RNaseH2 [5, 6] and the fourth gene TREX1 encodes the homodimeric 3′→5′ deoxyribonuclease [7–10]. A comprehensive molecular analysis of the gene mutations found in AGS, coupled with the clinical evaluations of these patients, has delineated a genotype-phenotype correlation of two distinguishable clinical presentations of AGS. An early-onset neonatal form with clinical features similar to congenital infection is seen mostly with TREX1 mutations, and a later onset form of presentation after a significant period of normal development is most frequently due to RNASEH2B mutations. These genetic defects in AGS are mostly inherited as recessive alleles, but dominant alleles have also been reported [11].
There is clinical and genetic overlap between AGS, familial chilblain lupus (FCL), systemic lupus erythematosus (SLE), and retinal vasculopathy and cerebral leukodystrophy (RVCL). Chilblain lesions have been reported in AGS patients [12], and these lesions are associated with mutations in the three RNaseH2 genes and in TREX1 [13]. Chilblain lesions have also been described in patients with dominant TREX1 mutations that cause AGS [11] and in dominant TREX1 mutations that cause an inherited form of lupus erythematosus called FCL [11, 14, 15]. Skin lesions are observed in SLE patients with TREX1 mutations [16], and diminished vasculature integrity in the retina and cerebral dysfunction has been described in patients with TREX1 C-terminal frame shift mutations that cause RVCL [17, 18]. These skin and blood vessel lesions characteristic of this spectrum of genetically related disorders point to a common molecular pathway related to dysfunctional nucleic acid processing leading to immune activation and interferon-induced inflammatory vasculopathy.
Nucleases process DNA and RNA polynucleotides to prevent these macromolecules from inappropriately activating the immune system. The connection between TREX1 and immune activation was first indicated in Trex1 null mice that develop inflammatory myocarditis consistent with autoimmune disease [19]. TREX1 acts with the NM23-H1 nuclease to degrade DNA in a cell death pathway [20]. We have proposed that TREX1 degrades DNA by acting at nicked double-stranded DNA [20, 21], and others have proposed that TREX1 degrades single-stranded DNA generated from the processing of aberrant replication intermediates [22] or derived from endogenous retroelements [23]. The RNase H enzymes recognize and cleave ribonucleotides present in RNA/DNA duplexes [24], but the precise RNA/DNA target of the human RNaseH2 has not been identified. The connection between RNaseH2 and TREX1 mutations and the apparent innate immune activation leading to autoimmune disease illustrates the importance of these nucleic acid degrading enzymes in the pathogenesis of inflammatory disease.
The functional consequences of genetic mutations identified in the RNASEH2A, RNASEH2B, RNASEH2C, and TREX1 genes of AGS patients is facilitated by biochemical analyses of the recombinant RNaseH2 and TREX1 mutant enzymes. We recently described the effects of TREX1 mutations found in autoimmune diseases on the 3′→5′ deoxyribonuclease activities of this enzyme, illustrating the challenges in correlating human genetic mutations to functional consequences in the encoded enzyme [10, 11, 15, 16, 21]. In order to determine the effects of genetic mutations identified in AGS patients on the nuclease activities of the RNaseH2 mutant enzymes, we cloned the RNASEH2A, RNASEH2B, and RNASEH2C genes and established a bacterial expression system to produce the human RNaseH2 heterotrimeric enzyme in sufficient quantities and purity to perform biochemical studies on wild-type and mutant RNaseH2 enzymes. Our data show that RNaseH2 mutant enzymes identified in AGS retain active ribonuclease H activity and indicate that RNaseH2 dysfunction in AGS is more likely mediated through altered interactions with unidentified nucleic acids or proteins.
Materials and methods
Materials
The synthetic 5′-FAM-DNA16-RNA4-DNA10,5′-FAM-RNA20-DNA10, and the complementary DNA 30-mer oligonucleotides were from Operon. The RNASEH2A (NM006397), RNASEH2B (BC010174), and RNASEH2C (NM032193) genes were commercially available clones (Open Biosystems). The pCDFDuet-1 and pET26b plasmids were from Novagen. Plasmid constructs and mutations were prepared by polymerase chain reaction [25], and constructs were verified by DNA sequencing.
Enzyme preparation
The human RNaseH2 heterotrimeric enzymes were generated in bacteria. Escherichia coli BL21 (DE3) Rosetta 2 cells (Novagen) were transformed with the pDuet-BC and pET-A plasmids (Fig. 1). Cells were grown to an A600=0.5 at 37°C and quickly cooled on ice to 17°C. After induction with 1 mM isopropyl-β-D-thiogalactopyranoside, the cells were allowed to grow for 15 h at 17°C. The RNaseH2 complex in cell extracts was bound to a nickel-NTA resin (Qiagen), washed, and eluted. The RNaseH2 was collected and purified to homogeneity using phosphocellulose and monoQ chromatography. Protein concentrations were determined by A280 using the molar extinction coefficient for human RNaseH2 complex ε=83,030 M−1 cm−1.
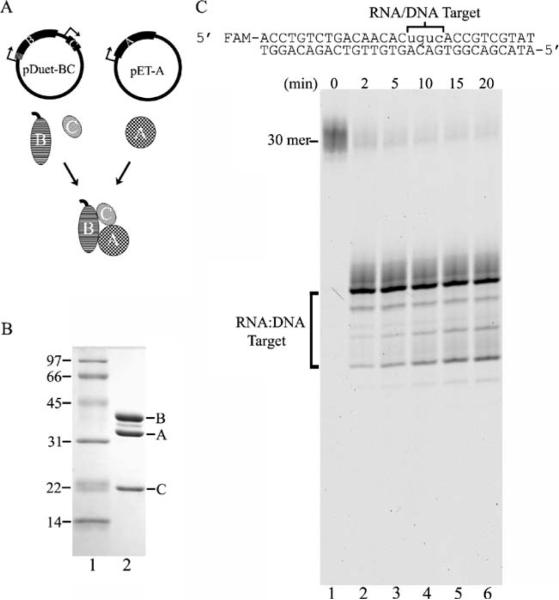
Fig. 1.
The human RNaseH2 is an active heterotrimeric ribonuclease H. A His6-tag was engineered at the N terminus (hatched marks)of the RNASEH2B gene and cloned upstream of the untagged RNASEH2C gene to yield the pDuet-BC plasmid (a). The untagged RNASEH2A gene was cloned on a separate plasmid to yield pET-A (a). Expression of each gene is controlled by a T7 promoter (arrows). The purified recombinant RNaseH2 three-subunit complex (6 μg) was subjected to 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was stained with Coomassie Brilliant Blue (b, lane 2). The positions of migration of the molecular weight standards (b, lane 1) and RNaseH2 subunits are indicated. An RNaseH2 (40 nM) reaction was prepared with the DNA16-RNA4-DNA10/DNA30 duplex, and samples were removed at the indicated times after incubation at 25°C. Reaction products were subjected to electrophoresis on a 23% urea-polyacrylamide gel (c). The positions of migration of the 30-mer and products (RNA/DNA Target) are indicated. The product sizes were determined by comparison to a ladder of fragments generated by TREX1 exonuclease digestion of the same oligomer and run in adjacent lanes (not shown)
Ribonuclease H assays
The RNaseH2 reactions contained 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM dithiothreitol, 50 μg/ml bovine serum albumin, 200 nM 5′-FAM-DNA16-RNA4-DNA10/DNA30 or 5−-FAM-RNA20-DNA10/DNA30 duplex (RNA/DNA target), and RNaseH2 as indicated in the figures. RNaseH2 enzymes were diluted on ice at ten times the final concentrations and added to reactions to yield the indicated final concentrations. After incubation at 25°C, samples were removed, quenched by addition of three volumes of cold ethanol, and dried in vacuo. Pellets were resuspended in 6 μl of formamide, heated to 95°C for 5 min, and separated on 23% denaturing polyacrylamide gels. Fluorescent-labeled bands were visualized and quantified using a Storm PhosphorImager (GE Healthcare) [10, 21, 26].
Results and discussion
The human RNaseH2 is an active heterotrimeric ribonuclease H
A strategy was developed to generate the human RNaseH2 heterotrimeric complex by co-expressing the RNASEH2A, RNASEH2B, and RNASEH2C genes in bacterial cells. The RNASEH2B gene, encoding the 312 amino acid subunit B, was cloned upstream of the RNASEH2C gene, which encodes the 164 amino acid subunit C to generate the pDuet-BC plasmid. The RNASEH2A gene, encoding the 299 amino acid subunit A, was cloned on a separate compatible plasmid to generate pET-A (Fig. 1a). A His6 affinity tag was engineered at the N terminus of only the RNaseH2 subunit B requiring subunits A and C to physically and stably associate with subunit B throughout the purification. In preliminary expression studies, the His6-tag was cloned at either the N or C terminus of each of the three RNaseH2 subunit genes, and maximal yields of the RNaseH2 three-subunit enzyme were recovered using the N-terminal His6-tag RNaseH2 subunit B (data not shown). The RNaseH2 complex from bacterial cell extracts was bound to a nickel-NTA resin by the His6-affinity tag present on subunit B in the initial step of purification, resulting in the isolation of the three-subunit complex. The RNaseH2 was purified further using phosphocellulose and monoQ chromatography, and a stable three-subunit enzyme complex of approximately 1:1:1 molar ratio is indicated by the staining intensities of the three RNaseH2 subunits (Fig. 1b). These data provide direct evidence for the heterotrimeric structure of the bacterially expressed human RNaseH2 enzyme, similar to that described for the mammalian cell expressed complex [5].
The human RNaseH2 complex is an active ribonuclease H with the greatest level of specificity for cleavage of the phosphodiester linkage between the two ribonucleotides nearest the RNA–DNA junction. The 5′-FAM-DNA16-RNA4-DNA10 substrate, containing four ribonucleotides flanked on the 5′ side with 16 deoxyribonucleotides and on the 3′ side by ten deoxyribonucleotides, was hybridized to the complementary DNA30 to generate the ribonuclease H substrate (Fig. 1c). The 5′-FAM-DNA16-RNA4-DNA10 30-mer migrates to a unique position in the urea-polyacrylamide gel (Fig. 1c, lane 1). To measure ribonuclease H activity, the purified RNaseH2 complex (4 nM, data not shown, or 40 nM, Fig. 1c) was incubated in time course reactions with the DNA16-RNA4-DNA10/DNA30 duplex. Upon incubation with RNaseH2, the DNA16-RNA4-DNA10 oligomer is cleaved with greater than 85% of the product migrating to a position 19 nucleotides in length with lesser amounts of product migrating to positions between 16 and 18 nucleotides in length (Fig. 1c, lanes 2–6). The major 19 nucleotide product (DNA16-RNA3) corresponds to phosphodiester cleavage of the DNA16-RNA4-DNA10 between the third and fourth ribonucleotides (rUra-rCyt) near the RNA-DNA junction. This precise position of cleavage was confirmed by recovering the 19 nucleotide RNaseH2-generated DNA16-RNA3 product, re-hybridizing it to the DNA30 template, and incubating the DNA16-RNA3:DNA30 partial duplex in four separate reactions with DNA polymerase and a single deoxyribonucleotide triphosphate. Polymerase extension of the DNA16-RNA3 is detected only in the reaction containing 2′-deoxycytidine 5′-triphosphate corresponding to elongation of the DNA16-RNA3 by insertion of deoxycytidine monophosphate opposite the template Gua (data not shown). These data also indicate that RNaseH2 cleavage generates a 3′-OH and 5′-phosphate. This position of cleavage by the human RNaseH2 is similar to that reported with the Saccharomyces cerevisiae RNaseH2 using a similar DNA-RNA-DNA oligomer [6, 27]. The detection of 16–19 nucleotide length products indicates that the RNaseH2 acts exclusively within the RNA/DNA duplex region confirming the ribonuclease H activity of the recombinant human RNaseH2 three-subunit complex.
The RNaseH2 mutants are active heterotrimeric ribonuclease Hs
Four RNaseH2 mutants were prepared and tested for ribonuclease H activity. The RNaseH2 mutants selected were based on the prevalence of these specific alleles in approximately 70% of AGS patients [13]. Plasmid constructs containing the appropriate nucleotide mutations to generate amino acid changes in subunit A (Gly37Ser), subunit B (Ala177Thr or Val185Gly), or subunit C (Arg69Trp) were prepared and transformed into bacteria with the wild-type genes of the other two RNaseH2 subunits. The mutant RNaseH2 enzymes were expressed and purified using the same procedure that was used for the preparation of the wild-type RNaseH2 enzyme resulting in similar yields and polypeptide compositions (Fig. 2a). These results indicate that the RNaseH2 heterotrimeric structures are retained in these mutant complexes containing these specific amino acid substitutions.
Fig. 2.
The RNaseH2 mutants are active heterotrimeric ribonuclease Hs. Site-directed mutations were introduced into the RNASEH2A, RNASEH2B, and RNASEH2C expression constructs, and the RNaseH2 mutant complexes were purified as described in “Materials and methods.” Approximately 6 μg of each RNaseH2 complex was subjected to 15% SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue (a, lanes 2–6). The positions of migration of the molecular weight standards (a, lane 1) and RNaseH2 subunits are indicated. RNaseH2 (40 nM) reactions containing the indicated mutant enzyme were prepared with the DNA16-RNA4-DNA10/DNA30 duplex, and samples were removed at the indicated times after incubation at 25°C. Reaction products were subjected to electrophoresis on a 23% urea-polyacrylamide gel (b). The positions of migration of the 30-mer and products (RNA/DNA Target) are indicated. The product sizes were determined by comparison to a ladder of fragments generated by TREX1 exonuclease digestion of the same oligomer and run in adjacent lanes (not shown)
The RNase H2 mutant complexes are active ribonuclease Hs. The purified RNaseH2 mutant complexes were incubated in time course reactions with the DNA16-RNA4-DNA10/DNA30 duplex, and products similar to those generated using wild-type RNaseH2 were observed (Fig. 2b). The products generated upon incubation with wild-type and mutant RNaseH2 enzymes are 16–19 nucleotides in length (Figs. 1c and 2b). However, the RNaseH2 subunit A (Gly37Ser) complex generates almost exclusively the 19 nucleotide product (Fig. 2b, lanes 2–6). These data confirm the presence of ribonuclease H activity in all of the human RNaseH2 mutant three-subunit complexes that were tested. Furthermore, cleavage specificity near the RNA–DNA junction is apparent for both wild-type and mutant RNaseH2 enzymes. The RNaseH2 subunit A (Gly37Ser) enzyme appears to exhibit a greater level of cleavage specificity near the RNA/DNA junction with little cleavage detected at other positions within the RNA/DNA target region (Fig. 2b, lanes 2–6).
Altered specificity in the RNaseH2 subunit A Gly37Ser mutant
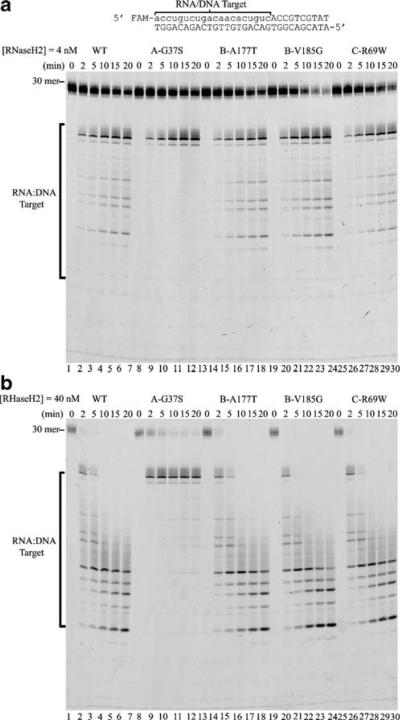
The RNaseH2 subunit A Gly37Ser mutant exhibits a greater level of preference for cleavage near a RNA–DNA junction within a polynucleotide that is not observed with wild-type RNaseH2 or with the other mutants tested. It has been proposed that RNaseH2 processes Okazaki fragments in a RNA primer removal pathway [6, 28]. The association of RNaseH2 with AGS might be explained if mutant enzymes failed to process these DNA replication intermediates. To test this hypothesis, the RNaseH2 mutants were incubated with a model Okazaki fragment substrate. A 5′-FAM-RNA20-DNA10 oligomer was hybridized to the complementary DNA30 oligomer to generate the RNA–DNA junction of an Okazaki fragment DNA replication intermediate (Fig. 3). The RNaseH2 enzymes at 4 nM (Fig. 3a) and 40 nM (Fig. 3b) concentrations were incubated in time-course reactions with the RNA20-DNA10/DNA30 duplex. The products generated by the wild-type RNaseH2 (Figs. 3a,b, lanes 2–6), subunit B Ala177Thr mutant (Figs. 3a,b, lanes 14–18), subunit B Val185Gly mutant (Figs. 3a,b, lanes 20–24), and subunit C Arg69Trp mutant (Figs. 3a,b, lanes 26–30) are nearly identical at both enzyme concentrations tested. In contrast, the subunit A Gly37Ser mutant (Figs. 3a,b, lanes 8–12) generates almost exclusively the 19 nucleotide product, demonstrating altered specificity in the ribonuclease H activity. The product banding pattern detected in this ribonuclease H analysis indicates that the wild-type RNaseH2, subunit B Ala177Thr mutant, subunit B Val185Gly mutant, and subunit C Arg69Trp mutant cleave the model Okazaki fragment substrate with the greatest specificity near the RNA–DNA junction and also cleave at additional RNA/DNA duplex regions less efficiently, but at measurable rates. The subunit A Gly37Ser mutant retains full cleavage activity near the RNA–DNA junction but exhibits drastically reduced activity within the RNA/DNA duplex regions. The mechanism for the observed altered specificity in the RNaseH2 subunit A Gly37Ser mutant enzyme will require further investigation.
Fig. 3.
Altered specificity in the RNaseH2 subunit A Gly37Ser mutant. RNaseH2 reactions containing the indicated mutant enzyme (a, 4 nM) and (b, 40 nM) were prepared with the RNA20-DNA10/DNA30 duplex and samples were removed at the indicated times after incubation at 25°C. Reaction products were subjected to electrophoresis on 23% urea-polyacrylamide gels. The positions of migration of the 30-mer and products (RNA/DNA Target) are indicated. The product sizes were determined by comparison to a ladder of fragments generated by TREX1 exonuclease digestion of the same oligomer and run in adjacent lanes (not shown)
The RNaseH2 mutants tested in this study do not support exclusively a mechanism for ribonuclease H loss of activity in AGS when measured using synthetic hybrid oligonucleotide substrates containing multiple ribonucleotides. The RNaseH2 subunits B and C mutants that were examined exhibit ribonuclease H activities indistinguishable from wild-type RNaseH2. The RNaseH2 subunit A Gly37Ser mutant retains the major catalytic activity exhibited near RNA–DNA junctions with some apparent altered nuclease specificity. Previous studies of the human RNaseH2 subunit A Gly37Ser using an oligonucleotide substrate containing a single ribonucleotide report greatly reduced catalytic activity [5]. Similarly, the equivalent S. cerevisiae RNaseH2 subunit A Gly42Ser mutant exhibits greatly reduced activities using oligonucleotide substrates containing single or multiple ribonucleotides [27]. The spectrum of possible RNaseH2 in vivo substrates is not known. Further experiments with additional quantification of potential RNaseH2 substrates that might reveal mutation causing alterations is clearly warranted. It is also possible that expression of these specific RNaseH2 mutations in vivo results in diminished activity in human cells that would not be revealed by these in vitro studies. Approximately 90% of the reported RNaseH2 mutants associated with AGS are located in subunits B and C, with the vast majority being the specific alleles tested in this paper [13]. Thus, our results indicate that the common AGS mutations are not likely to directly affect catalytic activity of the RNaseH2 complex. The subunit A is the catalytic component and the tightly associated subunits B and C likely affect the stability and/or cellular action of the catalytic subunit by some as yet determined mechanism.
Acknowledgments
This work was supported by grants National Institutes of Health GM069962 (FWP), Alliance for Lupus Research 67692 (FWP), and American Cancer Society RSG-04-187-01-GMC (TH).
References
- 1.Aicardi J, Goutieres F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal-fluid lymphocytosis. Ann Neurol. 1984;15:49–54. doi: 10.1002/ana.410150109. [DOI] [PubMed] [Google Scholar]
- 2.Goutieres F, Aicardi J, Barth PG, Lebon P. Aicardi–Goutieres syndrome: an update and results of interferon-alpha studies. Ann Neurol. 1998;44:900–907. doi: 10.1002/ana.410440608. [DOI] [PubMed] [Google Scholar]
- 3.Goutieres F. Aicardi–Goutieres syndrome. Brain Dev. 2005;27:201–206. doi: 10.1016/j.braindev.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Crow YJ, Hayward BE, Parmar R, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 cause Aicardi–Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 5.Crow YJ, Leitch A, Hayward BE, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi–Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 6.Jeong HS, Backlund PS, Chen HC, Karavanov AA, Crouch RJ. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004;32:1616–1616. doi: 10.1093/nar/gkh209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′→5′ exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 8.Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′→5′ exonucleases—characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 10.deSilva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J Biol Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 11.Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O'Hara A, Herrick AL, Bowden AP, Perrino FW, Lindahl T, Barnes DE, Crow YJ. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi–Goutieres syndrome. Am J Hum Genet. 2007;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolmie JL, Shillito P, Hughesbenzie R, Stephenson JBP. The Aicardi–Goutieres syndrome (familial, early-onset encephalopathy with calcifications of the basal ganglia and chronic cerebrospinal-fluid lymphocytosis) J Med Genetics. 1995;32:881–884. doi: 10.1136/jmg.32.11.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rice G, Patrick T, Parmar R, et al. Clinical and molecular phenotype of Aicardi–Goutieres syndrome. Am J Hum Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee-Kirsch MA, Gong ML, Schulz H, Ruschendorf F, Stein A, Pfeiffer C, Ballarini A, Gahr M, Hubner N, Linne M. Familial chilblain lupus, a monogenic form of cutaneous lupus erythematosus, maps to chromosome 3p. Am J Hum Genet. 2006;79:731–737. doi: 10.1086/507848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino FW, Lieberman J, Hubner N. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007;85:531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 16.Lee-Kirsch MA, Gong M, Chowdhury D, et al. Mutations in the gene encoding the 3′–5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 17.Richards A, van den Maagdenberg A, Jen JC, et al. C-terminal truncations in human 3′–5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 18.Kavanagh D, Spitzer D, Kothari PH, Shaikh A, Liszewski MK, Richards A, Atkinson JP. New roles for the major human 3′–5′ exonuclease TREX1 in human disease. Cell Cycle. 2008;7:1718–1725. doi: 10.4161/cc.7.12.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNaseIII) 3′–5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury D, Beresford PJ, Zhu PC, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J Biol Chem. 2008;283:31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hostomsky Z, Hostomska Z, Matthews DA. Ribonucleases H. In: Linn SM, Lloyd RS, Roberts RJ, editors. Nucleases. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 341–376. [Google Scholar]
- 25.Higuchi R. In: PCR protocols: a guide to methods and applications. Innis MA, Gelfand DH, Shinsky JJ, White TJ, editors. Academic; San Diego: 1990. pp. 177–183. [Google Scholar]
- 26.Perrino FW, Harvey S, McMillin S, Hollis T. The human TREX2 3′→5′-exonuclease structure suggests a mechanism for efficient nonprocessive DNA catalysis. J Biol Chem. 2005;280:15212–15218. doi: 10.1074/jbc.M500108200. [DOI] [PubMed] [Google Scholar]
- 27.Rohman MS, Koga Y, Takano K, Chon H, Crouch RJ, Kanaya S. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 2008;275:4836–4849. doi: 10.1111/j.1742-4658.2008.06622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu JZ, Qian Y, Frank P, Wintersberger U, Shen BH. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]