SUMMARY
Autophagy is an evolutionarily conserved intracellular process by which bulk cytoplasm is enveloped inside a double-membraned vesicle and shuttled to lysosomes for degradation. Within the last 15 years, the genes necessary for the execution of autophagy have been identified and the number of tools for studying this process has grown. Autophagy is essential for tissue homeostasis and development and defective autophagy is associated with a number of diseases. As intracellular parasites, during the course of an infection, viruses encounter autophagy and interact with the proteins that execute this process. Autophagy and/or autophagy genes likely play both anti-viral and proviral roles in the life cycles and pathogenesis of many different virus families. With respect to anti-viral roles, the autophagy proteins function in targeting viral components or virions for lysosomal degradation in a process termed xenophagy, and they also play a role in the initiation of innate and adaptive immune system responses to viral infections. Consistent with this anti-viral role of host autophagy, some viruses encode virulence factors that interact with the host autophagy machinery and block the execution of autophagy. In contrast, other viruses appear to utilise components of the autophagic machinery to foster their own intracellular growth or non-lytic cellular egress. As the details of the role(s) of autophagy in viral pathogenesis become clearer, new anti-viral therapies could be developed to inhibit the beneficial and enhance the destructive aspects of autophagy on the viral life cycle.
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is an intracellular process that serves important roles in both the maintenance of cellular homeostasis and in cell survival under stress conditions [1]. Classical autophagy is a catabolic process in which cytoplasmic proteins and organelles are sequestered within double- or multi-phospholipid membrane vesicles, known as autophagosomes, and shuttled to lysosomes for their degradation (Figure 1). Autophagy functions to maintain cellular homeostasis by removing damaged or unwanted cellular organelles and protein aggregates from the cytoplasm. Under conditions of metabolic stress, autophagy is strongly upregulated to provide an intracellular pool of energy and macromolecules through the catabolism of intracellular constituents to maintain essential cellular processes until normal growth conditions return. Perturbations resulting in too little or too much autophagy appear to play a role in numerous processes and disease conditions including ageing, neurodegeneration, heart diseases, cancer, inflammatory bowel disorders and infectious diseases [2].
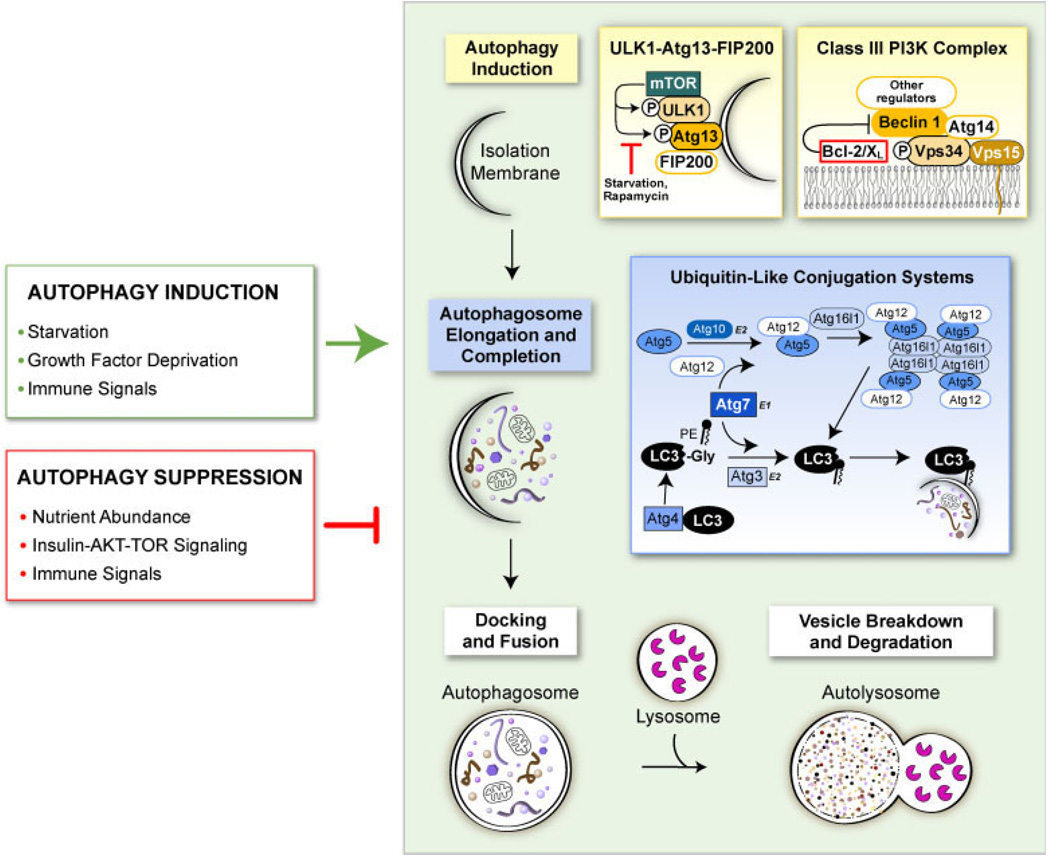
Figure 1.
Scheme of the autophagy pathway. The process of autophagy begins with the nucleation of an autophagic isolation membrane that eventually encloses the cytoplasmic and/or microbial cargo destined for lysosomal degradation. The mammalian target of rapamycin (mTOR) kinase plays a major role in suppressing autophagy induction by binding to ULK1-Atg13-FIP200 complexes and hyper-phosphorylating ULK1 and Atg13. Under autophagy promoting conditions such as starvation, mTOR does not associate with the ULK1-Atg13-FIP200 complex, leaving ULK1 and Atg13 hypo-phosphorylated, which allows the isolation membrane to expand. Vesicle nucleation is also promoted through the activation of the class III phosphatidylinositol 3-phosphate (PIP3) kinase Vps34 which contains the essential mammalian autophagy proteins Beclin 1 and Atg14. The Vps34-Beclin 1-Atg14 complex creates PIP3 residues on the isolation membrane that serve as docking sites for other autophagy-promoting proteins. Other proteins bind to this minimal class III PI3K complex to regulate autophagy and other membrane-trafficking processes, including Bcl-2 family members that function as important inhibitors of autophagy. The second major step of autophagy involves the elongation of the isolation membrane to form an autophagosome which has two phospholipid bilayer membranes with the selected cargo sequestered within the inner membrane. Two ubiquitin-like conjugation systems mediate this step by modifying two ubiquitin-like molecules, Atg5 and LC3, so that they can associate with the isolation membrane and promote its curvature and expansion. The last steps of autophagy involve the docking of completed autophagosomes with lysosomes and the lysosomal enzyme-mediated breakdown and degradation of the inner membrane of the autophagosome and its constituents. Autophagy is triggered and inhibited in cells in response to a diverse set of stimuli including nutrient availability, growth factor mediated-signaling and immune signals generated in response to microbes (green and red boxes)
As obligate intracellular parasites, the survival of viruses is intricately linked to their ability to subvert cellular anti-viral defences and to regulate cellular processes necessary for viral replication. An increasing body of research is finding that autophagy and/or the autophagy execution genes function in both anti-viral and pro-viral capacities in the life cycles of a broad range of viruses (Figure 2). Autophagy can serve as an intrinsic immune defence against viruses partly through xenophagy, a form of autophagy where foreign rather than self-molecules are targeted for degradation in autophagosomes. In addition to xenophagy, autophagy execution genes may function in non-autophagy- related processes to limit viral replication and spread. Autophagy genes also influence the innate and adaptive immune system responses to viral infections. To overcome the antiviral functions of the autophagy genes, a number of viruses have evolved mechanisms to hinder autophagy in infected cells. Despite the ability of autophagy to function as an anti-viral mechanism, some viruses appear to upregulate the process and/or subvert autophagic genes in order to enhance their replication. This review highlights how autophagy and autophagy genes function to limit viral replication and pathogenesis and how different viruses counteract and/or subvert the host autophagy machinery in order to enhance their own growth or survival.
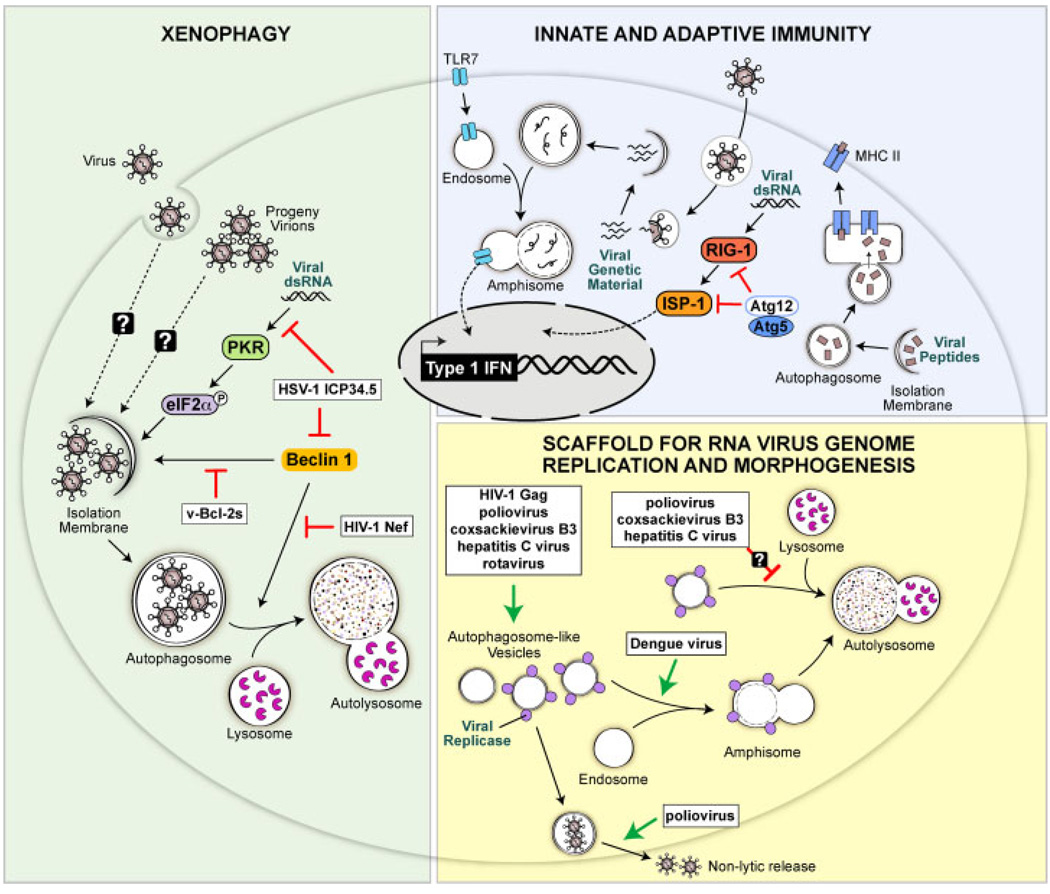
Figure 2.
Anti-viral and pro-viral functions of autophagy. (Green-shaded region) Autophagy may function as a cell-intrinsic anti-viral mechanism by directly capturing virions and/or viral components and targeting them for degradation in lysosomes, a process known as xenophagy. The PKR-eIF2α signaling pathway and the autophagy protein Beclin 1 function in promoting xenophagy, and some viruses have evolved to encode gene products to block the autophagy-promoting activities of these proteins. The HSV-1 ICP34.5 protein blocks both PKR- and Beclin 1-mediated activation of autophagy and viral Bcl-2 homologues found in γ-herpesviruses block Beclin 1-mediated autophagy. The Nef protein of HIV-1 blocks the ability of Beclin 1 to promote autophagosome maturation into autolysosomes. (Blue-shaded region) Autophagy and autophagy genes also exert anti-viral functions by collaborating with the innate and adaptive immune systems. Autophagy delivers viral genetic material from SV and vesicular stomatits virus to endosomes where the ssRNA-sensing toll like receptor 7 resides thus initiating anti-viral Type I interferon signaling. Autophagosomes also target viral peptides to MHC class II loading compartments, thereby enhancing CD4 T cell responses to these antigens. In contrast to promoting innate immunity, the autophagy proteins Atg5 and Atg12 may function to dampen innate immune responses to viruses by binding to RIG-I, a sensor of foreign genetic material, and blocking signaling downstream of RIG-I. (Yellow-shaded region) For some viruses, the autophagy machinery may function in a pro-viral capacity by creating double membrane structures that serve as scaffolds for the replication of viral genomes or as sites of virion morphogenesis. Several RNA viruses including poliovirus, coxsackievirus B3, hepatitis C virus and rotavirus have been shown to induce double-membraned, autophagosome like structures in infected cells, and replicases from these viruses associate with these structures. Poliovirus may also use these structures to promote the non-lytic release of new virions from cells. All of these viruses presumably have means to block the maturation of these structures into destructive autolysosomes. The Gag protein of HIV-1 also appears to be involved in forming early stage autophagosomes in macrophages which enhances Gag processing and HIV yields from macrophages through an unknown mechanism
MECHANISMS OF AUTOPHAGY
At least 30 Autophagy (Atg) genes have been identified in yeast that function in the regulation and execution of autophagy [3]. Many of these genes, but not all, have known homologues in mammalian cells. Classical autophagy can be divided into four major steps that are executed by specific Atg genes [2,3] (Figure 1). The first step involves vesicle regulation and nucleation. Many different signaling pathways and proteins appear to be involved in this first step, but it is not known if they work in concert or separately. The mammalian target of rapamycin (mTOR) kinase, a major controller of cap-dependent translation in cells, strongly inhibits autophagy. When nutrients are replete, mTOR associates with ULK1-Atg13-FIP200 complexes on autophagic isolation membranes and hyper-phosphorylates ULK1 and Atg13 which suppresses isolation membrane expansion [4]. Nutrient deprivation or the drug rapamycin inhibit the association of mTOR and ULK1 and/or mTOR kinase activity which keeps ULK1 and Atg13 hypo-phosphorylated and leads to the upregulation of autophagy in both mammalian cells and yeast. Other proteins and molecules that play a role in regulating this first step of autophagy include p53, c-jun-N-terminal kinase 1 (JNK1), eukaryotic initiation factor 2α (eIF2α), GTPases and intracellular calcium [2,5–8]. All these molecules and/or signaling pathways appear to promote autophagy by activating the class III phosphatidylinositol 3-kinase (PI3K) Vps34 that promotes the formation of phosphatidylinositol 3-phosphate (PIP3) on lipids. The activity of Vps34 in promoting autophagy depends on its association in a multi-protein complex that contains the essential mammalian autophagy protein Beclin 1 (homologue of yeast Atg6). Additional proteins such as Bcl-2, Ambra1, Bif-1, Atg14L, UVRAG and Rubicon can associate with the Vps34 and Beclin 1 minimal complex to form functionally distinct multi-protein complexes that regulate different stages of autophagosome development, endocytosis and perhaps vesicle transport [9–16]. The drug 3-methyladenine (3-MA) inhibits the kinase activity of class III PI3K and is often used to pharmacologically inhibit autophagy in cells [17]. One caveat that should be remembered for both rapamycin and 3-MA is that although they both influence autophagy, they also function to influence other cellular pathways and processes [18].
Two separate, evolutionarily conserved, ubiquitin-like conjugation systems mediate the second major step of autophagy, vesicle elongation and completion [2,3]. In the first system, the E1-like enzyme Atg7 and the E2-like enzyme Atg10 promote the covalent association of Atg12 to Atg5. Atg16 then associates with this complex to form an Atg16-Atg12–Atg5 heterotrimeric complex that associates primarily on the outer membrane of growing autophagosomes where it is hypothesised to mediate curvature of the growing membrane [3]. A dominant-negative form of Atg5 (that cannot conjugate to Atg12), Atg5 and Atg7 knockout mouse embryo fibroblasts (MEFs), and conditional Atg5 and Atg7 knockout mice have been developed to aid in studying autophagy [19–23]. The second ubiquitin-like conjugation system results in the cleavage of microtubule-associated protein, light chain 3 (LC3) (mammalian homologue of yeast Atg8) by Atg7 and the protease Atg4. After cleavage, the E2-like enzyme Atg3 adds phosphatidylethanolamine (PE) to a conserved glycine residue in the C terminus of the cleaved LC3 to create a species known as LC3-II or LC3-PE. LC3 is normally soluble and dispersed throughout the cytoplasm, but after cleavage and lipidation with PE, LC3 associates with both the outer and inner membranes of the growing autophagosome. Upon completion of the autophagosome, the LC3 associated with the outer membrane is recycled while the LC3 on the inner membrane is retained. As it is the only known protein to stably associate with completed autophagosomes, LC3 is the most commonly utilised marker for studying autophagy [18]. Autophagosome formation can be assessed biochemically by immuno-blotting for lipidated (LC3-II) and non-lipidated (LC3-I) forms of LC3. Additionally, plasmids encoding LC3 fused to green-fluorescent protein (GFP-LC3) can be transfected into cells and the activation of autophagy can be assessed by monitoring for the re-localisation of GFP-LC3 from a diffuse cytoplasmic distribution to distinct puncta using fluorescence microscopy. After the autophagosome is completed, Atg9 and Atg18 act to remove and recycle Atg16, Atg12, Atg5 and outer membrane LC3.
The third major step in autophagy is docking and fusing of the completed autophagosome with lysosomes to form autolysosomes [2,3]. This process is not well understood, but the lysosomal protein LAMP-2 and the GTP-binding protein Rab7 appear to aid in this process. Autophagosomes are also capable of fusing with endosomes to create structures known as amphisomes. It is still unclear whether amphisomes are distinct entities or precursors in the formation of autolysosomes. The last major step of autophagy is vesicle breakdown and degradation. Upon fusion of the outer membrane of the autophagosome with the lysosomal membrane, the lysosomal cathepsins B, D and L as well as other enzymes enter the autophagosome and degrade the constituents of the inner autophagosomal membrane. The byproducts of this degradation, amino acids and lipids, are exported from the autolysosome into the cytoplasm where they are recycled to generate new macromolecules [2,3].
An unclear area of autophagy research—that is particularly important for virology—is understanding how cytoplasmic cargo is selected for autophagic sequestration. In general, the mechanism(s) and proteins involved in selecting cargo for sequestration in autophagosomes are poorly defined in mammalian cells. However, recent studies indicate that, in addition to targeting proteins for degradation by the 26S proteasome, ubiquitin may also function in directing protein aggregates and oligomeric proteins to autophagosomes [24–26]. Two proteins, p62 (SQSTM1) and NBR1 (neighbour of BRCA1 gene 1), can function as adaptors to target ubiquitinated protein aggregates to autophagosomes through a common ability to bind both ubiquitin and LC3 [27–29]. It is not yet known whether similar mechanisms are involved in targeting viral proteins to the autophagosome or even whether the targeting of viral proteins is selective or represents a more random stochastic process.
Although Atg genes have fairly well defined roles in classical autophagy, it is not known if they all function similarly in xenophagy and if so, if they are regulated in a similar manner. In addition to classical autophagy, there are other morphologically distinct forms of autophagy that may occur in virus-infected cells including microautophagy, chaperone-mediated autophagy and various organelle specific forms of autophagy such as ribophagy (degradation of ribosomes) and mitophagy (degradation of mitochondria) [2]. Furthermore, the Atg genes potentially have roles independent of their role in autophagy that could potentially contribute to their ability to combat viral infections [30,31].
AUTOPHAGY AS A CELL-INTRINSIC ANTI-VIRAL IMMUNE DEFENCE
The first identified mammalian autophagy protein, Beclin 1, was found in a yeast two-hybrid screen for Bcl-2-interacting proteins and first studied in the context of CNS infection with Sindbis virus, a positive-stranded RNA virus in the alphavirus genus [32]. Enforced expression of Beclin 1 in mouse brains during Sindbis virus infection reduced viral titres, the number of Sindbis virus RNA-positive cells, and virus-induced neuronal apoptosis in parallel with protecting mice against fatal Sindbis virus encephalitis [32]. Further studies are needed to determine the mechanism by which Beclin 1 protects mice from Sindbis virus infection, but one hypothesis is that increased expression of Beclin 1 leads to increased xenophagy of Sindbis virus in infected neurons. Alternatively, Beclin 1 may protect against lethal encephalitis by reducing Sindbis virus-induced neuronal apoptosis either through the upregulation of autophagy or through an autophagy-independent function. As neurons are post-mitotic cells with limited regenerative capacity, apoptosis of infected neurons is not a practical anti-viral defence as it is in dividing cells, and thus the upregulation of classical autophagy and/or xenophagy may be a particularly important means of protecting neurons from viral infections.
A similar anti-viral role for autophagy/xenophagy is seen during infection of mouse brains with the α-herpesvirus, HSV-1, which has a double-stranded DNA (dsDNA) genome. Expression of the ICP34.5 gene product encoded by HSV-1 is essential for HSV-1-mediated neurovirulence in mice and humans [33–35]. The ability of ICP34.5 to promote fatal encephalitis in HSV-1-infected mice was initially attributed to its role in recruiting host protein phosphatase 1α (PP1α) to dephosphorylate eIF2α and reverse the shutdown of host protein translation mediated by phosphorylated eIF2α [36]. However, HSV-1 mutant viruses encoding mutated or non-functional ICP34.5 that retained the ability to reverse phosphorylated eIF2α-mediated host cell protein translation shutdown were also avirulent in vivo suggesting that other functions of ICP34.5 are necessary for HSV-1 neurovirulence [37,38]. Interestingly, one such additional function of ICP34.5 that is required for neurovirulence is the inhibition of autophagy through direct binding to Beclin 1 [39]. The deletion of amino acids 68–87 of ICP34.5 (ICP34.5Δ68–87) prevents ICP34.5 binding to Beclin 1 and inhibition of starvation-induced autophagy while maintaining ICP34.5-mediated recruitment of PP1α and dephosphorylation of eIF2α. A mutant HSV-1 virus that encodes ICP34.5 lacking these amino acids (HSV-1 34.5Δ68–87) behaves similarly to wild type HSV-1 in cultured cells in terms of viral replication and reversal of host cell protein shutdown, but causes less mortality in mice with HSV-1 encephalitis [39]. HSV-1 ICP34.5Δ68–87 induces an increased number of autophagosomes in cultured primary mouse neurons, and the mutant virus replicates to lower levels in brains of inoculated mice [40,41]. These data suggest that xenophagy may be an important means of controlling HSV-1 replication in the CNS, although this has not been formally proven in this model. In theory, the protective effects of autophagy could also relate to its pro-survival function or its other expanding roles in immunity, including the activation of Type I interferon production and/or antigen presentation [31]. An important open question is whether ICP34.5 mediated antagonism of autophagy not only contributes to acute HSV-1 pathogenesis (perhaps through promoting viral replication or antagonising host immunity), but also to chronic neurodegeneration [42,43].
Studies in model organisms, including plants and Drosophila, have also confirmed an important role for autophagy genes in defence against two single-stranded RNA (ssRNA) viruses, tobacco mosaic virus (TMV) and vesicular stomatitis virus (VSV), suggesting that the anti-viral function of autophagy is evolutionarily conserved. Plants rely on both intrinsic and innate immune mechanisms for protection against pathogens. They activate a form of programmed cell death (PCD) in response to a pathogen-triggered hypersensitive response (HR) to isolate the pathogen and prevent it from spreading to adjacent and distal healthy tissue. In Arabidopsis, the plant autophagy genes BECLIN 1, PI3K/VPS34, ATG3 and ATG7 function both in restricting HR PCD to the initial inoculation site after TMV infection and also in limiting TMV replication [44]. Whereas HR PCD is activated similarly in wild type and BECLIN 1-deficient plants in response to TMV infection, in BECLIN 1-deficient plants, the HR PCD response spreads to healthy uninfected tissue adjacent to the site of viral inoculation as well as to leaves distal to this site. Activation of HR PCD in wild type plants activates autophagy in both infected cells and uninfected cells as assessed by the formation of GFP-Atg8 puncta and this is reduced in plants lacking BECLIN 1. Given the increased levels of viral RNA in autophagy-gene silenced plants (without increased viral spread), it seems likely that autophagy in infected cells leads to increased degradation of TMV. The mechanism by which autophagy prevents the spread of death-promoting signals to uninfected cells still needs to be elucidated, but this study suggests a critical role for autophagy in protecting innocent bystander cells from undergoing cell death during a systemic immune response.
Similar to plants, Drosophila lack an adaptive immune system and rely on innate and intrinsic immune mechanisms to purge invading microbes. A recent study showed that autophagy is important in protecting Drosophila from infection by VSV [45]. Inhibition of autophagy in either adult flies or cultured Drosophila cells increased VSV replication while activation of autophagy suppressed replication. The surface glycoprotein, VSV-G, serves as a pathogen-associated molecular pattern (PAMP) that promotes autophagosome development in Drosophila cells without any additional virus components or viral genome replication. The authors propose that flies sense VSV infection, perhaps through the binding of VSV-G with an unidentified surface innate immune receptor, which then downregulates the autophagy suppressor PI3K/Akt signaling pathway. No formal demonstration of xenophagy of VSV was reported in this study, so further studies are needed to determine the mechanism by which the increased autophagy protects Drosophila and fly cell lines against VSV infection. While HSV-1 ultrastructural studies suggest that xenophagy eliminates primarily newly synthesised viruses and viral components (rather than incoming virus), the ability of UV-inactivated VSV to stimulate autophagy raises the possibility that xenophagy may also target viruses during cellular entry.
Although these in vivo studies (Table 1) provide compelling evidence for an evolutionarily conserved role of autophagy genes in combating infections by RNA and DNA viruses, further studies are needed to clarify the precise mechanism(s) by which autophagy or autophagy genes function in this manner. The observation of decreased viral titres in association with increased autophagosome formation suggests that xenophagy may serve as a means for controlling viral growth, but formal demonstration of this principle is still lacking. Autophagy gene-mediated protection may also be due to other effects of autophagy on the innate and adaptive immune systems that function to combat viral infections. Furthermore, non-autophagy related functions of beclin 1 or other autophagy genes may also contribute to the phenotypes observed in virus-infected autophagy gene-deficient organisms. For example, such a possibility has recently been proposed for Atg5 in the protection of mice against the protozoan T. gondii.; the primary protective role of Atg5 in this setting is postulated to result from recruitment of the immunity-related GTPase, Irga6, to the parasitophorous vacuole rather than from autophagosome formation [30].
Table 1.
Role of autophagy pathway and/or autophagy genes in protection against in vivo viral infection
| Virus | Autophagy gene(s) |
Effect(s) on host-pathogen interactions |
References |
|---|---|---|---|
| DNA viruses | |||
| Herpesviridae | |||
| HSV-1 | beclin 1 | Protects against lethal HSV-1 encephalitis in mice. Antagonised by the viral ICP34.5 gene product |
[39] |
| RNA viruses | |||
| Alphaviridae | |||
| Sindbis virus | beclin 1 | Overexpressed Beclin 1 in virus infected cells of the CNS protects mice from lethal Sindbis virus encephalitis |
[32] |
| Tobamoviruses | |||
| Tobacco mosaic virus |
BECLIN 1, ATG3, ATG7, VPS34 |
Deletion of autophagy genes causes spread of virus-induced hypersensitive response programmed cell death to healthy tissue beyond site of viral inoculation and increases viral replication at inoculation site |
[44] |
| Rhabdoviridae | |||
| Vesicular stomatitis virus |
Atg8, Atg7, Atg12, Atg18 |
Deletion of autophagy genes increases viral replication in Drosophila cell lines and decreases Drosophila survival |
[45] |
AUTOPHAGY ASSISTS IN INNATE AND ADAPTIVE IMMUNITY
In addition to serving as an intrinsic immune defence against viruses, autophagy genes are involved in the generation of both innate and adaptive immune responses to viruses. The innate immune system involves the function of pattern recognition receptors (PRRs) that are expressed on most if not all cell types [46,47]. The PRRs sample the environment for specific PAMPs and signal to initiate the translation of antimicrobial proteins. The PAMPs that activate the innate immune sensors are both proteins such as lipopolysaccharide (LPS) and flagellin as well as genetic material such as single- and double-stranded RNA and DNA. There are two major classes of PRRs, the toll-like receptors (TLRs) that reside primarily on the cell surface as well as in endosomes, and a family of helicases collectively known as retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) that reside primarily in the cytoplasm [47]. TLRs recognise both protein and nucleic acid PAMPs in the extracellular space while the RLRs recognise foreign genetic material in the cytoplasm. Both types of receptors initiate a signaling cascade that activates transcription factors such as NF-κB that aid in the transcription of antimicrobial proteins and activate other antimicrobial defences. Recent studies indicate that activation of specific TLRs initiates autophagy in certain cell types [48,49]. For example LPS, a common component of bacterial cell walls, activates signaling downstream of TLR4 and upregulates autophagy in macrophage cell lines but not in primary macrophages [48,50]. Two of the three TLRs that are most likely involved in the innate immune response to viruses, TLR7 and TLR3, initiate autophagy in vitro after binding their cognate ligand, ssRNA and dsRNA respectively [51,52]. The ability of the dsDNA sensing TLR9 or the RLRs to activate autophagy has not been reported. The signaling pathway(s) that activate autophagy downstream of TLRs is not known, but deletion of the TLR signaling adaptor proteins MyD88 and TRIF abrogates the activation of autophagy after TLR stimulation [51]. Autophagy activation after TLR stimulation presumably functions to mediate xenophagy of entering pathogens although this has not yet been demonstrated. Moreover, the upregulation of autophagy downstream of TLR stimulation may also enhance innate immune responses independently of xenophagy.
Autophagy and/or autophagy genes may also play a role in delivering ligands to TLRs for their activation. In plasmacytoid dendritic cells (pDCs), Atg5 is necessary for the delivery of VSV and Sendai virus (SV) genetic material from the cytoplasm into endosomes where the ssRNA sensor TLR7 resides [53]. Atg5-mediated delivery of VSV genetic material to pDC endosomes initiates signaling downstream of TLR7 resulting in the production of IFN-α and IFN-α production is suppressed in VSV-infected bone marrow cells by the autophagy-inhibitory drugs, wortmannin and 3-MA. These observations suggest that autophagic delivery of viral genetic material to endosomes is necessary for IFN-α production; however, the possibility of an autophagy-independent role of Atg5 in the generation of IFN-α cannot be excluded. If autophagy is involved, another interesting question is whether the autophagic machinery specifically recognises VSV and SV genetic material for sequestration in autophagosomes or if the viral genetic material is randomly acquired through autophagic sampling of the cytoplasm.
In contrast to activating innate immune signaling, at least two studies have found that autophagy genes can function to dampen innate immune responses to viruses. One study reported that the Atg5–Atg12 conjugate binds to RIG-I and suppresses the ability of MEFs to produce IFNs in response to stimulation by VSV nucleic acids; Atg5-deficient MEFs were resistant to VSV replication due to increased Type I IFN production in response to immunostimulatory VSV genetic material [54]. In another study, Atg5 deletion in MEFs enhanced signaling downstream of RLRs, increased IFN-α production, and conferred resistance to VSV infection [55]. The increased RLR signaling was attributed to increased reactive oxygen species (ROS) produced by the damaged mitochondria that are not cleared in autophagy defective, Atg5−/− cells. This study suggests that the homeostatic role of autophagy in eliminating old and damaged mitochondria impinges upon innate immune signaling. Furthermore, the question arises, does viral inhibition of autophagy (presumably designed to block anti-viral functions of autophagy) actually ‘backfire’, leading to enhanced anti-viral innate immune signaling? An important future area of research will be to determine the net balance of cell-intrinsic anti-viral effects of autophagy versus potential inhibitory effects of autophagy and/or autophagy genes on anti-viral innate immune signaling in the context of viral pathogenesis.
Adaptive immune responses mediated by antigen-specific T and B lymphocytes are initiated many hours after the onset of initial microbial infection, and autophagy may be involved in the generation of some of these responses. Peptides that associate with MHC II molecules were originally thought to derive only from lysosomal degradation of microbial material endocytosed from the extracellular environment. However, several recent studies have found that autophagy in antigen-presenting cells can promote the loading of self-proteins onto MHC II molecules [56,57]. Schmid et al. [58] observed constitutive autophagy in MHC II-positive cells, such as dendritic cells and B cells, and the fusion of autophagosomes in these cells with multi-vesicular MHC II-loading compartments. Furthermore, they found that the targeting of influenza virus matrix protein to autophagosomes by fusing it with LC3 increases antigen-specific CD4 T cell responses. An earlier study from the same group found that the EBV nuclear antigen EBNA1 can be presented on MHC II molecules through autophagy leading to increased recognition of EBNA1 positive cells by EBNA1-specific CD4 T-cells [59]. In contrast to MHC II, MHC I typically present shorter peptide fragments derived from proteasomal degradation of intracellular antigenic material. A recent study suggests that in later stages of HSV-1 infection, autophagy or an autophagy-like process may function in producing processed HSV-1 antigens for presentation onto MHC I [60]. Very little is known about the degradation process inside autolysosomes so it is unclear how peptides of the appropriate length are derived for loading onto either MHC II or MHC I. Further investigation into the role and mechanisms of autophagy in generating peptides for MHC II and MHC I presentation has the potential to improve anti-viral immune responses generated by vaccines.
FIGHTING BACK: VIRUSES THAT COUNTERACT AUTOPHAGY
Given the critical role of autophagy in anti-viral defence, it is not surprising that some viruses possess mechanisms to counteract autophagy (Table 2). A common trait shared by the two major virus families shown to encode means to inhibit autophagy is the ability to persist in their hosts. The herpesviruses are large, dsDNA viruses that have successfully co-evolved with their hosts such that they persist in a latent state for the life of their host after the initial lytic infection is cleared. To do this successfully, herpesviruses have evolved a number of mechanisms to counteract immune defences in their host [61]. Lentiviruses, which have ssRNA genomes, also persist in their hosts after the initial infection is cleared though the integration of their genome into their host’s genome. Lentiviral genomes have evolved from those of simple retroviruses to encode accessory genes that were initially thought to be dispensable for viral replication. It is now known that the protein products of these accessory genes play important roles in the replication and pathogenesis of lentiviruses in part by suppressing a variety of cell-intrinsic, innate and adaptive immune mechanisms designed to eliminate infected cells [62]. In support of the concept that autophagy is a major immune defence against viruses, all three subfamilies of herpesviruses (α-, β- and γ-) and lentiviruses, encode viral proteins and/or induce cellular signals to inhibit autophagy.
Table 2.
Virus autophagy evasion mechanisms
| Virus | Autophagy evasion mechanism |
Effects on host- pathogen interactions |
References |
|---|---|---|---|
| DNA viruses | |||
| Herpesviridae | |||
| HSV-1 | Viral ICP34.5 protein inhibits PKR signaling and directly binds to Beclin 1 to inhibit autophagy in primary neurons and MEFs |
Confers neurovirulence |
[5,39] |
| Kaposis sarcoma herpesvirus (KSHV), murine γ-HV68 (γMHV-86) |
Viral Bcl-2-like proteins bind directly to Beclin 1 and inhibit autophagy in multiple cell lines |
Unknown | [10,65,66] |
| Human Cytomegalovirus (HCMV) |
Infection inhibits autophagy in primary fibroblasts through unknown mechanism |
Unknown | [75] |
| RNA viruses | |||
| Retroviridae | |||
| HIV 1 | Infection inhibits autophagy in primary CD4 + lymphocytes and in U937 macrophage cell line possibly through reduction in Beclin 1 levels |
Unknown | [80] |
| Nef protein binds Beclin 1 and inhibits the maturation of autophagosome into autolysosomes. |
Unknown | [82] | |
| Simian immunodeficiency virus type 1 (SIV 1) |
Infection of microglia induces production of unknown molecule(s) that inhibit autophagy in neurons |
Proposed role in the accumulation of protein aggregates in neurons that potentially contribute to neurodegeneration |
[85] |
As mentioned earlier, the α-herpesvirus HSV-1 encodes the ICP34.5 protein that plays a major role in HSV-1 neuropathogenesis in part through antagonising autophagy by directly binding to Beclin 1 [39]. In addition, ICP34.5 possesses a second mechanism for inhibiting autophagy that is related to its ability to antagonise PKR signaling, presumably through its documented role in recruiting PP1α to dephosphorylate eIF2α [36]. In mammalian cells, two stress-induced eIF2α kinases, the interferon-inducible, double-stranded RNA-sensing kinase PKR and the ER stress-induced kinase PERK, as well as phosphorylated eIF2α, are required for autophagy induction [5,63]. Infection of wild type MEFs or neurons with HSV-1ΔICP34.5 (compared to wild-type HSV-1) results in a significant viral growth defect, accelerated viral protein degradation and increased numbers of virions inside autophagosomes and autolysosomes that is not observed in pkr−/− MEFs or neurons [5,40]. These data indicate a critical role for PKR/eIF2α in HSV-1-induced autophagy. The precise pathway by which PKR-mediated eIF2α phosphorylation leads to autophagy induction is not known; however, pkr deletion in mice restores the neurovirulence of HSV 34.5Δ68–87, placing PKR genetically upstream of Beclin 1 in autophagy induction in vivo [39]. The observation that ICP34.5 acts in at least two ways to inhibit autophagy induction in virus-infected cells strongly suggests an important role for autophagy in defence against HSV-1 infection. While HSV-1 ICP34.5-mediated inhibition of autophagy, rather than ICP34.5-mediated blockade of host cell shutoff, seems to be critical for HSV-1 ICP34.5’s role in neurovirulence, this function of ICP34.5 may not be important in controlling HSV-1 replication in certain cell types. The growth defect of HSV-1Δ34.5 is restored in pkr−/− MEFs but not in Atg5−/− MEFs, suggesting that ICP34.5-mediated inhibition of host cell protein translation shutdown may be more significant than inhibition of autophagy in the regulation of HSV-1 growth in some cultured cells [64].
The γ-herpesviruses all encode at least one, and in some cases two, viral Bcl-2 (v-Bcl-2) gene products; to date, two of these, KSHV vBcl-2 and murine γ-HV68 M11, have been shown to bind to Beclin 1 and to inhibit Beclin 1-mediated autophagy [10,65,66]. Similar to cellular Bcl-2, the v-Bcl-2s bind to a Bcl-2 homology 3 (BH3) domain in the N terminus of Beclin 1 to inhibit its autophagy function [10,65–68]. Although the significance of viral Bcl-2 inhibition of Beclin 1 function in viral replication and/or pathogenesis is not yet known, two lines of biochemical evidence suggest that viral Bcl-2 family members may have evolved to be more potent inhibitors of autophagy than their cellular counterparts. First, the γ-herpesvirus-encoded viral Bcl-2s may escape the physiological regulation that controls the binding of cellular Bcl-2 family members to Beclin 1. Cellular Bcl-2 contains a regulatory loop that is phosphorylated by JNK1 under starvation conditions resulting in the dissociation of cellular Bcl-2 from Beclin 1 and the activation of autophagy; however, this loop is lacking in viral Bcl-2s and starvation fails to result in the disruption of the viral Bcl-2/ Beclin 1 complex or autophagy activation in cells expressing viral Bcl-2 [7]. Second, the murine γ-HV68-encoded viral Bcl-2, M11, binds to the BH3 domain of Beclin 1 with significantly higher affinity than do cellular Bcl-2 family members [65,66]. Moreoever, M11 binds to the BH3 domain of Beclin 1 with higher affinity than to the BH3 domain of pro-apoptotic family members, raising the possibility that autophagy, rather than apoptosis, may be the primary target of this viral virulence factor [65,66]. Thus, although the significance of viral Bcl-2 evasion of autophagy in viral pathogenesis has not yet been investigated, these studies provide strong evidence that this family of viral proteins functions as potent inhibitors of autophagy. It should be noted that γ-HV68 M11 has been shown to be essential for many aspects of viral pathogenesis, including lytic replication, persistent infection and re-activation from latency and that virtually all γ-herpesviruses promote tumourigenesis in vivo [69–71]. Therefore, it will be important to investigate whether viral Bcl-2 inhibition of Beclin 1-mediated autophagy and/or of Beclin 1’s tumour suppressor activity contributes to γ-herpesvirus oncogenesis and/or other aspect of viral pathogenesis [72]. Some γ-herpesviruses also stimulate mTOR signaling which may serve as an additional mechanism for suppressing autophagy in infected cells [73,74].
The β-herpesvirus human cytomegalovirus (HCMV) suppresses autophagy induction in infected cells through an unknown mechanism [75]. HCMV does not encode known homologues of ICP34.5 or viral Bcl-2-like proteins. Surprisingly, HCMV inhibits autophagy even though it can induce phosphorylation of eIF2α through the induction of the unfolded protein response (UPR) at late times post-infection [76,77]. One possible mechanism by which HCMV may block autophagy in cells is through the stimulation of the mTOR signaling pathway which the virus does even in the presence of the mTOR inhibitor and the autophagy-enhancing drug rapamycin [75,78]. Since stimulation of mTOR signaling is also seen in many other virus infections it will be interesting to determine if this represents a general viral strategy for blocking autophagy [79].
The lentivirus HIV appears to regulate autophagy in complex ways to enhance progeny virion production. As will be discussed in the next section, HIV may in fact induce early stages of autophagy; however, multiple data indicate that HIV inhibits the destructive late stages of autophagy that could potentially suppress new virion production through xenophagy. HIV-1 infection of U937 (monocytic) cells or peripheral blood CD4 T cells in vitro was found to suppress the accumulation of the cleaved, lipidated form of LC3, LC3-II, reduce Beclin 1 protein and mRNA levels, and reduce the number of autophagosomes in cells 48 h after infection [80]. This HIV-mediated suppression of autophagosome formation could; however, be overcome by amino acid starvation or treating cells with rapamycin. Espert et al. [81] observed that HIV infection of monocyte-derived macrophages (MDM) resulted in either a low or high number of autophagosomes inside infected cells, and MDMs with high numbers of autophagosomes had fewer HIV particles inside them which would support the hypothesis that xenophagy can control HIV replication in MDMs. This hypothesis is strengthened by their observation that treatment of MDMs with the lysosomal inhibitor Bafilomycin A1 increased HIV yields from MDMs. While this group did not explore how HIV blocks autolysosome formation/activity in some infected MDMs, another study by Kyei et al. [82] found that a HIV accessory protein, negative factor (Nef), can bind to Beclin 1 and block Beclin 1-mediated maturation of autophagosomes into degradative autolysosomes. It will be critical to further dissect these diverse interactions between HIV and the autophagy pathway in the regulation of HIV replication in macrophages and CD4 T cells and in HIV pathogenesis.
The ability of HIV to regulate and inhibit autophagy may contribute to the development of HIV-induced encephalopathies (HIVE) and dementia that are often seen in patients with advanced stages of HIV infection. Autophagy plays a vital role in maintaining neuronal homeostasis, and defective neuronal autophagy correlates with the increased formation and accumulation of protein aggregates and subsequent neurodegeneration [83]. One study found that brains of HIV-infected individuals, and especially those of individuals with HIVE, contained more intraneuronal amyloid β (Aβ) aggregates than brains of uninfected individuals [84]. Interestingly, microglial cells infected with the related lentivirus, simian immunodeficieny virus (SIV), were found to secrete an unidentified molecule(s) that could inhibit autophagy in neurons in vitro, and the suppressive effects of this molecule on neuronal autophagy could be overcome by rapamycin [85]. Further work is needed to determine whether HIV Nef and/or other mechanisms by which HIV may inhibit autophagy in vitro, also function in vivo to inhibit autophagy and thereby contribute to the pathogenesis of HIVE. The ability of rapamycin to overcome HIV-mediated suppression of autophagy in multiple studies suggests that it might be a useful treatment to reduce or prevent HIVE in HIV-infected individuals. However, the potential beneficial effects of rapamycin must be carefully weighed against its potential detrimental effects, and the development of more specific, non-immunosuppressive, approaches to induce autophagy may be useful in the treatment not only of HIV and its complications, but also of other virus infections.
ABETING THE ENEMY: CAN AUTOPHAGY ENHANCE VIRUS REPLICATION?
Given the ability of autophagy genes to suppress viral replication and pathogenicity, a surprising number of viruses appear to induce autophagy or at least the formation of autophagosome-like structures during the course of infection (Table 3). In contrast to the studies discussed in the previous section describing mechanisms by which herpesviruses inhibit autophagy, at least two herpesviruses have been reported to induce autophagy, including VZV and EBV. VZV, an α-herpesvirus distantly related to HSV-1, induces autophagy in cultured cells presumably through the actions of viral immediate early and early genes, and autophagy is also seen in human zoster vesicles but not normal skin [86]. However, it is unknown whether VZV actively promotes autophagy in infected cells for its own benefit or if the observed increase in autophagosomes is an attempt by infected cells to limit VZV replication though xenophagy or some other autophagy gene-dependent function(s). VZV does not encode known homologues of ICP34.5 or v-Bcl-2s in its genome, so if the increased autophagy is a beneficial cellular response to combat VZV, it will be interesting to understand how the virus overcomes this host defence mechanism. The EBV-encoded latent membrane protein 1 (LMP1) is thought to induce cellular autophagy to regulate its own protein levels, as high levels of LMP1 result in cytostasis, inhibition of protein translation, and a decreased ability of LMP1-expressing cells to form colonies [87]. Low levels of LMP1 expression result in increased numbers of early stage autophagosomes while high levels of LMP1 increase the numbers of late stage autolysosomes. The mechanism by which LMP1 induces autophagy has not been elucidated, but it also has been reported that LMP1 promotes eIF2α phosphorylation, a potential trigger of autophagy [88,89]. As LMP1 is an integral membrane protein and not cytosolic, it is unclear how autophagy might specifically target this protein for degradation.
Table 3.
Viruses that promote or subvert autophagy
| Virus | Virus effect(s) on autophagy |
Effect(s) on host-pathogen interactions |
References |
|---|---|---|---|
| DNA viruses | |||
| Herpesviridae | |||
| VZV | Infection induces autophagy in cultured cells and human zoster vesicles |
Unknown | [86] |
| EBV | EBV latent membrane protein 1 (LMP1) induces autophagy possibly to regulate its own levels and block its cytostatic effects on B cells |
Unknown | [87] |
| Hepadnaviridae | |||
| Hepatitis B Virus | HBV × protein enhances Beclin 1 promoter activity and autophagy in hepatocytes |
Unknown | [102] |
| Papovaviridae | |||
| Human papillomavirus type 16 |
HPV16 E7 protein expression in human keratinocytes promotes 3-MA-sensitive accumulation of LC3-II |
Unknown | [103] |
| Parvoviridae | |||
| Human parvovirus B19 | B19 infection of erythroid cells induces mitochondrial autophagy |
Autophagy induction inhibits B19-mediated apoptosis. |
[101] |
| Polyomaviridae | |||
| Simian virus 40 | Small T antigen activates AMPK leading to mTOR inhibition and autophagy enhancement in HEK cells |
Protects cells from glucose deprivation |
[104] |
| RNA viruses | |||
| Retroviridae | |||
| HIV1 | HIV-1 Gag induces early stage autophagosomes and associates with LC3 |
Proposed role in Gag processing | [82] |
| HIV-1 Env protein expression on surface of infected cell induces autophagy gene- mediated cell death in adjacent cells by binding to CD4 and CXCR4. Fusogenic function of gp41 protein involved |
Proposed mechanism of CD4+ T cell depletion in HIV-infected individuals |
[92,93] | |
| Orthomyxoviridae | |||
| Influenza A virus | Infection increases autophagy and autophagic flux in vitro through unknown mechanism |
Increases viral replication and yield |
[94] |
| Flaviviridae | |||
| Dengue virus | Infection induces autophagy in vitro. Viral replication proteins associate with amphisomes. Potential role for autolysosomes in viral replication |
Autophagy increases viral yield |
[95,110,111] |
| Hepatitis C virus | Induction of early stages of autophagy in hepatocyte cell lines possibly through induction of unfolded protein response. Later stages of autophagy blocked by unknown mechanism |
Increases HCV replication | [96,97] |
| Picornaviridae | |||
| Poliovirus | Infection induces LC3- positive autophagosome-like structures with proposed roles as scaffold for viral genome replication and in non-lytic virus egress |
Increases viral replication and yields |
[106,108] |
| Coxsackievirus B3 (CVB3) | CVB3 induces autophagosome-like structures but blocks maturation to autolysosomes through unknown mechanism |
Increases viral replication and yields |
[107] |
| Coxsackievirus B4 (CVB4) | CVB4 induces autophagy in rat primary neurons through unknown mechanism involving activation of calpains |
Promotes calpain activity | [98] |
| Reoviridae | |||
| Rotavirus | Viral NSP4 co-localises with LC3 on viroplasms which are sites of nascent RNA replication |
Unknown | [109] |
In contrast to the data discussed in the previous section about the ability of HIV to suppress late stages of autophagy, some studies indicate that Atg genes and/or autophagy may benefit HIV infection and pathogenesis. A possible role for autophagy or Atg genes in supporting HIV infection was suggested by the identification of Atg7, Atg8, Atg12 and Atg16L2 in an siRNA screen in HeLa cells for genes important in HIV replication; however, it remains to be determined if these genes function in HIV infection in natural HIV target cells and in vivo [90]. The Atg genes likely enhance HIV infection through their functions in autophagosome formation, although non-autophagy functions of these genes could theoretically contribute to HIV replication. A beneficial role for the early stages of autophagy in HIV replication in MDMs was observed by Espert et al. [81] who found HIV infection of MDMs or the macrophage cell line THP1 induced autophagosomes and that inhibition of autophagy with 3-MA significantly reduced HIV yields from macrophages. Kyei et al. [82] found that the Gag protein of HIV induces the formation of early stage autophagosomes in macrophages which promotes maximal extracellular viral yields. The mechanism by which HIV Gag promotes autophagosome formation in macrophages is unclear, but Gag can directly associate with LC3, and the association of Gag with autophagosomes promotes processing of Gag, an important step in virion morphogenesis. Presumably, the actions of HIV Nef block the ability of the Gag-induced autophagosomes to mature into autolysosomes as discussed previously. Further investigation is needed to elucidate how early stage autophagosomes function to enhance HIV replication and morphogenesis. As will be discussed in the next section, some positive strand RNA viruses also induce early stage autophagosomes or autophagosome-like structures that serve as scaffolds for the replication of their genomes, but it is unlikely that HIV uses these structures in a similar manner as HIV replicates its genome in the nucleus. A role for these structures in the assembly and morphogenesis of new HIV virions cannot be excluded. The early stage autophagosomes may serve to target newly formed HIV virions to multi-vesicular bodies in macrophages that are believed to mediate the release of HIV from this cell type [91]. Besides regulating autophagy in its host cell, HIV also may induce autophagy in adjacent uninfected T cells. This induction is believed to be mediated through the interaction of HIV-1 envelope glycoproteins expressed on the surface of infected host cells with CD4 and CXCR4 molecules on neighbouring uninfected T cells [92]. This autophagy-inducing function of the HIV envelope protein has been further mapped to the fusogenic function of the HIV gp41 envelope component [93]. Both HIV viruses that utilise the CXCR4 and the CCR5 co-receptors can induce autophagy in bystander T cells [81]. Autophagy induction in innocent bystander cells is thought to trigger both apoptosis and autophagy gene-dependent cell death and thereby, is postulated to represent a mechanism by which uninfected CD4 T cells may be deleted in HIV-infected patients. However, the role of autophagy in CD4 T cell death in vivo is not yet established.
A number of viruses with RNA genomes have been reported to induce autophagy-like events and/or or require the formation of autophagosome-like structures in vitro but further investigation is needed to more precisely define the roles of autophagy proteins, double-membraned vesicles, and the classical autophagy pathway in these infections. A major unanswered question is whether the involvement of an autophagy protein in the formation of membranes involved in viral replication, virion morphogenesis or viral egress truly reflects the use of ‘autophagy’ for viral replication or rather, represents alternative functions of Atg proteins in the formation of membranous compartments involved in the viral life cycle. Influenza A, Dengue and HCV genotype 1a all have been reported to induce autophagy in vitro and pharmacological or genetic inactivation of autophagy results in lower viral yields [94–96]. HCV induces early stages of autophagy in part through activation of the UPR in infected cells but inhibits the maturation of autophagosomes to autolysosomes [97]. Furthermore, HCV proteins fail to co-localise with markers of the autophagosome, suggesting that the virus may possess mechanisms to avoid its own autophagic sequestration during the induction of cellular autophagy [96,97]. Coxsackievirus B4 (CVB4) induces autophagy by stimulating the action of calpains through an unknown mechanism, and autophagy induction by CVB4 facilitates its replication in cultured rat primary neurons [98]. Further studies will be needed to clarify which if any gene(s) in these viruses mediate the observed activation of autophagy/suppression of xenophagy, and whether the observed induction of autophagy is relevant for in vivo pathogenesis by these viruses. Other than Dengue virus, poliovirus and rotavirus, there is as yet no evidence for the association of proteins from any of these viruses with autophagosomes or markers of autophagosomes (e.g. LC3) in infected cells. One interesting hypothesis is that these viruses upregulate autophagy to inhibit anti-viral innate immune responses as discussed above.
At least one beneficial aspect of autophagy—either for the virus or the host cell depending on the context—may be the inhibition of virusinduced apoptosis. Classical autophagy provides energy and macromolecules that help cells to survive during stress conditions that might otherwise trigger apoptosis [99]. As virus infections dramatically upregulate metabolism in infected cells, the consequent depletion of energy and macromolecules may trigger cells to undergo apoptosis, thereby limiting the ability of infected cells to produce new progeny virions [100]. The human parvovirus B19 is a small single-stranded DNA virus that infects erythroid cells and induces mitophagy and autophagy in infected cells [101]. Pharmacologic inhibition of autophagy in infected cells results in increased B19-induced apoptosis. Therefore, one potential hypothesis is that autophagy induction has both a cytoprotective and a pro-viral role in B19-infected cells; by providing energy and macromolecules in infected cells, it functions to inhibit apoptosis and allow the continued production of new virions. It is also possible that the observed mitophagy represents a virus-induced mechanism to dampen innate immune responses to B19.
A few viral proteins have been shown to interact with autophagy regulators or trigger autophagy when expressed in isolation in vitro. For example, overexpression of hepatitis B virus × protein in hepatic cell lines can transactivate the beclin 1 promoter to increase beclin 1 mRNA and protein levels [102]. Expression of the E7 oncoprotein of the DNA virus human papillomavirus type 16 increases levels of LC3-II in transfected cells in a 3-MA-sensitive manner [103]. The SV40 small T antigen can enhance starvation-induced autophagy in cancer cells in vitro through activating the energy sensor AMP kinase (AMPK) which suppresses mTOR signaling [104]. Although these studies indicate a role for these viral proteins in regulating autophagy in vitro, further investigation is needed to determine whether these genes regulate autophagy in in vitro or in vivo infection models in the context of their complete genomes.
SUBVERSION OF AUTOPHAGY FOR VIRAL REPLICATION
All positive-stranded RNA viruses induce membrane rearrangements inside infected cells to serve as scaffolds for these viruses to replicate their genomes [105]. The origin of these virus-induced membranes is still unknown. Some studies indicate that certain virus-induced membrane rearrangements share characteristics with autophagosomes including a double-membraned architecture and colocalisation with LC3. However, most of these rearrangements are smaller in size than traditional autophagosomes and it is not clear what, if any, cargo is contained inside them. Two members of the picornavirus family, poliovirus and coxsackievirus B3, have both been shown to induce autophagosome-like structures in infected cells, and replicases from both viruses associate with these structures [106,107]. With both of these viruses, pharmacologic or genetic inhibition of autophagy reduces viral yields while pharmacologic or starvation-induced stimulation of autophagy increases viral yields, suggesting that these viruses may subvert the classical autophagy machinery to create scaffolds for replication. The expression of poliovirus protein 2BC by itself can induce formation of the cleaved and lipidated form of LC3, LC3-II, but the expression of both 2BC and poliovirus protein 3A are needed for formation of the double-membraned structures seen in poliovirus-infected cells [106,108]. In addition to serving as a scaffold for the replication of the poliovirus genome, it is also possible that poliovirus utilises autophagosome-like structures for the non-lytic release of progeny virions as depletion of autophagy genes was found to have a more significant effect on extracellular viral release than on viral replication [106]. If these poliovirus-and coxsackievirus-induced structures are indeed ‘autophagosomes’, it is not known how either virus induces their formation or blocks their maturation into autolysosomes. The non-structural protein 4 (NSP4) of another ssRNA virus, rotavirus, localises to LC3-positive vesicular structures when expressed in HEK cells, and during rotavirus infection, NSP4 co-localises with LC3 in viroplasms which are sites of nascent RNA replication [109]. The replication machinery of both Dengue virus 2 and 3 (DENV-2 and DENV-3) associate with amphisomes in HepG2 cells, but only DENV-2 blocks further maturation of the amphisome into an autolysosome [110]. Dengue virus 3 does not block autolysosome formation suggesting a role for this structure in the DENV-3 life cycle [111]. Further investigation is needed to determine what viral or virus-induced factors account for the different requirements for autolysosomes in DENV-2 and DENV-3 infections.
Subversion of autophagy or autophagosomal components does not appear to be a universal mechanism for the replication of positive-stranded RNA viruses. Another human picornavirus, human rhinovirus type 2, was not found to induce LC3-positive vesicles in infected cells and its replication was not affected by drugs that promote (tamoxifen and rapamycin) or inhibit (3-MA) autophagy [112]. In addition, the replication of a Drosophila picornavirus was unaffected by siRNA against several different autophagy genes [113]. The nidovirus family, which includes coronoviruses and arteriviruses, is another family of single-stranded RNA viruses that are known to induce double-membraned structures in infected cells for the purpose of replicating their genomes [114]. Unlike certain picornaviruses, nidovirus replication seems to be unaffected by the absence of an intact autophagosomal pathway. The murine hepatitis virus (MHV), a coronovirus, replicates equally well in wild type or Atg5−/− bone marrow-derived macrophages and MEFs, but it is not known what cellular mechanisms govern MHV-induced formation of double-membraned vesicles [115]. An ultrastructural analysis of membrane vesicles induced by the severe acute respiratory syndrome coronovirus suggested that these vesicles are most likely derived from the endoplasmic reticulum [116]. Thus, it is possible that some of these RNA virus-induced double-membraned vesicles are derived from other intracellular membranes and do not require the autophagy machinery.
The utilisation of autophagosomes or autophagy proteins for DNA virus replication or morphogenesis has not been reported. Vaccinia virus is a large double-stranded DNA virus that replicates exclusively in the cytoplasm and immature vaccinia virus virions are surrounded by double-membraned structures that appear similar to autophagosomes. However, vaccinia virus maturation and the formation of double-membraned structures around immature virions was normal in Atg5−/− MEFs and beclin 1−/− embryonic stem cells [117]. Furthermore, vaccinia virus growth was not affected either positively or negatively in these cell lines that lack autophagy execution genes. As most DNA viruses replicate their genomes in the nucleus, they are not expected to require the autophagy machinery for their replication, but a role for autophagy or autophagy genes in the assembly of new DNA virions or in the acquisition of outer phospholipid membranes cannot be ruled out.
CONCLUSION
Autophagy is a vital cellular process that maintains homeostasis by removing damaged or superfluous organelles and that protects cells during stress by providing a source of energy and macromolecules to maintain essential cellular processes until normal growth conditions return. Given this primordial role of autophagy in removing harmful components of the cytoplasm, it is not surprising that autophagy may be used as a cell-intrinsic defence against viruses. In turn, given the ability of autophagy and autophagy genes to regulate diverse processes in cells and organisms, it is not surprising that viruses have evolved to interact with this process to promote their own survival. Looking forward, it will be necessary to further define the mechanism(s) by which autophagy and other autophagy gene-dependent processes affect the outcome of different viruses in relevant cell types in vitro and in vivo. A better understanding of how different viruses manipulate autophagy and interact with autophagy genes to enhance replication and pathogenesis may lead to the development of a new class of specific antiviral therapies that antagonise the pro-viral and promote the anti-viral functions of autophagy.
ACKNOWLEDGEMENTS
The authors thank Angela Diehl for her expert medical illustration. The work in the authors’ laboratory was supported in part by an Ellison Medical Foundation Senior Scholars Award in Infectious Diseases (B.L.) and NIH grants ROI A151367 (B.L.) and CA109618 (B.L.). We apologise to the authors whose work could not be included.
Abbreviations used
- Aβ
intraneuronal amyloid β
- AMPK
AMP kinase
- Atg
autophagy
- CVB4
Coxsackievirus B4
- DENV-2
dengue virus 2
- DENV-3
dengue virus 3
- dsDNA
double-stranded DNA
- eIF2α
eukaryotic initiation factor 2α
- GFP-LC3
green-fluorescent protein-light chain 3
- HIVE
HIV-induced encephalopathies
- HR
hypersensitive response
- JNK1
c-jun-N-terminal kinase 1
- LC3-I
non-lipidated form of LC3
- LC3
microtubule-associated protein, light chain 3
- LC3-II
lipidated form of LC3
- LMP1
latent membrane protein 1
- LPS
lipopolysaccharide
- 3-MA
3-methyladenine
- MDM
monocyte-derived macrophages
- MHV
murine hepatitis virus
- mTOR
mammalian target of rapamycin
- Nef
negative factor
- NSP4
nonstructural protein 4
- PAMP
pathogen-associated molecular pattern
- PCD
programmed cell death
- PDCs
plasmacytoid dendritic cells
- PE
phosphatidylethanolamine
- PI3K
phosphatidylinositol 3-kinase
- PIP3
phosphatidylinositol 3-phosphate
- PP1α
protein phosphatase 1α
- PRRs
pattern recognition receptors
- RIG-I
retinoic acid-inducible gene I
- RLRs
retinoic inducible gene-like receptors
- ROS
reactive oxygen species
- SIV
simian immunodeficiency virus
- ssRNA
single-stranded RNA
- SV
Sendai virus
- TLRs
toll-like receptors
- TMV
tobacco mosaic virus
- UPR
unfolded protein response
- VSV
vesicular stomatitis virus
REFERENCES
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talloczy Z, Jiang W, Virgin HW, 4th, et al. Regulation of starvation- and virus-induced autophagy by the eIF2a kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99(1):190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12 Suppl 2:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 7.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 10.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C, Lee JS, Inn KS, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152(4):657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurode-generative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurode-generation in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 24.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 25.Korolchuk VI, Menzies FM, Rubinsztein DC. A novel link between autophagy and the ubiquitin-proteasome system. Autophagy. 2009;5(6):862–863. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- 26.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 28.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(4):517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, Fux B, Goodwin M, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host and Microbe. 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10(5):461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolovan CA, Sawtell NM, Thompson RL. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn + are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J Virol. 1994;68(1):48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kesari S, Lasner TM, Balsara KR, et al. A neuroattenuated ICP34.5-deficient herpes simplex virus type 1 replicates in ependymal cells of the murine central nervous system. J Gen Virol. 1998;79(Pt 3):525–536. doi: 10.1099/0022-1317-79-3-525. [DOI] [PubMed] [Google Scholar]
- 35.Liu BL, Robinson M, Han ZQ, et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10(4):292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 36.He B, Gross M, Roizman B. The g(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1a to dephosphorylate the a subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markovitz NS, Baunoch D, Roizman B. The range and distribution of murine central nervous system cells infected with the g(1)34.5- mutant of herpes simplex virus 1. J Virol. 1997;71(7):5560–5569. doi: 10.1128/jvi.71.7.5560-5569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr I, Sternberg D, Ward S, Leib D, Mulvey M, Gluzman Y. A herpes simplex virus type 1 g34.5 second-site suppressor mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J Virol. 2001;75(11):5189–5196. doi: 10.1128/JVI.75.11.5189-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orvedahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host and Microbe. 2007;1(1):23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Talloczy Z, Virgin HW, 4th, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2(1):24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 41.Smith JD, de Harven E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. III. Cytochemical localization of lysosomal enzymes in infected cells. J Virol. 1978;26(1):102–109. doi: 10.1128/jvi.26.1.102-109.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventruti A, Cuervo AM. Autophagy and neurodegeneration. Curr Neurol Neurosci Rep. 2007;7(5):443–451. doi: 10.1007/s11910-007-0068-5. [DOI] [PubMed] [Google Scholar]
- 43.Itzhaki RF, Cosby SL, Wozniak MA. Herpes simplex virus type 1 and Alzheimer’s disease: the autophagy connection. J Neurovirol. 2008;14(1):1–4. doi: 10.1080/13550280701802543. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30(4):588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 47.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8(12):911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27(1):135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol. 2009;21(1):53–62. doi: 10.1016/j.coi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitoh T, Fujita N, Jang MH, et al. Loss of autophagy protein Atg16L1 enhances endotoxin-induced IL-1b production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 51.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283(48):33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27(7):1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 54.Jounai N, Takeshita F, Kobiyama K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104(35):14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106(8):2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nimmerjahn F, Milosevic S, Behrends U, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33(5):1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 57.Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102(22):7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26(1):79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paludan C, Schmid D, Landthaler M, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307(5709):593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 60.English L, Chemali M, Duron J, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10(5):480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vossen MT, Westerhout EM, Soderberg-Naucler C, Wiertz EJ. Viral immune evasion: a masterpiece of evolution. Immunogenetics. 2002;54(8):527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- 62.Malim MH, Emerman M. HIV-1 accessory proteins-ensuring viral survival in a hostile environment. Cell Host and Microbe. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 63.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2a phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14(2):230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 64.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007;81(22):12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4(8):989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ku B, Woo JS, Liang C, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gammaherpesvirus 68. PLoS Pathog. 2008;4(2):e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 68.Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarakanova VL, Kreisel F, White DW, Virgin HW., 4th Murine gammaherpesvirus 68 genes both induce and suppress lymphoproliferative disease. J Virol. 2008;82(2):1034–1039. doi: 10.1128/JVI.01426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gangappa S, van Dyk LF, Jewett TJ, Speck SH, Virgin HW., 4th Identification of the in vivo role of a viral bcl-2. J Exp Med. 2002;195(7):931–940. doi: 10.1084/jem.20011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Damania B. Oncogenic gamma-herpesviruses: comparison of viral proteins involved in tumorigenesis. Nat Rev Microbiol. 2004;2(8):656–668. doi: 10.1038/nrmicro958. [DOI] [PubMed] [Google Scholar]
- 72.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446(7137):745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 73.Wang L, Damania B. Kaposi’s sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res. 2008;68(12):4640–4648. doi: 10.1158/0008-5472.CAN-07-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sodhi A, Chaisuparat R, Hu J, et al. The TSC2/ mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10(2):133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 75.Chaumorcel M, Souquere S, Pierron G, Codogno P, Esclatine A. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy. 2008;4(1):46–53. doi: 10.4161/auto.5184. [DOI] [PubMed] [Google Scholar]
- 76.Isler JA, Skalet AH, Alwine JC. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol. 2005;79(11):6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xuan B, Qian Z, Torigoi E, Yu D. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J Virol. 2009;83(8):3463–3474. doi: 10.1128/JVI.02307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol. 2004;78(20):11030–11039. doi: 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchkovich NJ, Yu Y, Zampieri CA, Alwine JC. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6(4):266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22(6):695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Espert L, Varbanov M, Robert-Hebmann V, et al. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS ONE. 2009;4(6):e5787. doi: 10.1371/journal.pone.0005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kyei GB, Dinkins C, Davis AS, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186(2):255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winslow AR, Rubinsztein DC. Autophagy in neurodegeneration and development. Biochim Biophys Acta. 2008;1782(12):723–729. doi: 10.1016/j.bbadis.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Achim CL, Adame A, Dumaop W, Everall IP, Masliah E. Increased accumulation of intraneuronal amyloid beta in HIV-infected patients. J Neuroimmune Pharmacol. 2009;4(2):190–199. doi: 10.1007/s11481-009-9152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alirezaei M, Kiosses WB, Flynn CT, Brady NR, Fox HS. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS ONE. 2008;3(8):e2906. doi: 10.1371/journal.pone.0002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi MN, Jackson W, Laird DT, et al. Varicella-zoster virus infection induces autophagy in both cultured cells and human skin vesicles. J Virol. 2009;83(11):5466–5476. doi: 10.1128/JVI.02670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27(20):2833–2842. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 88.Lam N, Sandberg ML, Sugden B. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J Virol. 2004;78(4):1657–1664. doi: 10.1128/JVI.78.4.1657-1664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee DY, Sugden B. The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood. 2008;111(4):2280–2289. doi: 10.1182/blood-2007-07-100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319(5865):921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 91.Carter CA, Ehrlich LS. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol. 2008;62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 92.Espert L, Denizot M, Grimaldi M, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116(8):2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Denizot M, Varbanov M, Espert L, et al. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4(8):998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- 94.Zhou Z, Jiang X, Liu D, et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5(3):321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- 95.Lee YR, Lei HY, Liu MT, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374(2):240–248. doi: 10.1016/j.virol.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82(5):2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48(4):1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoon SY, Ha YE, Choi JE, et al. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol. 2008;82(23):11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 100.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2(12):e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nakashima A, Tanaka N, Tamai K, et al. Survival of parvovirus B19-infected cells by cellular autophagy. Virology. 2006;349(2):254–263. doi: 10.1016/j.virol.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 102.Tang H, Da L, Mao Y, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of Beclin 1 expression. Hepatology. 2009;49(1):60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- 103.Zhou X, Munger K. Expression of the human papillomavirus type 16 E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology. 2009;385(1):192–197. doi: 10.1016/j.virol.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kumar SH, Rangarajan A. SV40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J. Virol. 2009 doi: 10.1128/JVI.00603-09. doi:10.1128/JVI.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jackson WT, Giddings TH, Jr, Taylor MP, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wong J, Zhang J, Si X, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82(18):9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taylor MP, Kirkegaard K. Modification of cellular autophagy protein LC3 by poliovirus. J Virol. 2007;81(22):12543–12553. doi: 10.1128/JVI.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006;80(12):6061–6071. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Panyasrivanit M, Khakpoor A, Wikan N, Smith DR. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009;90(Pt 2):448–456. doi: 10.1099/vir.0.005355-0. [DOI] [PubMed] [Google Scholar]
- 111.Khakpoor A, Panyasrivanit M, Wikan N, Smith DR. A role for autophagolysosomes in DEN-3 production in HepG2 cells. J Gen Virol. 2009;90(pt 5):1093–1103. doi: 10.1099/vir.0.007914-0. [DOI] [PubMed] [Google Scholar]
- 112.Brabec-Zaruba M, Berka U, Blaas D, Fuchs R. Induction of autophagy does not affect human rhinovirus type 2 production. J Virol. 2007;81(19):10815–10817. doi: 10.1128/JVI.00143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2(10):e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de Haan CA, Reggiori F. Are nidoviruses hijacking the autophagy machinery? Autophagy. 2008;4(3):276–279. doi: 10.4161/auto.5241. [DOI] [PubMed] [Google Scholar]
- 115.Zhao Z, Thackray LB, Miller BC, et al. Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3(6):581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 116.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang H, Monken CE, Zhang Y, et al. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2(2):91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]