Abstract
This article addresses some of the questions relating to how hepatitis δ virus (HDV), an agent so far unique in the animal world, might have arisen. HDV was discovered in patients infected with hepatitis B virus (HBV). It generally makes HBV infections more damaging to the liver. It is a subviral satellite agent that depends upon HBV envelope proteins for its assembly and ability to infect new cells. In other aspects of replication, HDV is both independent of and very different from HBV. In addition, the small single-stranded circular RNA genome of HDV, and its mechanism of replication, demonstrate an increasing number of similarities to the viroids – a large family of helper-independent subviral agents that cause pathogenesis in plants.
Keywords: hepatitis B virus, hepatitis δ virus, retroviroid, reverse transcription, ribozyme, splicing, viroid, virusoids
Hepatitis δ virus (HDV) was first detected 33 years ago in patients with a more severe form of hepatitis B virus (HBV) infection [1]. It was identified in both liver biopsies and the serum from such patients via a novel antigen, designated the δAg. Later, HDV was found to be an infectious agent separable from HBV. However, it is dependent upon HBV for the provision of envelope proteins so as to achieve particle assembly, and for new rounds of infection. For both viruses, the only susceptible cells are liver hepatocytes [2]. Thus, a complete and natural cycle of HDV infection and the emergence of new virus particles demand that both HBV and HDV infect the same hepatocyte. HDV, similarly to HBV, is a blood-borne infection, and may be transmitted by parenteral routes. For example, both are readily transmitted by contamination on needles shared in intravenous drug use. Unlike HBV, HDV is not typically a sexually transmitted disease, although such transmission may sometimes occur [3].
Hepatitis δ virus infections have now been detected in many parts of the world, always in association with HBV [4,5]. Lookback studies of patient sera reveal that even prior to 1977 there were significant life-threatening HDV infections in the Amazon basin of South America [6]. Currently, many nucleotide sequences have been obtained for HDV isolates from around the world, and are available in the Subviral RNA Database [101]. They differ by no more than 30 nucleotides in length but as much as 30% in sequence [5]. They have been divided into eight clades, based partly on nucleotide sequence and partly on geographic associations, although such geographic associations can sometimes be blurred, for example by population movements.
As mentioned previously, HDV is dependent upon HBV infection for its propagation. HBV is a member of a family of viruses known as Hepadnaviridae, which replicate via reverse transcription. Most retroviruses package an RNA genome and after infection use reverse transcription to make a DNA provirus. By contrast, the hepadnaviruses package an RNA ‘pregenome’, which is partially reverse transcribed before being released as particles from the infected cell. Thus, the genome of the hepadnavirus is a partially dsDNA species with a relaxed circular conformation. During infection this genome is completed as a covalently closed dsDNA that acts in the nucleus as a template for the transcription by the host RNA polymerase II (RNAP II) [7].
Hepatitis B virus infections can progress from an acute phase to a chronic phase (defined as one lasting at least 6 months). HDV infections can arise as a ‘coinfection’ with HBV or as a ‘super-infection’ in a patient already chronically infected with HBV. Approximately 2 billion individuals worldwide have come into contact with HBV and potentially 400 million of these have progressed to a chronic infection. Such chronic HBV infections can cause significant liver damage, and over a period of 10–30 years have a 25–40% risk of progressing to cirrhosis and hepatocellular carcinoma. Co- and super-infections with HDV greatly increase the risk of fulminant hepatitis during the acute phase. If the HBV infection becomes chronic, so does the HDV, and this can greatly increase the liver damage relative to HBV alone, leading to a life-threatening loss of liver function or more rapid progression to liver cancer [3].
As indicated in Figure 1, the HDV genome is a small ssRNA of approximately 1700 nucleotides in length that is circular in conformation [8]. This RNA can fold using approximately 74% base pairing to form an unbranched rod-like structure. The genome sequence shows no homology to the 3300-nucleotide genome of a hepadnavirus. Furthermore, the mode of replication of the HDV genome is fundamentally different.
Figure 1. Comparison of hepatitis δ virus RNAs with those of a viroid.
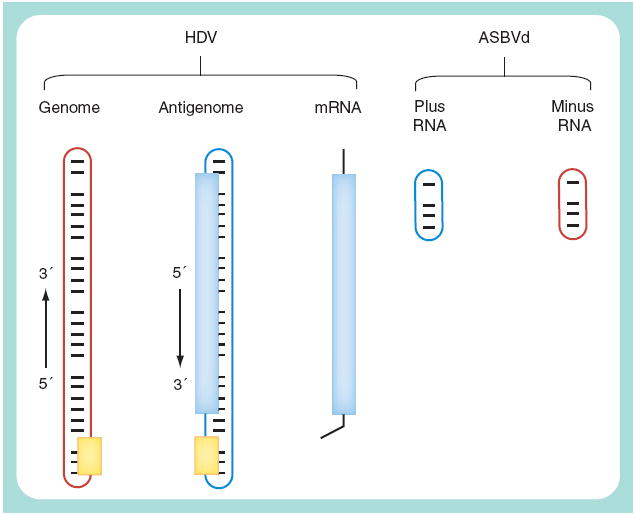
There are three stable HDV RNAs that accumulate during replication. The genome, which is the RNA assembled into new virus particles, and its exact complement, the antigenome, are approximately 1700 nucleotides in length, with a circular conformation and the ability, based on approximately 74% intramolecular base pairing, to fold into an unbranched rod-like structure. Both genome and antigenome contain their own ribozyme domain, of approximately 85 nucleotides (yellow box). The antigenome contains the open reading frame for the δ antigen (blue box), but this protein is actually translated from a third RNA, the mRNA, which is linear, less than full length, 5′-capped and 3′-polyadenylated. Also indicated are the RNAs of a plant viroid, ASBVd. This approximately 250-nucleotide long RNA and its exact complement are indicated as plus and minus, even though there is no coding capacity. As with HDV, RNAs can be circular and have the ability to fold intramolecularly. While the RNAs of ASBVd may be almost rod-like, the RNAs of other viroids can have more complicated folding. For viroids such as potato spindle tuber viroid, the circular conformation is only found for the plus RNA. ASBVd: Avocado sunblotch viroid; HDV: Hepatitis δ virus.
Hepatitis δ virus is considered to replicate through a symmetrical rolling-circle mechanism [9] that involves only RNA intermediates [8]. Replication of the HDV genome involves accumulation of new genomes and complementary RNA species known as antigenomes. As many as 300,000 molecules of genome accumulate per infected cell, along with approximately 100,000 copies of the antigenome [10]. It is thought that the genomic and antigenomic RNA circles act as templates for the generation of the multimeric strands of both polarities, which are greater than the 1700-nucleotide unit length. These are processed to a unit length since both the genome and antigenome contain a sequence of approximately 85 nucleotides that can act as a site-specific ribozyme [11]. After such ribozyme cleavage, the unit-length RNAs are ligated, possibly via a host ligase [12], to form new circular RNA species. Since HDV does not encode its own replicase and can replicate autonomously in its host, one or more host RNAP is redirected for its replication [8].
A third RNA species that is only approximately 900 nucleotides long and of antigenomic polarity is also produced at approximately 500 copies per cell [13]. As indicated in Figure 1, this mRNA is processed with a 5′-cap structure and is polyadenylated at its 3′-end because, similar to host mRNA precursors, it contains essential features, such as a polyadenylation site AAUAAA and an appropriately placed CA sequence, which acts as a polyadenylation cleavage and acceptor site [14]. The open reading frame encodes a protein that is 195 amino acids long and is referred to as the small δAg. This protein is an RNA-binding protein essential for the accumulation of HDV genomic and antigenomic RNA [15], although it is still controversial as to whether it is required for the transcription process [8]. During replication, an adenosine deaminase that acts on dsRNA (ADAR) converts an adenosine located in the termination codon of the small δAg on antigenomic RNA to an inosine [16]. Such action leads to the generation of an mRNA where the termination codon now encodes tryptophan, and thus to the production of a second viral protein species that is 19 amino acids longer at the C-terminus, referred to as the large δAg [16]. The large δAg is unable to support HDV RNA accumulation; however, in terms of the HDV lifecycle it plays an essential role in that it not only binds the HDV RNA, but then makes an interaction with the envelope proteins of HBV, leading to the assembly and release of new infectious HDV [15,17]. The unique 19 amino acid extension of large δAg contains a cysteine that undergoes farnesylation, and this is a necessary step in the functioning to achieve virion assembly [18].
Theories of origin
Similarities to plant viroids
Since they have remarkable similarities to HDV in terms of their genome structures and mechanisms of replication, much attention has been given to viroids to explain the origin of HDV [8,19,20]. Viroids were first detected owing to their obvious pathogenic effects in plants, and given names based upon their observed pathogenesis [21]. For example, potato spindle tuber viroid (PSTVd) was first discovered in 1971 as the causative agent of potato spindle tuber disease [22]. Another agent is avocado sunblotch viroid (ASBVd) [23]. There are now more than 30 species of viroids that can be distinguished according to their nucleotide sequences and modes of replication. Figure 1 shows a comparison of the ASBVd RNAs with those of HDV.
Viroid RNA was characterized long before it was realized in 1986 that the HDV RNA genome was also circular in conformation [24]. Viroids have small single-stranded circular RNA genomes in the range of 250 to 400 nucleotides [25-29]. This size is even smaller than HDV, but consistent with the fact that viroid genomes have no known coding capacity. Similar to HDV, some viroids (i.e., avsunviroids) are replicated by host RNAP(s) via a ‘symmetrical rolling-circle mechanism’ that involves only RNA intermediates, and are post-transcriptionally processed to unit length by viroid ribozyme domains (i.e., hammerhead) [30]. However, most known viroids (i.e., pospiviroids) replicate by an ‘asymmetrical rolling-circle mechanism’. This pathway involves the generation of linear multimeric head-to-tail RNAs of negative polarity from circular RNAs of positive polarity by host RNAP(s). Instead of being processed, the multimeric RNAs serve directly as templates for the synthesis of linear positive multimeric RNAs, which are then cleaved and ligated into monomeric positive circular RNAs (possibly using a host RNase and a host ligase [31]). Although it is generally considered that circular HDV RNAs of both polarities can act as the templates for the generation of the multimeric strands, it is also possible that linear multimeric RNA templates might be used during HDV replication, in a similar way to that reported in pospiviroids.
The nucleotide sequences of viroids have been compared with HDV, and while one study suggested they were related [32], a subsequent study suggested the opposite [33]. However, given the rates of nucleotide change for both infectious agents, and the apparent requirements for intramolecular folding, the lack of detected sequence homology does not preclude evolutionary relatedness. In addition, unlike viroids, HDV had to evolve a protein and RNA features to undergo assembly by proteins from its helper virus (i.e., HBV) and to release infectious virus particles.
The similarities between HDV and viroids are not limited to common RNA features and their modes of replication, but also include the proteins that their RNA genomes interact with. Both subviral RNA species are reported to interact with proteins involved with RNA-processing pathways and with translation machinery, and at least one of those proteins is common to both species (i.e., eEF1α1) [34,35]. More importantly, both HDV and viroids are able to divert RNAP(s) for the replication of their own RNA genomes. It is generally accepted that HDV RNA and most viroids (i.e., pospiviroids) are transcribed by host RNAP II [8,36]. In addition, it has been reported that part of HDV replication might involve another host polymerase, possibly RNAP I [37]. It is considered that there might even be different sites – nucleoplasmic versus nucleolar – for transcription of different HDV RNAs [38,39], but the issue remains controversial [8,40].
For some viroids, such as ASBVd, there is evidence that replication does not occur in the nucleus using RNAP II but in the chloroplast, using either a plastid-encoded RNAP or a nuclear-encoded RNAP [41]. In addition, studies have demonstrated that specific regions of HDV RNA can interact not only with mammalian RNAP II [42], but also with RNAP I and III [43]. Similarly, only one polarity of the PSTVd RNA was found to stably associate in vitro with RNAP II [Pelchat M et al., Unpublished Data] and PSTVd RNA was reported to interact with transcription factor IIIA both in vitro and in vivo [Ding B, Pers. Comm.]. Furthermore, several viroid-derived RNAs were shown to be recognized and used as templates by other polymerases, such as Escherichia coli RNA polymerase [44]. Such seeming promiscuity suggests that given an appropriate situation, the RNA genomes of viroids and HDV can make use of different polymerases. However, in vivo, the situation might be more complicated as not only does it involve redirection of host polymerases to achieve transcription, but also requires survival and accumulation of at least some of the RNA transcripts. Nevertheless, these similarities imply the evolutionary relationship of these RNA species and suggest that they might use common proteins during their lifecycle.
The analogy between HDV and viroids has been carried to another level by four recent findings [Pelchat M et al., Unpublished Data]. First, when multimers of HDV RNA, as transcribed in vitro, were inoculated into the leaves of tomato seedlings under specific conditions, HDV genome replication was achieved as well as spreading within the plant and obvious cytopathic effects, including inhibition of plant growth. Second, following transfection of multimers of PSTVd RNA into animal cells, accumulation of new RNA was detected, but only when δAg was also expressed. Third, the genome of HDV RNA was engineered to reduce its size to 342 nucleotides, leaving only the ribozyme domains and small regions from the opposite ends of the rod-like structure reported to interact with the three human RNAPs [42,43]. After this, modified RNA was transfected into animal cells, and new RNA accumulated, but again only in the presence of expressed δAg. Fourth, when this same 342-nucleotide RNA was inoculated into tomato leaves, spreading and accumulation of newly synthesized RNA species was detected.
While these new findings cement the relationship between the structure and replication of HDV and plant viroid RNAs, they still leave open the question of whether they have a distinct origin with convergent evolution, or whether one somehow acted as a precursor for the other. This will be difficult to resolve since even for the many species of viroids there is the issue of whether they are evolutionarily related [41]. If some of the viroids arose independently and converged to common structures and replication mechanisms, then it is equally possible that HDV did the same.
A recent study has proposed that recombination may be the only possible route to evolutionary innovation in viroids [45]. In this respect, it should be noted that intermolecular recombination between different HDV genomes has been reported [46] and template switching, the presumed mechanism of recombination, has been demonstrated as an intramolecular event that can initiate HDV genome replication [47,48].
Evolution from a virusoid or a retroviroid
Although there are strong structural and functional similarities between viroids and HDV, other species of plant agents are structurally and functionally similar to HDV. Some plant RNA viruses are known to accumulate, other than their genomes, relatively smaller RNA sequences that can modulate the disease symptoms associated with the infection [49]. At one time, such agents were referred to as ‘virusoids’, a name given to indicate a possible relationship to viroids [50]. These smaller RNAs may contain little or no detectable homology to the genome of their helper virus. In some cases, these smaller sequences can achieve a circular conformation, and this no doubt contributes to their ability to accumulate. They can even contain ribozyme domains, consistent with a post-transcriptional processing to circles. However, these agents use the RNA polymerase of the helper virus, instead of host RNAP. Nevertheless, while one can suggest that plant viroids and HDV might have evolved from virusoid RNAs that became able to utilize a host polymerase, there is no supporting evidence.
There are two examples of retroviroids in plants, where small circular RNAs are associated with either DNA templates or reverse transcription. Daros and coworkers in a study of a viroid-like RNA detected association of that RNA with its cDNAs [51]. No further examples of this have since been reported. The second example comes from the Varkud small plasmid that is not known to encode any protein and replicates in the mitochondria of Neurospora, in the presence of a larger plasmid that encodes at least a reverse transcriptase [52,53]. This agent is derived from DNA-directed RNA transcripts. It has no RNA-directed component. However, the DNA-directed RNA transcripts do undergo post-transcriptional processing that is ribozyme mediated and leads to the accumulation of small ssRNA circles.
There is no question at this time that most viroids and HDV replicate their RNAs without reverse transcription and/or a DNA intermediate. However, one should not discard the possibility that the origin of these RNAs might have involved reverse transcription, because HDV likely arose in an HBV-infected cell; that is, during HBV replication dependent upon reverse transcription.
Evolution from a host mRNA precursor & associated ribozymes
Another possibility that cannot be excluded is that host RNA might have been used as a template for replication to generate an HDV-like species. Studies of the processing of nascent human β-globin mRNAs have shown that downstream of the poly(A) signal and CA acceptor site there is a ribozyme domain [54]. The authors cite the conservation of this domain in other primate globin genes and other examples occur in myxomycetes, indicating an evolutionary conservation of such associations, which importantly are also present on HDV antigenomic RNA (Figure 1). The HDV ribozymes are faster than other ribozymes, such as the hammerhead ribozyme, and recent studies show that the insertion of the δ ribozyme into nascent RNAP II transcripts can dramatically inhibit other cotranscriptional RNA processing events, including splicing and ADAR editing [55]. Furthermore, HDV studies have focused on how the polyadenylation and ribozyme affect the processing of nascent antigenomic RNA transcripts [56,57]. That is, there is important alternative processing directed by these signals present on the HDV RNA.
In 2006, a ribozyme RNA was found in animal cells, which had similarities in structural folding and biochemical properties to the δ ribozymes [58]. This ribozyme has limited nucleotide sequence similarity to the HDV ribozymes and is encoded in an intron of a known gene, CPEB3, one which has no relationship to δAg. However, in a recent follow-up study using a structure-based search, many HDV-like ribozymes were found [59]. These occurred in diverse eukaryotic species, such as eels, sharks, sea urchins, worms and mosquitoes, and even in an insect virus. Some of these ribozymes have been confirmed as active in vitro and some have proved to be transcribed in vivo. Furthermore, from the locations of these ribozymes within the host genomes there are provocative indications of relevance to retrotransposition and the selection of biologically relevant regulation of host genes.
Some time ago it was also speculated that HDV RNA might have arisen as a consequence of an aberrant splicing event [20]. In fact, the RNA was examined for similarities to self-splicing introns [60]. Alternatively, a process known as mis-splicing, which produces RNA circles, was proposed; this occurs when a splice donor acts on a splice acceptor located 5′ rather than 3′ [61]. However, to also explain the base pairing of the HDV RNA, one might have to presume a prior event of RNA-primed hairpin formation.
δ antigens, both large & small
As previously mentioned, we know that both forms of the δAg are essential for the lifecycle of HDV. However, at this point we do not know where δAg came from. One study identified an animal cell protein that interacted with δAg, designated δ-interacting protein A (DIPA) [62]. This protein, based on its size and possible sequence homology, was considered as a candidate. This led to speculation that some form of viroid-like RNA might have transduced DIPA–RNA coding sequences to create the HDV genome. However, the homology (DIPA) to δAg was limited and the interpretation controversial [63,64]. While others have gone on to study DIPA, there have been no further assertions of its relation to δAg [65].
It has been proposed that the role of δAg in HDV replication is to displace the cellular negative elongation factor, thereby stimulating HDV transcription by RNAP II [66]. Since putative negative elongation factor orthologs have not been found in several species, including plants, it is possible that a plant viroid-like RNA might have captured the coding region for a cellular protein (maybe related to DIPA), and that δAg is an evolutionary adaptation of HDV that was necessary for it to replicate in the new mammalian host. However, the homology of DIPA to δAg was limited.
Unbranched rod-like folding
In addition to its circularity, which offers protection against host nucleases that are predominantly exo- rather than endonucleases [67], the HDV RNA genome folds on itself to form what is referred to as an unbranched rod-like folding (Figure 1). This folding is predicted to include 74% of all nucleotides [68] and from electron microscope observations, the short rod-like appearance has been demonstrated [69].
During viral RNA replication, there is evidence that δAg binds to the rod-like folding of HDV RNAs and can facilitate the accumulation of processed HDV RNAs, even when these are transcribed by RNAP II from DNA templates [70]. Some studies have shown that insertion of short sequences to disrupt the rod-like folding can block replication competence [71]. Other studies have shown that insertion of sequences as long as 1 kb onto the HDV RNA can nevertheless lead to the selection of genomes that have removed the sequence and are replication competent [47]. For such removal, template switching during RNA-directed transcription by host RNAP(s) was suggested, as this process was shown to occur with HDV RNA [48].
In addition to δAg binding, the level of base pairing in the HDV RNAs is sufficient to attract an adenosine deaminase that acts on dsRNA, an ADAR that converts adenosine to inosine, to produce the essential large δAg [16]. ADAR editing at other sites is also a major source of nucleotide sequence changes that can accumulate on HDV RNAs [72]. Other changes might arise via nucleotide misincorporation during RNA-directed transcription.
How did the rod-like folding arise? A likely explanation would be to invoke what could be termed back-priming (or fold-back priming). If the 3′-end of an RNA template is used as a primer for transcription that folds back on the RNA as a template, this will ultimately generate a 100% double-stranded hairpin molecule that will be a chimera of the template and its complementary sequence.
Numerous examples of back-priming have been reported for in vitro transcription, such as with poliovirus RNA and its polymerase or various RNAs with either HCV polymerase or with the polymerase of phage phi6 [73]. As represented in Figure 2, studies with phi6 polymerase have shown the ability to use back-priming on HDV RNA templates to produce such hairpin chimeric RNAs that are double the size of the template [Taylor J et al., Unpublished Data]. This is not to say that HDV RNA replication necessarily continues to use back-priming. In fact, other in vitro studies with less than full-length HDV RNAs and RNAP II have reported leader-priming [74,75], as represented in Figure 2.
Figure 2. Possible primed transcription of hepatitis δ virus RNA.
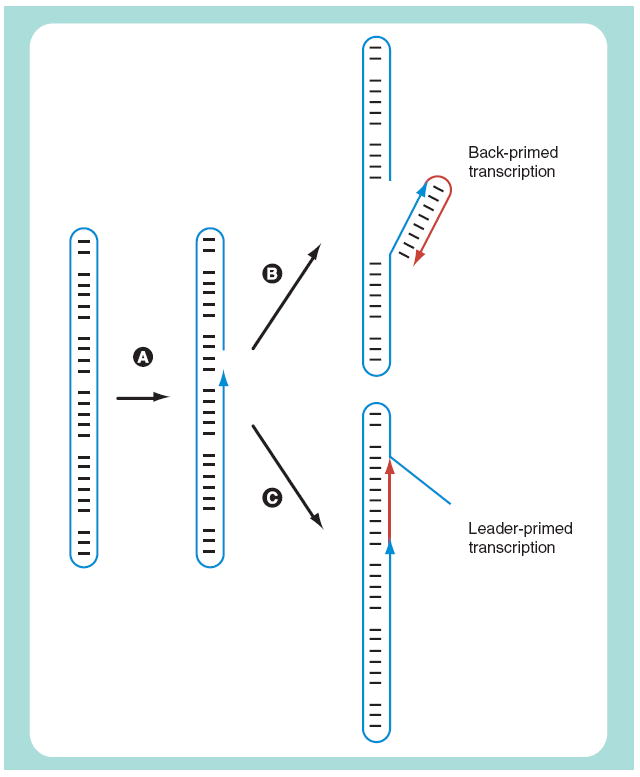
The example shown is for an antigenomic RNA template. (A) Indicates cleavage, which may be via a cellular exonuclease or even polymerase II in association with transcription factor IIS [75]. (B) The 3′-OH produced by the cleavage allows back-priming (or fold-back priming). This occurs in vitro with phage phi6 polymerase [Taylor J et al., Unpublished Data]. (C)Indicates leader-priming (or in situ priming), which has been reported in vitro with polymerase II [74,75]. Note that (B & C) produce different chimeras of genomic RNA transcript linked to antigenomic RNA template.
Independent of how HDV RNA is currently replicated, it may have arisen along with its rodlike folding, by back-primed transcription on the 3′-end of a host RNA. Furthermore, if the RNA were an mRNA precursor (especially one encoding the putative ancestor of the δAg) that contained a ribozyme domain downstream of the poly(A) signal and CA acceptor site, this might somehow lead to a processed RNA that acted as a template for further RNA-directed transcription. Mutations would then have had to occur rapidly to decrease the amount of base pairing. This is because RNAs with high levels of pairing would activate host innate responses, such as those involving the dsRNA-dependent protein kinase PKR and RNA silencing, and lead to repression of HDV replication [76,77]. Accordingly, the observed 74% amount of base pairing no doubt reflects a selective pressure to allow interaction with proteins necessary for the HDV lifecycle and yet avoid recognition by host innate responses.
A recent study has shown that back-priming can indeed occur in vivo and with significant biological consequences. Maida et al. discovered that a small 267-nucleotide noncoding nucleolar RNA, RMRP, redirects telomerase to perform RNA-directed RNA synthesis and back-priming to create a dsRNA [78]. Some of this species is converted by the nuclease DICER to produce siRNAs that negatively regulate RMRP. Of relevance here is not only the back-priming but the ability of a structured RNA to redirect a host polymerase.
HBV as the helper virus
Finally, an important question one should ask when considering the origin of HDV is how this agent developed in relation to its helper virus, HBV. We do know that in the serum of HBV-infected patients, one can detect the 42-nm diameter infectious HBV that contains a 27-nm diameter nucleocapsid, along with an excess of subviral particles (SVPs) that are empty, and exist as 25-nm diameter spheres and 22-nm diameter filaments of variable length. The SVP can be present in 100- to 100,000-fold excess relative to the infectious HBV particles [7]. Thus, redirection of SVP formation could easily be part of HDV assembly.
How did the current dependence of HDV on HBV arise? A recent study reported that humans (and most likely other animals) are able to act as vehicles for the dissemination of certain plant viruses through their digestive tracts [79]. One possibility is that a viroid-like genome (possibly from plants) infected the liver of an HBV-infected primate, not necessarily human, leading to assembly by HBV envelope proteins. Another possibility is that in cells undergoing HBV replication, a chance event(s) other than infection led to the origin and accumulation of HDV-like RNAs, and that the replication and accumulation of such RNAs allowed the selection of species capable of assembly by the HBV envelope proteins.
Based upon nucleotide sequence analyses it has been possible to separate HBV isolates into eight genotypes and HDV into eight clades [5]. Only to a limited extent is there evidence of a correlation between a HBV genotype and a HDV clade; in such situations, parallel evolution of the two viruses might have occurred.
Why has HDV remained?
There is no evidence that HDV RNA and the replication strategy it has adopted reflect anything more than a successful selfish RNA, whose replication interferes with that of its helper virus [80] and enhances the liver damage relative to HBV alone [81]. HDV might even be more selfish than this. For example, while we do not know to what extent HDV infection of hepatocytes precedes or follows HBV infections of individual hepatocytes, we realize that prior arrival of HDV will lead to genome replication, but no assembly or release, unless subsequent HBV infection occurs. Thus, an untested speculation is that HDV infection might somehow enhance the susceptibility of the cell to achieve the necessary second infection by HBV.
Conclusion & future perspective
This article offers a first look at some surprising findings linking HDV and viroids, and yet we cannot assert that somehow one is a precursor to the other. It is equally possible that the two arose by convergent evolution, by a process involving aberrant RNA-directed RNA transcription coupled with an RNA processing event. Also relevant and provocative are the new data that host telomerase can act as an RNA-directed RNA polymerase on a structured RNA template, and that this is achieved by back-priming. Furthermore, discovery of the widespread occurrence of HDV-like ribozymes and their possible relationship to retrotransposition further demonstrates that some of what appeared to be unique properties of HDV and viroids are barely the tip of a major biological iceberg. This article also raises speculation regarding the origin of the dependence of HDV upon HBV as a helper virus. Certainly, we will have to consider the possibility of new infectious agents of humans that might act alone or in conjunction with helper viruses as replicating RNAs, almost totally dependent upon redirection of host functions. Such agents might be new or currently unrecognized sources of pathogenesis. However, we might also envision harnessing such replication-competent RNAs for positive means, for example to deliver therapeutic signals.
Executive summary.
Hepatitis δ virus is unique relative to other human pathogens
-
▪
Hepatitis δ virus (HDV) is subviral in that it depends upon hepatitis B virus (HBV) for envelope proteins.
-
▪
HDV has an RNA genome smaller than that of any animal virus.
-
▪
The RNA genome and its complement, the antigenome, have a circular conformation, and are able to form by base pairing, into an unbranched rod-like structure.
-
▪
These RNAs are transcribed by redirection of host RNA polymerase activity.
-
▪
HDV has just one open reading frame encoding two proteins, the small and large δ antigen, which are essential for RNA accumulation and propagation.
-
▪
RNA editing by an adenosine deaminase that acts on dsRNA during replication leads to translation of a longer protein, large δ antigen, which together with HBV envelope proteins is essential for virion assembly.
HDV shows distinct similarities to certain pathogens of plants
-
▪
The viroids are noncoding RNAs, roughly five-times smaller than HDV.
-
▪
Like HDV RNA, the viroid RNAs are often circular in conformation and some possess ribozyme activity.
-
▪
Like HDV RNA, they replicate by redirection of host RNA polymerases.
-
▪
New experimental studies demonstrate that HDV RNAs can accumulate in plant tissues and that viroid RNAs can accumulate in human cells.
Theories of HDV origin must explain multiple aspects
-
▪
The essential HBV helper function implies HDV arose in the liver of an HBV-infected patient
-
▪
The source of the genetic information for δ antigen remains unsolved.
-
▪
The unbranched rod-like folding might best be explained by a fold-back transcription event.
-
▪
New data strengthen the analogy between HDV and the plant viroids but do not resolve whether one is a precursor to the other or whether they represent convergent evolution.
-
▪
While virusoid-like agents might be a progenitor of viroids, we cannot envision such a role for HDV.
-
▪
While only one example of a retroviroid is known, we cannot ignore that HDV, at one point in its evolution, was present in a cell expressing the reverse transcriptase of HBV.
-
▪
A most promising contributing component to HDV evolution might be recent studies that show host mRNAs where the polyadenylation signals are followed by ribozyme domains, an arrangement also present on HDV antigenomic RNA.
-
▪
Overall, one must ask whether HDV is no more than a selfish RNA or whether it provides some function, negative or positive, in relation to its essential helper virus, HBV.
Future studies
-
▪
Not just future HDV studies, but also cell biology, will be greatly affected by the linkage of HDV to viroids, the new example of in vivo redirection of a host polymerase to become an RNA-directed RNA polymerase, the evidence of in vivo back-priming, and the revelation of multiple and widespread examples of HDV-like ribozymes.
-
▪
Other HDV- or viroid-like RNAs might be discovered in animal cells, with or without associated helper viruses.
-
▪
Clarifying HDV-associated pathogenesis and gaining an understanding of the requirements for self-replicating HDV- and viroid-like RNAs will be key steps in advancing RNA biology and may open the way for valuable experimental and therapeutic applications.
Acknowledgments
William Mason, Carolina Alves, Dorota Sikora and Erica Schissel, together with two anonymous reviewers, provided constructive comments on this manuscript. John Taylor acknowledges unpublished observations from Carolina Alves, Severin Gudima and Xing-Cao Nie. Martin Pelchat similarly acknowledges observations by Teodora Dimitrijevic, Dorota Sikora and Erica Schissel.
Footnotes
Financial & competing interests disclosure
John Taylor was supported by grants AI-256522 and CA-06927 from the NIH and by an appropriation from the Commonwealth of Pennsylvania. Martin Pelchat was supported by a grant from the Canadian Institutes of Health Research (CIHR). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
John Taylor, Chase Cancer Center, PA 19111, USA, Tel.: +1 215 728 2436, Fax: +1 215 728 2412, john.taylor@fccc.edu.
Martin Pelchat, Department of Biochemistry, Microbiology & Immunology, Faculty of Medicine, University of Ottawa, Ottawa, ON K1H 8M5, Canada, Tel.: +1 613 562 5800 ext. 8846, Fax: +1 613 562 5452, mpelchat@uottawa.ca.
Bibliography
Papers of special note have been highlighted as:
-
▪
of interest
-
▪▪
of considerable interest
- 1.Rizzetto M, Canese MG, Arico J, et al. Immunofluorescence detection of a new antigen–antibody system associated to the hepatitis B virus in the liver and in the serum of HBsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizzetto M, Canese MG, Gerin JL, London WT, Sly DL, Purcell RH. Transmission of the hepatitis B virus-associated δ antigen to chimpanzees. J Infect Dis. 1980;141:590–602. doi: 10.1093/infdis/141.5.590. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JM, Farci P, Purcell RH. Hepatitis D (δ) virus. In: Knipe DM, editor. Fields Virology. Lippincott Williams & Wilkins; PA, USA: 2007. pp. 3031–3046. [Google Scholar]
- 4.Radjef N, Gordien E, Ivaniushina V, et al. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis δ virus, suggesting a δ virus genus of at least seven major clades. J Virol. 2004;78:2537–2544. doi: 10.1128/JVI.78.5.2537-2544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deny P. Hepatitis δ virus genetic variability: from genotypes I, II, III to eight major clades. In: Casey JL, editor. Hepatitis δ Virus. Springer; Heidelberg, Germany: 2006. pp. 151–171. ▪▪ Defines the many clades of hepatitis δ virus HDV.
- 6.Casey JL, Niro GA, Engle RE, et al. Hepatitis B virus/hepatitis D virus (HDV) coinfection in outbreaks of acute hepatitis in the Peruvian Amazon basin: the roles of HDV genotype III and HBV genotype F. J Infect Dis. 1996;174:920–926. doi: 10.1093/infdis/174.5.920. [DOI] [PubMed] [Google Scholar]
- 7.Seeger C, Zoulim F, Mason WS. Hepadnaviruses. In: Knipe DM, editor. Fields Virology. Lippincott Williams & Wilkins; PA, USA: 2007. pp. 2977–3030. [Google Scholar]
- 8.Taylor JM. Replication of the hepatitis δ virus RNA genome. Adv Vir Res. 2009;74:102–121. doi: 10.1016/S0065-3527(09)74003-5. ▪▪ Reviews how HDV RNAs are transcribed.
- 9.Branch AD, Robertson HD. A replication cycle for viroids and small infectious RNAs. Science. 1984;223:450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- 10.Chen P-J, Kalpana G, Goldberg J, et al. Structure and replication of the genome of hepatitis δ virus. Proc Natl Acad Sci USA. 1986;83:8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat Struct Mol Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 12.Reid CE, Lazinski DW. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc Natl Acad Sci USA. 2000;97:424–429. doi: 10.1073/pnas.97.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudima S, Wu S-Y, Chiang C-M, Moraleda G, Taylor J. Origin of the hepatitis δ virus mRNA. J Virol. 2000;74:7204–7210. doi: 10.1128/jvi.74.16.7204-7210.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh S-Y, Taylor J. Regulation of polyadenylation of HDV antigenomic RNA. J Virol. 1991;65:6438–6446. doi: 10.1128/jvi.65.12.6438-6446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chao M, Hsieh S-Y, Taylor J. Role of two forms of the hepatitis δ virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casey JL, Gerin JL. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69:7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang FL, Chen PJ, Tu SJ, Chiu MN, Wang CJ, Chen DS. The large form of hepatitis δ antigen is crucial for the assembly of hepatitis δ virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordier BB, Ohkanda J, Liu P, et al. In vivo antiviral efficacy of prenylation inhibitors against hepatitis δ virus. J Clin Invest. 2003;112:407–414. doi: 10.1172/JCI17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor JM. Replication of human hepatitis δ virus: influence of studies on subviral plant pathogens. Adv Virus Res. 1999;54:45–60. doi: 10.1016/s0065-3527(08)60365-6. [DOI] [PubMed] [Google Scholar]
- 20.Diener TO. The viroid: biological oddity or evolutionary fossil? Adv Virus Res. 2001;57:137–184. doi: 10.1016/s0065-3527(01)57003-7. [DOI] [PubMed] [Google Scholar]
- 21.Owens RA, Hammond RW. Viroid pathogenicity: one process, many faces. Viruses. 2009;1:298–316. doi: 10.3390/v1020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diener TO. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology. 1971;45:411–428. doi: 10.1016/0042-6822(71)90342-4. [DOI] [PubMed] [Google Scholar]
- 23.Daros J, Marcos J, Hernandez C, Flores R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme procesing. Proc Natl Acad Sci USA. 1994;91:12813–12817. doi: 10.1073/pnas.91.26.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K-S, Choo Q-L, Weiner AJ, et al. Structure, sequence and expression of the hepatitis δ viral genome. Nature. 1986;323:508–513. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 25.Tsagris EM, Martinez de Alba AE, Gozmanova M, Kalantidis K. Viroids. Cell Microbiol. 2008;10:2168–2179. doi: 10.1111/j.1462-5822.2008.01231.x. [DOI] [PubMed] [Google Scholar]
- 26.Tabler M, Tsagris M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004;9:339–348. doi: 10.1016/j.tplants.2004.05.007. ▪▪ Reviews the many plant viroid RNAs.
- 27.Daros JA, Elena SF, Flores R. Viroids: an Ariadne’s thread into the RNA labyrinth. EMBO Rep. 2006;7:593–598. doi: 10.1038/sj.embor.7400706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocheleau L, Pelchat M. The Subviral RNA Database: a toolbox for viroids, the hepatitis δ virus and satellite RNAs research. BMC Microbiol. 2006;6:24. doi: 10.1186/1471-2180-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores R, Gas M-E, Molina-Serrano D, et al. Vioroid replication: rolling-circles, enzymes and ribozymes. Viruses. 2009;1:317–334. doi: 10.3390/v1020317. ▪▪ Reviews the replication of viroids RNAs.
- 30.Flores R, Gas ME, Molina D, Hernandez C, Daros JA. Analysis of viroid replication. Methods Mol Biol. 2008;451:167–183. doi: 10.1007/978-1-59745-102-4_12. [DOI] [PubMed] [Google Scholar]
- 31.Branch AD, Robertson HD, Greer C, Gegenheimer P, Peebles C, Abelson J. Cell-free circularization of viroid progeny RNA by an RNA ligase from wheat germ. Science. 1982;217:1147–1149. doi: 10.1126/science.217.4565.1147. [DOI] [PubMed] [Google Scholar]
- 32.Elena SF, Dopazo J, Flores R, Diener TO, Moya A. Phylogeny of viroids, viroidlike satellite RNAs, and the viroid-like domain of hepatitis δ virus RNA. Proc Natl Acad Sci USA. 1991;88:5631–5634. doi: 10.1073/pnas.88.13.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins GM, Woelk CH, Rambaut A, Holmes EC. Testing the extent of sequence similarity among viroids, satellite RNAs, and hepatitis δ virus. J Mol Evol. 2000;50:98–102. doi: 10.1007/s002399910011. [DOI] [PubMed] [Google Scholar]
- 34.Sikora D, Greco-Stewart VS, Miron P, Pelchat M. The hepatitis δ virus RNA genome interacts with eEF1A1, p54(nrb), hnRNP-L, GAPDH and ASF/SF2. Virology. 2009;390:71–78. doi: 10.1016/j.virol.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 35.Dube A, Bisaillon M, Perreault JP. Identification of proteins from prunus persica that interact with peach latent mosaic viroid. J Virol. 2009;83:12057–12067. doi: 10.1128/JVI.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindler I-M, Muhlbach H-P. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 1992;84:221–229. [Google Scholar]
- 37.Li YJ, Macnaughton T, Gao L, Lai MM. RNA-templated replication of hepatitis δ virus: genomic and antigenomic RNAs associate with different nuclear bodies. J Virol. 2006;80:6478–6486. doi: 10.1128/JVI.02650-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng C-H. Lai MMC: Hepatitis δ virus RNA replication. Viruses. 2009;1:818–831. doi: 10.3390/v1030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tseng CH, Jeng KS, Lai MM. Transcription of subgenomic mRNA of hepatitis δ virus requires a modified hepatitis δ antigen, distinct from antigenomic RNA synthesis. J Virol. 2008;82:9409–9416. doi: 10.1128/JVI.00428-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greco-Stewart VS, Pelchat M. Coercion of host cellular proteins by the hepatitis δ virus. Viruses. 2010 doi: 10.3390/v2010189. In press. ▪ Demonstrates that all three mammalian polymerases will bind to HDV RNAs.
- 41.Flores R, Gas M-E, Molina-Serrano D, et al. Viroid replication: rolling-circles, enzymes, and ribozymes. Viruses. 2009;1:317–334. doi: 10.3390/v1020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greco-Stewart VS, Miron P, Abrahem A, Pelchat M. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis δ virus RNA genome. Virology. 2007;357:68–78. doi: 10.1016/j.virol.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Greco-Stewart VS, Schissel E, Pelchat M. The hepatitis δ virus RNA genome interacts with the human RNA polymerases I and III. Virology. 2009;386:12–15. doi: 10.1016/j.virol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Pelchat M, Perreault JP. Binding site of Escherichia coli RNA polymerase to an RNA promoter. Biochem Biophys Res Commun. 2004;319:636–642. doi: 10.1016/j.bbrc.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 45.Elena SF, Gomez G, Daros J-A. Evolutionary constraints to viroid evolution. Viruses. 2009;1:241–254. doi: 10.3390/v1020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao M. RNA recombination in hepatitis δ virus: implications regarding the abilities of mammalian RNA polymerases. Virus Res. 2007;127:208–215. doi: 10.1016/j.virusres.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Gudima SO, Chang J, Taylor JM. Reconstitution in cultured cells of replicating HDV RNA from pairs of less than full-length RNAs. RNA. 2005;11:90–98. doi: 10.1261/rna.7164905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang J, Taylor J. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 2002;21:157–164. doi: 10.1093/emboj/21.1.157. ▪ Demonstrates that template switching can occur during HDV RNA transcription.
- 49.Simon AE, Roossinck MJ, Havelda Z. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Ann Rev Phytopathol. 2004;42:415–437. doi: 10.1146/annurev.phyto.42.040803.140402. [DOI] [PubMed] [Google Scholar]
- 50.Symons RH, Randles JW. In: Encapsidated Circular Viroid-Like Satellite RNAs (Virusoids) of Plants. Symons RH, Randles JW, editors. Springer-Verlag; Berlin, Germany: 1999. [DOI] [PubMed] [Google Scholar]
- 51.Daros J, Flores R. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci USA. 1995;92:6856–6860. doi: 10.1073/pnas.92.15.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones FD, Ryder SP, Strobel SA. An efficient ligation reaction promoted by a Varkud satellite ribozyme with extended 5′- and 3′-termini. Nucl Acids Res. 2001;29:5115–5120. doi: 10.1093/nar/29.24.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuiper MT, Sabourin JR, Lambowitz AM. Identification of the reverse transcriptase encoded by the Mauriceville and Varkud mitochondrial plasmids of Neurospora. J Biol Chem. 1990;265:6936–6943. [PubMed] [Google Scholar]
- 54.Teixeira A, Tahiri-Alaoui A, West S, et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. ▪ Reports a ribozyme downstream of globin mRNA.
- 55.Fong N, Ohman M, Bentley DL. Fast ribozyme cleavage releases transcripts from RNA polymerase II and aborts co-transcriptional pre-mRNA processing. Nat Struct Mol Biol. 2009;16:916–922. doi: 10.1038/nsmb.1652. ▪ Demonstrates that ribozyme cleavage can regulate other RNA processing events.
- 56.Nie X, Chang J, Taylor JM. Alternative processing of hepatitis δ virus antigenomic RNA transcripts. J Virol. 2004;78:4517–4524. doi: 10.1128/JVI.78.9.4517-4524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown AL, Perrotta AT, Wadkins TS, Been MD. The poly(A) site sequence in HDV RNA alters both extent and rate of self-cleavage of the antigenomic ribozyme. Nucl Acids Res. 2008;36:2990–3000. doi: 10.1093/nar/gkn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 59.Webb C-HT, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. ▪▪ Reports many HDV-like ribozymes.
- 60.Dinter-Gottlieb G. Designing a ribozyme from the hepatitis δ virus. In: Dinter-Gottlieb G, editor. The Unique Hepatitis δ Virus. RG Landes, Co; Austin, TX, USA: 1995. pp. 33–46. [Google Scholar]
- 61.Nigro J, Cho K, Fearon E, et al. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 62.Brazas R, Ganem D. A cellular homolog of hepatitis δ antigen: implications for viral replication and evolution. Science. 1996;274:90–94. doi: 10.1126/science.274.5284.90. [DOI] [PubMed] [Google Scholar]
- 63.Brazas R, Ganem D. δ-interacting protein A and the origin of hepatitis δ antigen. Science. 1997;276:824–825. doi: 10.1126/science.276.5313.824. [DOI] [PubMed] [Google Scholar]
- 64.Long M, de Souza SJ, Gilbert W. δ-interacting protein A and the origin of hepatitis δ antigen. Science. 1997;276:824–825. doi: 10.1126/science.276.5313.824. [DOI] [PubMed] [Google Scholar]
- 65.Du X, Wang Q, Hirohashi Y, Greene MI. DIPA, which can localize to the centrosome, associates with p78/MCRS1/MSP58 and acts as a repressor of gene transcription. Exp Mol Pathol. 2006;81:184–190. doi: 10.1016/j.yexmp.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi Y, Filipovska J, Yano K, et al. Stimulation of RNA polymerase II elongation by hepatitis δ antigen. Science. 2001;293:124–127. doi: 10.1126/science.1057925. [DOI] [PubMed] [Google Scholar]
- 67.van Hoof A, Parker R. The exosome: a proteosome for RNA? Cell. 1999;12:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 68.Kuo MY-P, Goldberg J, Coates L, Mason W, Gerin J, Taylor J. Molecular cloning of hepatitis δ virus RNA from an infected woodchuck liver: sequence, structure, and applications. J Virol. 1988;62:1855–1861. doi: 10.1128/jvi.62.6.1855-1861.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (δ) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 70.Lazinski DW, Taylor JM. Expression of hepatitis δ virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J Virol. 1994;68:2879–2888. doi: 10.1128/jvi.68.5.2879-2888.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H-W, Wu H-L, Chen D-S, Chen P-J. Identification of the functional regions required for hepatitis D virus replication and transcription by linker-scanning mutagenesis of viral genome. Virology. 1997;239:119–131. doi: 10.1006/viro.1997.8818. [DOI] [PubMed] [Google Scholar]
- 72.Chang J, Gudima SO, Taylor JM. Evolution of hepatitis δ virus RNA genome following long-term replication in cell culture. J Virol. 2005;79:13310–13316. doi: 10.1128/JVI.79.21.13310-13316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ranjith-Kumar CT, Kao CC. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA. 2006;12:303–312. doi: 10.1261/rna.2163106. ▪ Demonstrates that RNA polymerases can initiate transcription on circular RNAs.
- 74.Filipovska J, Konarska MM. Specific HDV RNA-templated transcription by pol II in vitro. RNA. 2000;6:41–54. doi: 10.1017/s1355838200991167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450:445–449. doi: 10.1038/nature06290. [DOI] [PubMed] [Google Scholar]
- 76.Katze MG, Fornek JL, Palermo RE, Walters KA, Korth MJ. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat Rev Immunol. 2008;8:644–654. doi: 10.1038/nri2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maida Y, Yasukawa M, Furuuchi M, et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:E3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerritzen A, Brackmann H, van Loo B, et al. Chronic δ hepatitis in haemophiliacs. J Med Virol. 1991;34:188–190. doi: 10.1002/jmv.1890340311. [DOI] [PubMed] [Google Scholar]
- 81.Taylor JM. Viral hepatitis δ. In: Monga P, editor. Molecular Pathology of Liver Diseases. Springer; NY, USA: 2009. In press. [Google Scholar]
Website
- 101.Subviral RNA Database. http://subviral.med.uottawa.ca.


