Abstract
Members of the bone morphogenetic protein (BMP) subfamily of cytokines control many aspects of metazoan development including patterning and organogenesis. Despite the recognition that schistosomes possess key components of a BMP signaling pathway, a BMP-like ligand in the parasitic flatworm Schistosoma mansoni remained elusive. Here, we describe the cloning and characterization of an S. mansoni BMP (SmBMP). SmBMP is most closely related to BMP homologues from the free-living flatworms Schmidtea mediterranea and Dugesia japonica, with 51% and 47% identity at the amino acid level, respectively. Based on reverse transcription-PCR, SmBMP is expressed throughout the mammalian life cycle of the parasite in both male and female schistosomes. In support of these results, antibodies to SmBMP successfully immunoprecipitated the protein in adult male and female antigen preparations with more protein detected in male parasites. Immunofluorescent studies localized SmBMP to the protonephridia of adult parasites, and SmBMP was identified in the excretory/secretory products of adult male parasites via immunoprecipitation. With the previous description of a TGF-β subfamily homologue in S. mansoni, ligands representing both arms of the TGF-β superfamily have now been described in this trematode.
Keywords: Trematode, Schistosome, Transforming growth factor-beta, Bone morphogenetic protein, Protonephridia
1. Introduction
Over 600 million people are at risk of infection with trematode parasites of the genus Schistosoma (Chitsulo et al., 2000). Schistosomiasis is one of the nine neglected tropical diseases that have received considerable attention over the last several years as the causes of extensive morbidity (Hotez et al., 2006). While seven of these neglected diseases are caused by nematode infections, only schistosomiasis is caused by a platyhelminth.
TGF-β signaling in mammals and model metazoan organisms such as Caenorhabditis elegans and Drosophila melanogaster, are known to mediate a large number of physiological processes including growth and differentiation, cell death, tissue repair and developmental patterning (Massague et al., 2000). Members of the TGF-β superfamily can be split into two main subfamilies based on sequence homology and the different downstream pathways they activate, namely the TGF-β/activin/nodal subfamily and the bone morphogenetic protein/growth and differentiation factor/Muellerian inhibiting substance (BMP/GDF/MIS) subfamily (Shi and Massague, 2003). The basic TGF-β signaling mechanism involves binding of the extracellular TGF-β homologue to a heterodimeric receptor complex resulting in the activation of specific cytoplasmic proteins (Smads) that eventually translocate to the nucleus to influence transcriptional responses. In mammals, it is known that members of the two subfamilies activate distinct classes of Smad proteins: TGF-β subfamily members activate Smad2 and Smad3 homologues while BMP subfamily members activate Smad1, Smad5 and Smad8 homologues. Several components of TGF-β signaling have been characterized from S. mansoni including TGF-β receptors SmRK1 (also known as SmTβRI) (Davies et al., 1998) and SmRKII (also known as SmTβRII) (Forrester et al., 2004; Osman et al., 2006), several Smad proteins (Beall et al., 2000; Osman et al., 2001, 2004; Carlo et al., 2007), and one homologue of the TGF-β subfamily, SmInAct (Freitas et al., 2007).
Previous work elucidating the role of TGF-β signaling in S. mansoni has been focused on components of the TGF-β subfamily, where this signaling pathway has been implicated in host-parasite interactions, parasite reproductive development and embryogenesis (Davies et al., 1998; Forrester et al., 2004; Osman et al., 2006; Freitas et al., 2007). The existence of a BMP signaling pathway in S. mansoni has been assumed since the cloning and characterization of two S. mansoni Smad1 homologues (SmSmad1 and SmSmad1b) (Beall et al., 2000; Carlo et al., 2007), however a ligand for the pathway has not been described. Here, we describe the cloning of an S. mansoni BMP homologue, SmBMP, and characterize its expression in various stages of parasite development.
2. Materials and methods
2.1. Parasites and animals
The Puerto Rican/Naval Medical Research Institute (NMRI) strain of S. mansoni was used in all experiments. Cercariae were collected by exposing infected Biomphalaria glabrata to light for 1 h. Adult parasites were recovered by hepatic-portal perfusion of C57BL/6 female mice (Jackson Laboratory, Bar Harbor, ME) infected 8 weeks previously via percutaneous exposure to ~ 60 cercariae. Schistosoma mansoni eggs were collected from the livers of infected mice as previously described (MacDonald et al., 2001). The use of mice in this study was approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania.
2.2. SmBMP isolation
The C-terminal deduced amino acid sequence of the D. melanogaster Decapentaplegic (DmDPP, NP 477311, amino acids 487–588) was used in a tblastn search of the Wellcome Trust’s Sanger Institute’s S. mansoni genome scaffolds version 3.1. (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/s_mansoni). A scaffold (Smp_scaff000116) with sequence showing homology to BMP-like ligands was identified and the corresponding putative coding sequence was named SmBMP. The 5′ and 3′ ends of SmBMP were isolated using total RNA (1 μg) from adult parasites and the Superscript III Generacer RACE kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. An SmBMP-specific primer was designed for isolating the 3′ end (5′-TTGGGTTATTGCACCACAAG-3′) and was used with the Generacer 3′ primer in reverse transcription (RT)-PCR as per the manufacturer’s instructions. The resulting amplicon was cloned into the TA-TOPO cloning vector (Invitrogen, Carlsbad, CA) and sequenced. An in silico analysis of the S. mansoni genome aided in the identification of the 5′ end of SmBMP. The Wellcome Trust’s Sanger Institutes S. mansoni predicted coding sequence databases (Augustus3, GlimmerHMM, and Twinscan2) were searched for SmBMP by performing a blastn search of each database with the isolated 3′ end of SmBMP. Each algorithm suggested different start sites and forward primers were designed corresponding to each (Augustus3 5′-ATGAAATATGCAAATGTCAGTT-3′; GlimmerHMM 5′-ATGTTTAAATTACATGAACGTCATA-3′; Twinscan2 5′-ATGGGGGAAAAGTCACTTACTCT-3′). Each forward primer was used individually with an SmBMP-specific reverse primer (5′-TGGAAATGGACATTGACCTAAACA-3′) in an RT-PCR using cercariae cDNA as template. Only the combination of the GlimmerHMM forward primer with the SmBMP-specific reverse primer amplified a product of predicted size which was confirmed to be SmBMP upon sequencing. Further in silico analysis revealed a long open reading frame (1,027 bp) upstream of the predicted GlimmerHMM 5′ end. To isolate the 5′ end of SmBMP, two SmBMP-specific reverse primers were designed within the GlimmerHMM amplified region and were used in 5′ Rapid Amplification of cDNA ends (RACE) (5′-CTGGTTCAAAAATGGCTGCGGTTTG-3′) and nested 5′ RACE (5′-TGCTGCAACTAATTTTTCATTTGAAGGT-3′) reactions using the Generacer RACE kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The resulting amplicon was cloned and was confirmed by sequencing to be SmBMP (GenBank accession number EU684544).
2.3. Sequence analysis
Sequence comparisons between the deduced amino acid sequence of SmBMP and other TGF-β superfamily members were determined using the ClustalW algorithm and the Align 2 sequences (bl2seq) program at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi) using the final ~ 100 amino acid residues of each sequence. An unrooted dendrogram was drawn using the final ~ 100 amino acids within the conserved domain of SmBMP and other members of the TGF-β superfamily, and distances were drawn using the Jones-Taylor-Thornton matrix and neighbor joining algorithm in the PHYLIP software package developed by J. Felsenstein (University of Washington, Seattle, Washington). Percentages at branch points were based on 1,000 bootstrap runs.
2.4. RNA extraction and RT-PCR
Total RNA was extracted from parasite material using Qiagen’s RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Contaminating genomic DNA was removed using Turbo DNA-free endonuclease (Applied Biosystems, Foster City, CA). First strand cDNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Briefly, 200 ng of total RNA from S. mansoni eggs, cercariae, adult males and adult females were reverse transcribed using Superscript II Reverse Transcriptase primed with oligo dT. RT-minus controls were preformed to confirm the absence of any contaminating genomic DNA (data not shown).
Presence of an SmBMP transcript in the various parasite life stages was determined via conventional RT-PCR using Invitrogen’s Recombinant taq DNA polymerase according to the manufacturer’s instructions with 1 μl of cDNA from each stage tested as template or water as a negative control. SmBMP primers were: forward 5′-GGTTGGGCTGGTTGGGTTAT-3′ and reverse 5′-TGGAAATGGACATTGACCTAAACA-3′. Paramyosin primers were: forward 5′-CGTGAAGGTCGTCGTATGGT-3′ and reverse 5′-GACGTTCAAATTTACGTGCTTG-3′. The cycling program used was 94°C for 2 min, then 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, followed by 72°C for 5 min. Amplified products were resolved on a 2% ethidium bromide stained agarose gel.
2.5. Recombinant SmBMP expression and antisera generation
EcoRI (forward) and XhoI (reverse) tagged primers were designed to amplify the region corresponding to the last 179 amino acids of SmBMP (forward 5′-GGAATTCAGTCAAGATCGTTATTATTA-3′ and reverse 5′-CCGCTCGAGTTAACGACAAGCACAACTTT-3′). The cycling conditions were 1 cycle of 94 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 65 °C for 30 s, and 68 °C for 1 min, ending with one cycle of 68 °C for 5 min using Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA). The amplified product was digested with EcoRI and XhoI and cloned into the bacterial expression vector pET28a (Novagen, Madison, WI), also digested with EcoRI and XhoI. Sequence analysis confirmed the absence of any mutations. Expression of recombinant SmBMP was induced in Escherichia coli BL21(DE3) by addition of 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG) when cultures reached an O.D.600 of 0.5 at 37 °C followed by further shaking at 37 °C for 3 h. Recominant SmBMP was expressed in the bacteria as insoluble inclusion bodies and was purified via nickel column chromatography under denaturing conditions (6 M urea) as per the manufacturer’s suggestions (Novagen, Madison, WI). Antiserum to SmBMP was generated by Cocalico Biologicals (Cocalico Biologicals, Inc., Reamstown, PA) by s.c. inoculation of 100 μg of purified recombinant SmBMP in FCA, followed by three boosts of 50 μg in Freund’s incomplete adjuvant on days 14, 21 and 49, followed by exsanguination on day 64.
2.6. Immunoprecipitation and Western blot analysis
Antigen from adult male and female parasites was extracted by boiling worms in lysis buffer (1% SDS, 200 mM NaCl, 40 mM Tris pH8.8) and sheared via repeated passage through a 23 G needle. Protein concentration was determined by bicinchoninic acid (BCA) Protein Assay (Pierce, Rockford, IL). For immunoprecipitation of SmBMP, 80 μg of male and female antigen were diluted with 7 vol. of 1% NP40 and pre-cleared with 10 μl of anti-Rabbit IgG beads from the Rabbit IgG TrueBlot kit (eBioscience, San Diego, CA) for 1 h on a rocking platform at 4°C. Supernatant was removed and incubated with 3 μg of either pre-immune rabbit IgG or anti-SmBMP IgG at 4°C for 16 h on a rocking platform. Antibodies were precipitated by adding 15 μl of anti-Rabbit IgG beads to the antigen-IgG mixtures and incubated for 1 h at 4°C on a rocking platform. Precipitated antibody-antigen complexes were washed three times with 1% NP40 and prepared for SDS-PAGE analysis by boiling in a reducing SDS lysis buffer (50 mM Tris pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol) in a total volume of 25 μl. Samples were separated by 10% SDS-PAGE, electroblotted onto Immobilon nylon membrane (Millipore, Billerica, MA), and probed with anti-SmBMP antiserum (1:10,000). Bound rabbit antibodies were detected using Rabbit IgG TrueBlot antibodies (eBioscience, San Diego, CA) according to the manufacturer’s instructions. As a loading control, 20 μg of each adult antigen preparation used in the immunoprecipitation were also separated via 10% SDS-PAGE and electroblotted. This blot was probed with a monoclonal antibody (4B1) against paramyosin (1:10,000) and bound antibodies were detected using affinity purified horseradish peroxidase (HRP)-conjugated horse anti-mouse IgG (Cell Signaling Technology, Danvers, MA). Secondary antibodies were detected using ECL reagents as directed by the manufacturer (GE Healthcare, Piscataway, NJ).
Excretory/secretory products from adult male parasites were collected by culturing 10 worms in each well of a six well plate in a total volume of 4 ml of M199 (Invitrogen, Carlsbad, CA), 10% FCS, 1% Antibiotic/Antimycotic (Gibco), and 1% HEPES in a 37 °C/5% CO2 atmosphere for 48 h. Harvested culture supernatant was pre-cleared with 30 μl of anti-Rabbit IgG beads for 1 h on a rocking platform at 4°C, split into equal volumes and incubated with 3 μg of either pre-immune rabbit IgG or anti-SmBMP IgG at 4°C for 16 h on a rocking platform. Antibodies were precipitated and antibody-antigen complexes were detected as described above.
2.7. Immunolocalization
To localize SmBMP within adult parasites, 5 μm sections of worms were treated with xylene and rehydrated through an ethanol series ending in PBS. Slides were blocked with 2% normal goat serum (Sigma, St. Louis, MO) for 1 h at room temperature and probed with either rabbit pre-immune serum or anti-SmBMP serum (1:100) overnight at 4°C. Sections were washed three times in PBS and bound rabbit antibodies were detected by probing with a fluorescein-conjugated F(ab′)2 goat anti-rabbit IgG (1:50) (Jackson Immunoresearch, West Grove, PA) for 1 h at room temperature. Slides were washed three times in PBS, dehydrated in ethanol and mounted. Worm sections were photographed using a Leica DMIRB microscope and DC500 camera (Leica, Germany).
3. Results
3.1. Cloning and sequence analysis of SmBMP
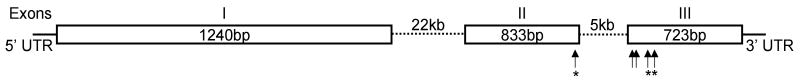
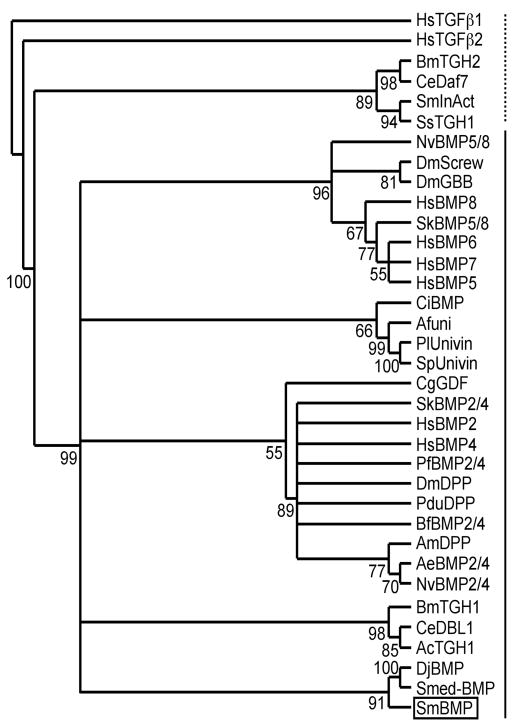
SmBMP was identified in the Wellcome Trust’s Sanger Institute’s S. mansoni genome sequence database through a tblastn search using the last 100 amino acids of the D. melanogaster decapentaplegic (DPP) protein. The 5′ and 3′ ends of SmBMP were isolated via 5′ and 3′ RACE using SmBMP-specific primers designed from within the predicted coding sequence according to the genome sequence and adult parasite cDNA as template. The full-length transcript is 3,013 bp including a 124 bp 5′ untranslated region (UTR) and a 102 bp 3′UTR with a poly-A tail. The deduced amino acid sequence of SmBMP is 931 residues and contains many of the molecular characteristics of a BMP homologue including several putative proteolytic cleavage sites at positions 660 (RKPR), 700 (RYKR), 703 (RLQR), 738 (RSRR) and 748 bp (RHNR) where the bioactive, C-terminal domain (268–180 amino acids) could be enzymatically cleaved from the N-terminal pro-domain (Fig. 1). Seven invariant cysteine residues are predicted in SmBMP along with other invariant residues (proline 851 and glycine 861) necessary for proper tertiary structure and dimerization of a functional BMP homologue (Fig. 2). Interestingly, signal sequence prediction software suggests the N-terminus of SmBMP does not contain a secretion signal. Within the C-terminal conserved domain (the last ~ 100 amino acids), SmBMP is 51% and 47% identical to BMP homologues from the free-living flatworms Schmidtea mediterranea and Dugesia japonica, respectively (Fig. 2). Furthermore, it is 46% identical to D. melanogaster DPP and 49% identical to human BMP2 at the amino acid level (Fig. 2). Phylogenetic analysis of SmBMP among other members of the TGF-β superfamily groups this homologue with members of the BMP subfamily (Fig. 3) and most closely with BMP-like ligands from S. mediterranea and D. japonica (Smed-BMP and DjBMP). This close clustering is expected considering all three worms are members of the phylum Platyhelminthes. Based on this analysis, it cannot be determined whether SmBMP is most closely related to members of the BMP2/4 or BMP5/8 subfamilies.
Fig. 1.
Genomic organization of SmBMP. Boxes represent exons with Roman numerals above each box indicating the exon number. Dashed lines represent introns. Exon length is indicated within each box while intron length is indicated above each dashed line. The solid lines flanking the coding region of SmBMP represent the 5′ and 3′ untranslated region (UTR) of the transcript. Arrows indicate potential cleavage sites (RXXR) where asterisks specify those sites with experimental evidence.
Fig. 2.
Multiple sequence alignment of the deduced amino sequence of the C-terminal region of SmBMP and other members of the BMP/GDF/MIS subfamily of TGF-β proteins. Shaded residues represent the invariant cysteine, proline and glycine moieties of all BMP homologues. Numbers to the right indicate the position of the last amino acid in the row within each sequence. In the case of DjBMP, a full-length sequence is not available, therefore the positions are not applicable. Asterisks under the alignment indicate exact matches across all five sequences, “:” represents strong similarity, “.” represents weak similarity, and a lack of a symbol indicates no similarity among residues.
Fig. 3.
Phylogenetic representation of SmBMP among other members of the TGF-β superfamily. SmBMP (boxed) is shown clustering among members of the BMP/GDF/MIS subfamily (solid line) and not with members of the TGF-β subfamily (dashed line). Conserved amino acids in the C-terminal region (the last ~100 amino acids) of each protein was used in the analysis. Percentages at branch points are based on 1,000 bootstrapping events. Accession numbers for sequences used in the analysis are: Acropora millepora decapentaplegic (AmDPP), AAM54049; Actinia equina BMP2/4 (AeBMP2/4), AAK20912; Amphiura filiformis afuni (Afuni), AAX54512; Ancylostoma caninum TGH-1 (AcTGH1), AAX36081; Branchiostoma floridae BMP2/4 (BfBMP2/4), AAC97488; Brugia malayi TGH-1 (BmTGH1), AF012878; B. malayi TGH-2 (BmTGH2), AF104016; Caenorhabditis elegans DAF-7 (CeDAF7), NP_497265; C. elegans DBL-1 (CeDBL1), NP_504709; Ciona intestinalis TGF-β superfamily signaling ligand (CiBMP), NP_001072008; Crassostrea gigas GDF (CgGDF), CAA10268; Drosophila melanogaster decapentaplegic (DmDPP), NP_477311; D. melanogaster glass bottom boat (DmGBB), NP_477340; D. melanogaster Screw (DmScrew), NP_524863; Dugesia japonica BMP (DjBMP), BAA32087; Homo sapiens BMP-2 (HsBMP2), NP_001191; H. sapiens BMP-4 (HsBMP4), NP_001193; H. sapiens BMP-5 (HsBMP5), NP_066551; H. sapiens BMP-6 (HsBMP6), AAH99625; H. sapiens BMP-7 (HsBMP7), NP_001710; H. sapiens BMP-8 (HsBMP8), XP_001716617; H. sapiens TGF-β1 (HsTGFβ1), CAA29283; H. sapiens TGF-β2 (HsTGFβ2), NP_003229; Nematostella vectensis BMP2/4 (NvBMP2/4), AAR13362; N. vectensis BMP5-8 (NvBMP5/8), ABC88372; Paracentrotus lividus univin (PlUnivin), ABG00200; Platynereis dumerilii decapentaplegic (PduDPP), CAJ38807; Ptychodera flava BMP2/4 (PfBMP2/4), BAA89012; Saccoglossus kowalevskii BMP2/4 (SkBMP2/4), ABD97262; S. kowalevskii BMP5/8 (SkBMP5/8), ABD97263; S. mansoni TGF-β (SmInAct), ABI64156; Schmidtea mediterranea BMP (Smed-BMP), ABV04322; Strongylocentrotus purpuratus (SpUnivin), NP_999793; Strongyloides stercoralis (SsTGH1), AY662390.
3.2. SmBMP transcript and protein expression and localization
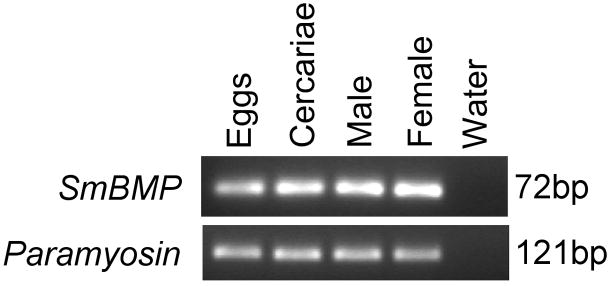
To determine the expression profile of the SmBMP transcript, conventional RT-PCR was performed on cDNA from eggs, cercariae, adult male and adult female parasites. As seen in Fig. 4, SmBMP is expressed across all four stages tested compared with the reference gene paramyosin. This expression pattern was reproducible between separate RT-PCR experiments with different RNA/cDNA preparations. SmBMP was not identified in blast searches of the Sanger Institute’s S. mansoni expressed sequence tag (EST) databases.
Fig. 4.
Expression of SmBMP transcript. First strand cDNA from various Schistosoma mansoni stages was used in reverse transcription-PCR. Ethidium bromide-stained gel of products after 35 cycles of amplification. Numbers to the right of each gel represent the amplicon length for each transcript. The life-cycle stage is indicated at the top of each lane.
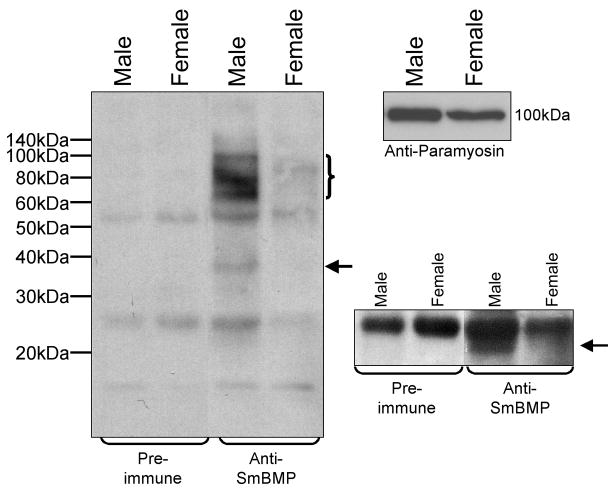
Immunoprecipitation and Western blot analyses using polyclonal antibodies against recombinant SmBMP were performed to investigate the expression of SmBMP protein. Here, we focused on SmBMP expression in adult male and female parasites. An ~ 100 kDa band was precipitated using anti-SmBMP antibodies in male antigen preparations which, based on the predicted molecular weight of the full-length transcript (109 kDa), corresponds to the preprocessed form of SmBMP (Fig. 5). This band was not immunoprecipitated using equivalent amounts of pre-immune antibodies. A smaller band was precipitated from female antigen (~ 85 kDa) as well as a smear of proteins under the 100 kDa band in the male antigen preparation. It is unclear at this time whether these are degraded products or represent alternative forms of SmBMP. A band representing a putative C-terminal, bioactive form of SmBMP with a molecular weight of ~ 37 kDa was also precipitated from male antigen while overexposing the same blot revealed a second potential bioactive form of SmBMP with a molecular weight of ~ 22 kDa. The ~ 37 kDa band could be the result of a cleavage event at the 660 bp predicted site while the ~ 22 kDa band may be formed by cleavage at either position 738 or 748. Based on the intensities of the pre-processed SmBMP bands, male parasites appear to express more SmBMP than female parasites.
Fig. 5.
Expression of native SmBMP protein. Antigen extracts of adult male and female parasites were immunoprecipitated with antibodies from either rabbit pre-immune serum or antibodies from rabbit serum after immunization with recombinant SmBMP. SmBMP was only precipitated when anti-SmBMP antibodies were used. The bracket indicates the pre-processed form of SmBMP (~ 100 kDa) while the arrow points to a processed form of SmBMP (~ 37 kDa). Heavy and light chains of the rabbit IgG molecules used in immunoprecipitation are seen at ~ 50 and 25 kDa, respectively. This blot was over-exposed and the region surrounding ~ 25 kDa is shown to the right with an arrow pointing to a second potential processed form of SmBMP (~ 22 kDa). Equivalent parasite antigen was loaded on a separate gel and probed with anti-paramyosin antibodies.
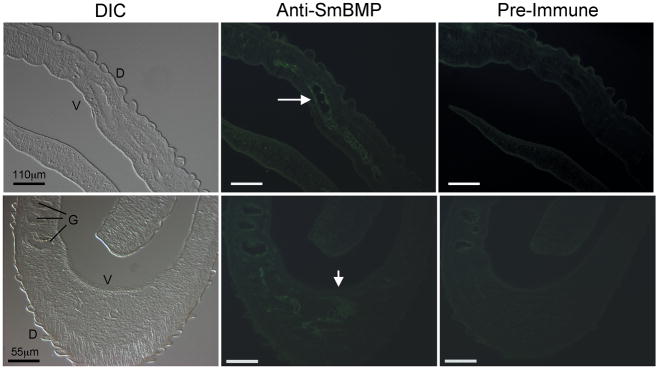
To determine where SmBMP is expressed in the worm, we performed immunofluorescence localization assays on sections of adult parasites. Anti-SmBMP antibodies localized SmBMP to the protonephridia of adult male parasites (Fig. 6). Intense staining was observed in the main tubules of the protonephridia that run laterally on either side of the worm (panel C) as well as within branches of the system (panel D). No positive staining was detected in parasite sections using pre-immune serum.
Fig. 6.
Immunofluorescence localization analyses. Panels A and B are differential interference contrast (DIC) images and panels C–F are fluorescence images of adult male sections. Arrows point to the localization of SmBMP in the main tubules (panel C) and branches of the protonephridia (panel D). G, gut; D, dorsal; V, ventral. Scale bar length indicated in panels A and B are the same as panels C and E, and D and F, respectively. Pictures shown are representative of two separate experiments.
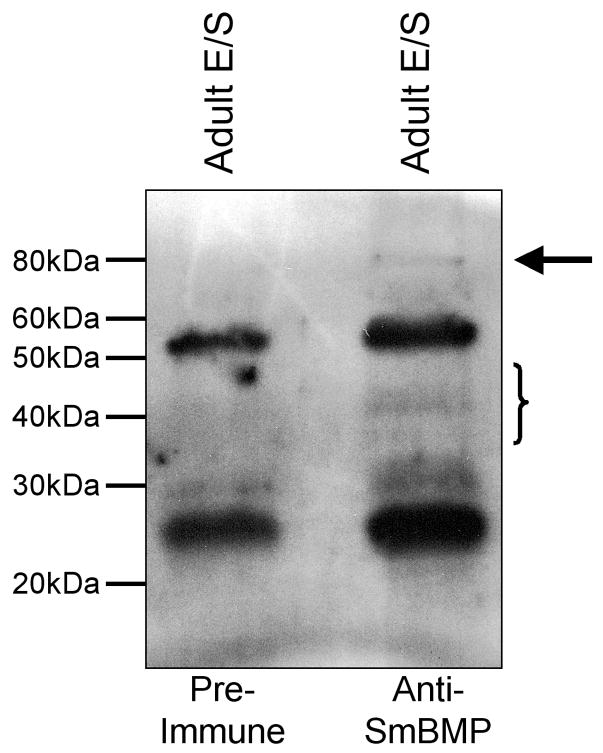
Using culture supernatant of adult male parasites, we performed immunoprecipitation and Western blot analyses to determine whether or not SmBMP is secreted from the worm. Anti-SmBMP antibodies immunoprecipitated several bands from media exposed to adult males including an ~ 80 kDa band and bands around 40 kDa in size. These bands were not immunoprecipitated using equivalent amounts of pre-immune antibodies.
4. Discussion
It is well established that S. mansoni possesses multiple components of active TGF-β signaling, however little focus has been given to the BMP subfamily in this parasitic flatworm. In this study, we report the identification and characterization of SmBMP, a BMP homologue in S. mansoni. SmBMP is expressed in medically important stages of the parasite, namely the egg, cercariae and adult parasites, and localizes to the protonephridia of the adult worm. A recent review by Loverde and colleagues alludes to a BMP homologue in the S. mansoni genome (Loverde et al., 2007). Assuming the gene described in this review is that found in Genbank under accession number EF028064, we believe the BMP homologue characterized in this study is the same as that described by Loverde et al. with two exceptions: (i) our clone’s deduced amino acid sequence is 402 amino acids longer at the N-terminus and, (ii) the putative cleavage site is not 129 amino acids upstream from the stop codon, but rather 272 amino acids upstream, with a second and/or third potential cleavage site(s) at either 194 or 184 amino acids upstream of the stop codon. Multiple cleavage sites are not atypical of BMP homologues (Cui et al., 2001). This would make the molecular weight of the SmBMP active dimer to be approximately 74 kDa (or ~44 kDa from either of the downstream sites), rather than the previously described 23 kDa (Loverde et al., 2007).
The deduced amino acid sequence of SmBMP is longer than all other full-length BMP homologues analyzed in this study. For example, SmBMP is 530 residues longer than its most related homologue, Smed-BMP, and 535 residues longer than human BMP-2. Most of this increased length is attributed to a longer N-terminus of SmBMP; 659 amino acids lie upstream of the first cleavage site of SmBMP (737 and 747 residues are upstream of the second and third potential cleavage sites, respectively) while 287 make up the N-terminus of Smed-BMP. Functions of the pro-domain in BMP homologues include directing the ligand to the extracellular matrix (Gregory et al., 2005) and in protein stability (Constam and Robertson, 1999). Furthermore, the pro-domain is believed to be essential for proper dimerization and folding of nearly all TGF-β superfamily members (Gray and Mason, 1990). It remains possible that, because different stages of S. mansoni live at different temperatures (~24°C in the intermediate snail host and environment and at 37°C in the definitive host), this longer pro-domain may aid in proper folding of the protein regardless of temperature.
One function of BMP signaling is to establish the dorsalventral (DV) axis in the development of both vertebrates and invertebrates (reviewed in De Robertis and Kuroda, 2004). Recently, RNA interference (RNAi) studies have shown that BMP signaling plays a role in specifying the DV axis in the free-living planaria D. japonica and S. mediterranea (Molina et al., 2007; Orii and Watanabe, 2007). Considering SmBMP is most closely related to the BMP homologues isolated from these free-living flatworms and that, while S. mansoni is a trematode and D. japonica and S. mediterranea are free-living flatworms, they are nonetheless closely related phylogenetically, it is likely that SmBMP plays a similar role in S. mansoni. We treated adult parasites as well as growing schistosomula with double-stranded RNA (dsRNA) corresponding to SmBMP, however, while ~40% knockdown was observed using quantitative RT-PCR (data not shown), immediate phenotypes were not observed. This could be due to insufficient knockdown compared with that obtained in D. japonica and S. mediterranea. It is appealing to speculate that the increase expression of SmBMP in male parasites is a reflection of a potential role for SmBMP in establishing the DV axis. The dorsal and ventral surfaces of male parasites are anatomically and physiologically much more distinct than in females with many tubercles on the dorsal surface and the formation of the gynacophoral canal on the ventral surface, for example.
The localization of SmBMP in S. mansoni is different than that observed of the two other platyhelminth BMP homologues where SmBMP is expressed in the protonephridia while DjBMP and Smed-BMP are found along the dorsal midline (Orii et al., 1998; Molina et al., 2007). The protonephridia is believed to serve as a primitive kidney, functioning to regulate the composition of fluids in the organism and in the clearance of metabolic wastes (Wilson and Webster, 1974). Other protein processing and secreting functions have also been proposed for this organ (Finken-Eigen and Kunz, 1997; Skelly and Shoemaker, 2001). One possible explanation for the discrepancy in localization could be the methods employed. We used immunfluorescence on worm sections to localize SmBMP while whole-mount in situ hybridization was used to localize both DjBMP and Smed-BMP in the free-living worms. Therefore, we could be detecting cells that have simply bound and are responding to SmBMP as opposed to those actually producing it. Additionally, considering the potential of the protonephridia to serve as a means of protein excretion/secretion, localization of SmBMP here may be explained by impending release of SmBMP in excretory/secretory products. To this end, we isolated proteins in the excretory/secretory products of adult male parasites using our antibodies against SmBMP, suggesting SmBMP is secreted by the parasite. This is an interesting finding in that secreted forms of SmBMP will have the opportunity to interact with host receptors. Lastly, BMP signaling is known to play an important role in the development of the vertebrate kidney (reviewed in Cain et al., 2008). Thus, it remains possible that SmBMP functions in the development and/or maintenance of the trematode protonephridia.
To date, two Smad1 homologues have been characterized in S. mansoni, SmSmad1 and SmSmad1b (Beall et al., 2000; Carlo et al., 2007). The expression of SmSmad1 has not been localized to specific tissues of the worm while SmSmad1b has been localization to the vitellaria, reproductive ducts and subtegumental tissues of the adult female and in subtegumental and an unidentified tissue of the adult male (Carlo et al., 2007). If SmBMP ligation to its receptor leads to the phosphorylation of both of these Smad1 homologues, SmBMP may have distinct functions in cells that express SmSmad1, SmSmad1b, or a combination of the two, by mediating different transcriptional responses. To our knowledge, only one Smad1 homologue has been identified in free-living flatworms (Molina et al., 2007). Future experiments will be necessary to determine if SmBMP is capable of leading to the phosphorylation of either S. mansoni Smad1 homologue.
With the isolation of a BMP homologue from S. mansoni, we now have representatives from each major subfamily of the TGF-β superfamily in this parasitic flatworm. The current version of the S. mansoni genome sequence suggests SmBMP and SmInAct are the only two TGF-β-like ligands present. TGF-β signaling is believed to be present in all metazoans providing a pathway that helps to regulate aspects of development key to being a multicellular organism, such as cell proliferation, differentiation, organization and death (Massague and Gomis, 2006). Considering D. melanogaster has seven TGF-β-like ligands and the nematode C. elegans has four, it will be interesting to determine how many functions the two TGF-β homolgues in the platyhelminth S. mansoni may fulfill.
Fig. 7.
Secretion of SmBMP from adult male parasites. Culture supernatants from male parasites were immunoprecipitated using antibodies from either rabbit pre-immune serum or antibodies from rabbit serum after immunization with recombinant SmBMP. The arrow points to an 80 kDa band specifically immunoprecipitated with anti-SmBMP antibodies while the bracket corresponds to bands ~ 35–42 kDa in size also immunoprecipitated with anti-SmBMP antibodies.
Acknowledgments
Schistosome life stages used in this research were supplied by the National Institute of Allergy and Infectious Diseases (NIAID) Schistosomiasis Resource Center at the Biomedical Research Institute (Rockville, Maryland, United States) through NIAID Contract NO1-AI-30026. This work was supported by NIH grant R01AI075226 to E.J.P. T.C.F was supported by grant number F32AI071417 from the National Institute of Allergy and Infectious Disease.
Footnotes
Note: Nucleotide sequence data reported in this paper is available in the GenBank database under accession number EU684544.
References
- Beall MJ, McGonigle S, Pearce EJ. Functional conservation of Schistosoma mansoni Smads in TGF-beta signaling. Mol Biochem Parasitol. 2000;111:131–142. doi: 10.1016/s0166-6851(00)00307-8. [DOI] [PubMed] [Google Scholar]
- Cain JE, Hartwig S, Bertram JF, Rosenblum ND. Bone morphogenetic protein signaling in the developing kidney: present and future. Differentiation. 2008 doi: 10.1111/j.1432-0436.2008.00265.x. [DOI] [PubMed] [Google Scholar]
- Carlo JM, Osman A, Niles EG, Wu W, Fantappie MR, Oliveira FM, LoVerde PT. Identification and characterization of an R-Smad ortholog (SmSmad1B) from Schistosoma mansoni. Febs J. 2007;274:4075–4093. doi: 10.1111/j.1742-4658.2007.05930.x. [DOI] [PubMed] [Google Scholar]
- Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/s0001-706x(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ. Regulation of bone morphogenetic protein activity by pro domains and proprotein convertases. J Cell Biol. 1999;144:139–149. doi: 10.1083/jcb.144.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hackenmiller R, Berg L, Jean F, Nakayama T, Thomas G, Christian JL. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15:2797–4802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SJ, Shoemaker CB, Pearce EJ. A divergent member of the transforming growth factor beta receptor family from Schistosoma mansoni is expressed on the parasite surface membrane. J Biol Chem. 1998;273:11234–11240. doi: 10.1074/jbc.273.18.11234. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finken-Eigen M, Kunz W. Schistosoma mansoni: gene structure and localization of a homologue to cysteine protease ER 60. Exp Parasitol. 1997;86:1–7. doi: 10.1006/expr.1996.4123. [DOI] [PubMed] [Google Scholar]
- Forrester SG, Warfel PW, Pearce EJ. Tegumental expression of a novel type II receptor serine/threonine kinase (SmRK2) in Schistosoma mansoni. Mol Biochem Parasitol. 2004;136:149–156. doi: 10.1016/j.molbiopara.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Freitas TC, Jung E, Pearce EJ. TGF-beta signaling controls embryo development in the parasitic flatworm Schistosoma mansoni. PLoS Pathog. 2007;3:e52. doi: 10.1371/journal.ppat.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray AM, Mason AJ. Requirement for activin A and transforming growth factor--beta 1 pro-regions in homodimer assembly. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- Gregory KE, Ono RN, Charbonneau NL, Kuo CL, Keene DR, Bachinger HP, Sakai LY. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J Biol Chem. 2005;280:27970–27980. doi: 10.1074/jbc.M504270200. [DOI] [PubMed] [Google Scholar]
- Hotez P, Ottesen E, Fenwick A, Molyneux D. The neglected tropical diseases: the ancient afflictions of stigma and poverty and the prospects for their control and elimination. Adv Exp Med Biol. 2006;582:23–33. doi: 10.1007/0-387-33026-7_3. [DOI] [PubMed] [Google Scholar]
- Loverde PT, Osman A, Hinck A. Schistosoma mansoni: TGF-beta signaling pathways. Exp Parasitol. 2007;117:304–317. doi: 10.1016/j.exppara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald AS, Straw AD, Bauman B, Pearce EJ. CD8- dendritic cell activation status plays an integral role in influencing Th2 response development. J Immunol. 2001;167:1982–1988. doi: 10.4049/jimmunol.167.4.1982. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Molina MD, Salo E, Cebria F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol. 2007;311:79–94. doi: 10.1016/j.ydbio.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Orii H, Kato K, Agata K, Watanabe K. Molecular cloning of bone morphogenetic protein (BMP) gene from the planarian Dugesia japonica. Zoological Science. 1998;15:871–877. [Google Scholar]
- Orii H, Watanabe K. Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev Growth Differ. 2007;49:345–349. doi: 10.1111/j.1440-169X.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- Osman A, Niles EG, LoVerde PT. Identification and characterization of a Smad2 homologue from Schistosoma mansoni, a transforming growth factor-beta signal transducer. J Biol Chem. 2001;276:10072–10082. doi: 10.1074/jbc.M005933200. [DOI] [PubMed] [Google Scholar]
- Osman A, Niles EG, LoVerde PT. Expression of functional Schistosoma mansoni Smad4: role in Erk-mediated transforming growth factor beta (TGF-beta) down-regulation. J Biol Chem. 2004;279:6474–6486. doi: 10.1074/jbc.M310949200. [DOI] [PubMed] [Google Scholar]
- Osman A, Niles EG, Verjovski-Almeida S, LoVerde PT. Schistosoma mansoni TGF-β Receptor II: Role in Host Ligand-Induced Regulation of a Schistosome Target Gene. PLoS Pathog. 2006;2:e54. doi: 10.1371/journal.ppat.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Skelly PJ, Shoemaker CB. Schistosoma mansoni proteases Sm31 (cathepsin B) and Sm32 (legumain) are expressed in the cecum and protonephridia of cercariae. J Parasitol. 2001;87:1218–1221. doi: 10.1645/0022-3395(2001)087[1218:SMPSCB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Wilson RA, Webster LA. Protonephridia. Biol Rev Camb Philos Soc. 1974;49:127–160. doi: 10.1111/j.1469-185x.1974.tb01572.x. [DOI] [PubMed] [Google Scholar]