Abstract
Gene sequences for seven glycoproteins from 20 independent isolates of rhesus monkey rhadinovirus (RRV) and of the corresponding seven glycoprotein genes from nine strains of the Kaposi’s sarcoma-associated herpesvirus (KSHV) were obtained and analyzed. Phylogenetic analysis revealed two discrete groupings of RRV gH sequences, two discrete groupings of RRV gL sequences and two discrete groupings of RRV gB sequences. We called these phylogenetic groupings gHa, gHb, gLa, gLb, gBa and gBb. gHa was always paired with gLa and gHb was always paired with gLb for any individual RRV isolate. Since gH and gL are known to be interacting partners, these results suggest the need of matching sequence types for function of these cooperating proteins. gB phylogenetic grouping was not associated with gH/gL phylogenetic grouping. Our results demonstrate two distinct, distantly-related phylogenetic groupings of gH and gL of RRV despite a remarkable degree of sequence conservation within each individual phylogenetic group.
Keywords: RRV glycoprotein, variation, phylogenetic grouping
Introduction
Rhesus monkey rhadinovirus (RRV; Macacine herpesvirus 5) is a natural infectious agent of rhesus macaques (Macaca mulatta) that is closely related to the human Kaposi’s sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) (Desrosiers et al., 1997). KSHV is a causative factor in the development of Kaposi’s sarcoma and certain lymphoproliferative disorders including body cavity-based lymphomas and multicentric Castleman’s disease (Cesarman et al., 1995; Chang et al., 1994; Huang et al., 1995; Moore and Chang, 1995; Soulier et al., 1995). The overall organization of the genomes of these two viruses is very similar, with greater than 95% of the reading frames correspondingly placed in a co-linear fashion (Alexander et al., 2000; Searles et al., 1999). Both viruses are classified in the Rhadinovirus genus of the Gammaherpesvirinae subfamily (family Herpesviridae, order Herpesvirales). The Rhadinovirus genus also includes Ateline herpesvirus 2 (Herpesvirus ateles), Bovine Herpesvirus 4, Murid Herpesvirus 4 (murine herpesvirus 68), and Saimiriine Herpesvirus 2 (herpesvirus saimiri) (McGeoch, 2001).
Receptor-mediated viral entry initiated by viral envelope glycoproteins is a critical first step for the replication of any virus. With herpes simplex virus (HSV), fusion to susceptible target cells requires the joint presence of glycoprotein B (gB), gD and the gH-gL complex (Muggeridge, 2000; Pertel et al., 2001; Turner et al., 1998). Similarly, KSHV gB, gH and gL can mediate cell fusion using Chinese hamster ovary and human embryonic kidney cells (Pertel, 2002). KSHV K8.1, one of the most antigenic KSHV products (Chandran et al., 1998; Huang et al., 1995; Lang et al., 1999), is known to interact with heparin sulfate (Wang et al., 2001) but it is dispensable for viral entry into 293 cells (Luna et al., 2004). The envelope glycoprotein M (gM), which has counterparts in all herpesviruses, is essential for lytic replication of Murine Herpesvirus 68 (May, Colaco, and Stevenson, 2005; May et al., 2008). With KSHV, gM and gN form a complex and, when co-expressed, they can inhibit the fusion with 293 cells (Koyano et al., 2003). Thus, herpesviruses appear more complex than other viral families in terms of the numbers of glycoproteins needed to achieve virus entry.
Antibodies capable of neutralizing viral infectivity are directed to the viral-encoded envelope glycoproteins on the surface of virions. Neutralizing antibodies are one potential source of selective pressure for sequence change. Among the herpesviruses, there is little information linking glycoprotein sequence variation with selective pressure from neutralizing antibodies. In the case of human cytomegalovirus (CMV), a betaherpesvirus, Klein et al. have noted a strain specificity to the neutralizing antibody response (Klein et al., 1999). Although discrete phylogenetic groupings of gN of CMV have been documented (Pignatelli, Dal Monte, and Landini, 2001), it is not known to what extent this may be related to the strain specificity of the neutralizing activity.
For RRV, Bilello et al (2006) have presented evidence for a relative strain specificity to the neutralizing antibody response. Of all the monkeys tested so far, monkeys infected with the RRV prototype strain 26-95 exhibited the highest neutralizing antibody titers against this same strain (Bilello et al., 2006). Some monkeys with high antibody-binding titers to whole virus by ELISA showed quite weak neutralizing titers to this same RRV strain 26-95. Two complete RRV genomes have been sequenced to date (Alexander et al., 2000; Searles et al., 1999). Although there was very high sequence identity between almost all genes of these two isolates, very high divergence was observed in the gH and gL reading frames (Alexander et al., 2000; Searles et al., 1999). One question resulting from this observation is whether there is a continuum of sequence divergence in RRV gH and gL reading frames, or whether there are discrete phylogenetic groupings. A continuum of sequence divergence would be consistent with the possibility of ongoing selective pressure from neutralizing antibodies and potential linkage to the observed strain specificity of the neutralizing antibody response.
In the current study, we obtained 20 new isolates of RRV and derived sequences from the seven glycoprotein genes. These seven glycoprotein genes are gB, gH, gL, gM, gN, R8.1 and orf68. Since K1 is the most variable of the KSHV genes (Meng et al., 2001; Nicholas et al., 1998; Zong et al., 1999; Zong et al., 1997), we also sequenced the R1 reading frames from the 20 RRV isolates. Analysis of the sequences revealed interesting, surprising patterns of sequence variation.
Results
We determined 8700 nucleotide positions per RRV isolate (174,000 total) and 8874 per KSHV isolate (88,740 total). Sequences from the 20 new RRV isolates were compared to each other and to the sequences previously published for RRV strains 26-95 and 17577 (Alexander et al., 2000; Searles et al., 1999) (GenBank Accession numbers AF210726 and NC_003401). Sequences from the nine KSHV-positive lines were compared to each other and to published sequences for the GK18 isolate (Glenn et al., 1999) (NCBI reference sequence NC_009333).
Breeding groups of Indian-origin rhesus monkeys were initially formed at the New England Primate Research Center (NEPRC) in the late 1960s from a diversity of sources. Indian-origin rhesus monkeys from external sources have occasionally been added to the breeding groups since that time. Nineteen of the 20 new RRV isolates used for our study were obtained from rhesus monkeys born at NEPRC. None of these 19 had the same parents and all were raised in different social groups in order to minimize the chances of obtaining RRV isolates that were closely linked epidemiologically. Seven of the 19 were born in 2004 (indicated by the last two numbers after the dash in the animal number, e.g. 102-04). One of the monkeys (373-03) was obtained from a supplier (Covance) at 15 months of age in August 2003. This monkey was already RRV-positive upon arrival at that time.
RRV gH and gL
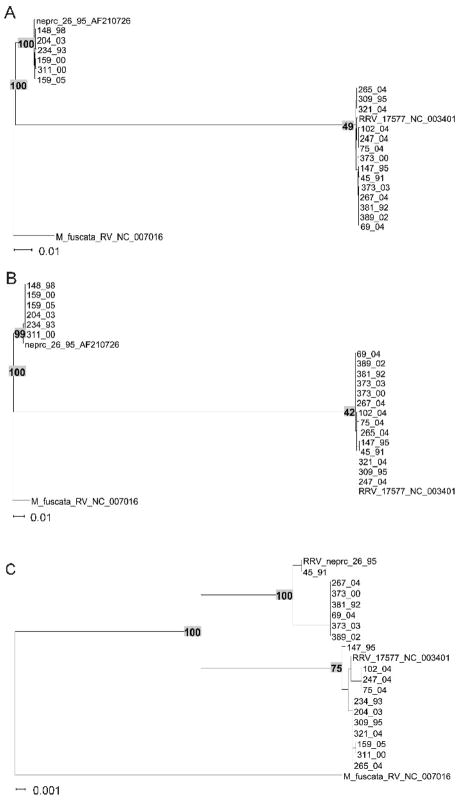
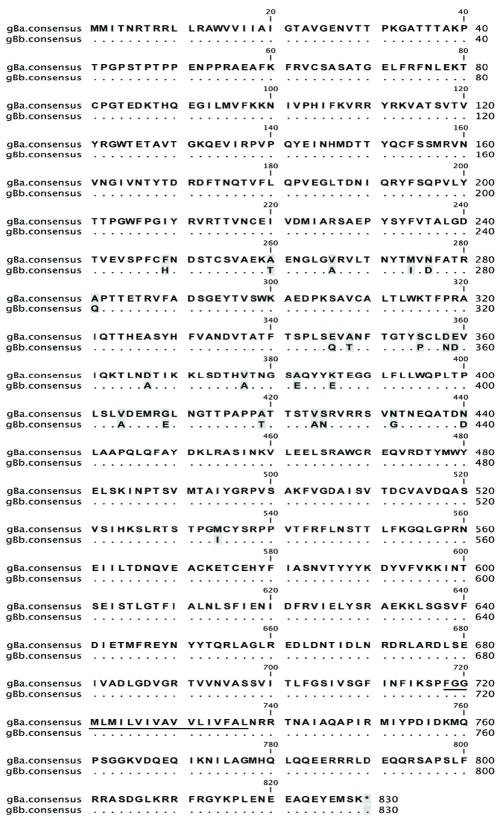
Sequences of the gH gene from the twenty new RRV isolates very clearly fell into two distinct groupings (Fig. 1, Fig. 2, Fig. 3A and Table 1). Six of them were very similar to the gH sequences present in the original RRV isolate 26-95 from the New England Primate Research Center (Alexander et al., 2000) and fourteen of them were very similar to RRV isolate 17577 from the Oregon National Primate Research Center (Searles et al., 1999). We refer to the former as gHa and the latter as gHb. gHa is extremely different from gHb in the extracellular portion of this membrane-spanning protein (Fig. 2). The consensus amino acid sequence of gHa differs from that of gHb at 201 positions of the 726 amino acids that comprise the full stretch of gHa protein. Sequence differences were confined to the N-terminal portion of the extracellular domain (Fig. 2). gHa and gHb consensus sequences were identical over the C-terminal portion of the extracellular domain, the membrane-spanning domain, and the cytoplasmic domain (Fig. 2). There is one stretch of 10 identical amino acids within positions 105-138 and another 10 identical amino acids within positions 207-240 in the extracellular domain, but in general there is little similarity in sequence between gHa and gHb over the first 440-460 amino acids. There is enough similarity, however, to suggest that these indeed are related sequences, one connected to the other through some evolutionary lineage.
Figure 1.
Phylogenetic trees of RRV glycoproteins with greatest variation. A) gH; B) gL; C) gB. The trees were obtained with the Neighbor-Joining algorithm using the Kimura 2 parameter distance correction. Numbers on branches indicate bootstrap supports. Branch lengths are proportional to the number of nucleotide substitutions per aligned site. The scale bars correspond to 0.01 (A,B) or 0.001 (C) substitutions per alignment position.
Figure 2.
Comparison of consensus amino acid sequences of gHa and gHb of RRV. Identical residues are indicated with dots. Gaps in the sequence are indicated by dashes. Shading indicates variable positions. The sequences were aligned using the Mafft program with default parameters. The putative membrane-spanning domain is underlined.
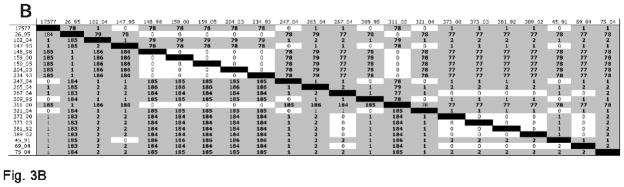
Figure 3.

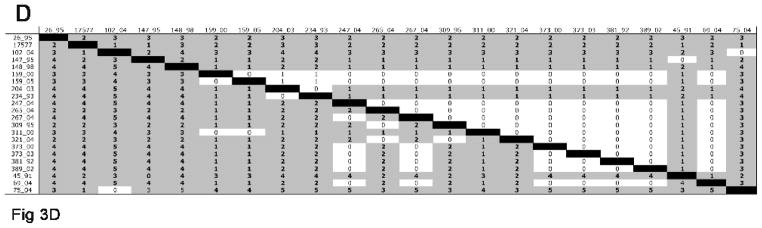
Pairwise comparison of amino acid and nucleotide differences in selected RRV reading frames. A) gH; B) gL; C) gB; D) R1. Numbers above the black boxes are the numbers for pairwise amino acid differences. Numbers below the black boxes are the numbers for pairwise nucleotide differences.
Table 1.
Summary of pair-wise amino acid differences and non-synonymous to synonymous substitutions (dN/dS) within the individual RRV genes that were analyzed.
| Glycoprotein | mean1 | median | range | dN/dS | length2 |
|---|---|---|---|---|---|
| gB | 11.5 | 19 | 0–24 | 0.17 | 829 |
| gBa | 1.7 | 0 | 0–4 | 0.03 | 829 |
| gBb | 0.3 | 0 | 0–1 | 0.20 | 829 |
| gH | 93.5 | 5 | 0–205 | 0.31 | 715 |
| gHa | 1.6 | 1 | 0–4 | 0.72 | 726 |
| gHb | 3.0 | 3 | 0–7 | 0.85 | 704 |
| gL | 35.7 | 2 | 0–79 | 0.22 | 166 |
| gLa | 0.0 | 0 | - | 0.00 | 163 |
| gLb | 0.9 | 1 | 0–2 | 1.11 | 169 |
| R8.1 | 0.7 | 0 | 0–3 | 0.08 | 275 |
| gM | 0.5 | 1 | 0–2 | 0.37 | 378 |
| gN | 0.0 | 0 | - | 0.00 | 104 |
| orf68 | 0.1 | 0 | 0–1 | 0.10 | 457 |
| R1 | 1.2 | 1 | 0–4 | 0.12 | 423 |
Mean number of amino acid differences in pair-wise comparisons among 20 (gB, R8.1, gM, gN, and orf68) or 22 (gH, gL, and R1) strains.
Number of amino acids (gB, gBa, gBb, gHa, gHb, gLa, gLb, R8.1, gM, gN, orf68, R1) or mean number of amino acids (gH and gL).
Despite the dramatic difference between gHa and gHb sequences, there was very little variation within gHa sequences, and very little variation within gHb sequences (Fig. 3A and Table 1). Among gHa sequences, median pairwise nucleotide differences were only 2 (range 0 to 5) for the 2178 nucleotides of gHa coding sequence and median pairwise amino acid differences were only 1 (range 0 to 4) for the 726 amino acids of gHa. Among gHb sequences, median pairwise nucleotide differences were only 5 (range 0 to 9) for the 2112 nucleotides of gHb coding sequence and median pairwise amino acid differences were only 3 (range 0 to 7) for the 704 amino acids of gHb.
Sequences of the gL gene from the twenty new RRV isolates also very clearly fell into two discrete groupings (Fig. 1, Fig. 3B, Fig. 4 and Table 1). Six of them were very similar to the gL sequence present in the original RRV isolate 26-95 from NEPRC (Alexander et al., 2000) and fourteen of them were very similar to RRV isolate 17577 from the ONPRC (Searles et al., 1999). We refer to the former as gLa and the latter as gLb. gLa is also extremely different from gLb, differing at 78 of the 163-169 amino acid positions across the full length of gL in their consensus sequences (Fig. 4). Again, there is enough similarity to suggest that these indeed are related sequences, one connected to the other through some evolutionary lineage.
Figure 4.

Comparison of consensus amino acid sequences of gLa and gLb of RRV. Identical residues are indicated with dots. Gaps in the sequence are indicated by dashes. Shading indicates variable positions. The sequences were aligned using the Mafft program with default parameters.
Despite the dramatic differences between gLa and gLb sequences, there was again very little variation within each group, i.e. within gLa and within gLb. Among gLa sequences, median pairwise nucleotide differences were zero (range 0 to 2) for the 489 nucleotides of gLa coding sequence and median pairwise amino acid difference was zero (range 0) for the 163 amino acids of gLa. Among gLb sequences, median pairwise nucleotide differences were only 1 (range 0 to 2) for the 507 nucleotides of gLb coding sequence and median pairwise amino acid differences were only 1 (range 0 to 2) for the 169 amino acids of gLb.
Interestingly, gHa was always paired with gLa and gHb was always paired with gLb.
We tested the interaction of gH and gL by co-immunoprecipitation using different tagged versions of each protein. Tagged gH protein of 26-95 was transiently expressed together with alternatively tagged versions of gL protein from strain 26-95 or 17577 in HEK 293T cells. Both gL proteins showed readily detectable, efficient interaction with the gH protein from 26-95 (Fig. 5). This ability to cross-interact does not rule out functional defects that may occur when gHa is paired with gLb.
Figure 5.
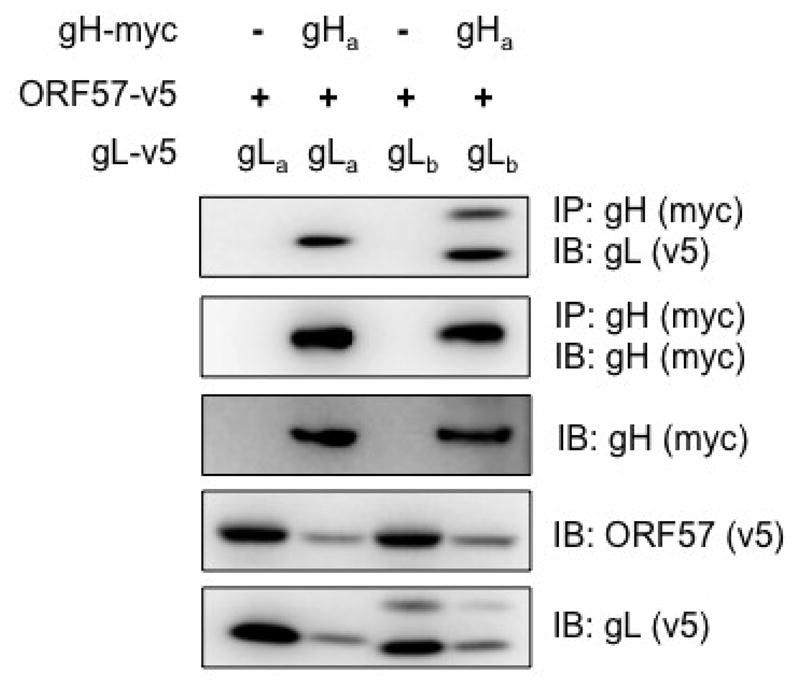
Interaction of gHa with gLa and gLb. RRV gHa, gLa, and gLb were transiently expressed in HEK 293T cells. RRV ORF57 was included in all transfections to allow expression of the glycoproteins. The expression of each gene in whole cell lysates was evaluated by immunoblot (IB) analysis using anti-myc (gHa) or anti-v5 (gLa, gLb, and ORF57) antibodies. Anti-myc antibody was used for immunoprecipitation (IP) of gHa protein. Co-immunoprecipitated gLa or gLb protein was detected using anti-v5 antibody.
KSHV gH and gL
Distinct phylogenetic groupings were not observed for gH or gL of KSHV among our ten KSHV lines analyzed (Fig. 6A and B), nor when two additional KSHV gH and gL sequences (U40377 and U93872) in the sequence database were included in the analysis (data not shown). The greatest divergence from the rest of the KSHV sequences were noted for VG-1 and BC-3 gH sequences (Fig. 6A) and for VG-1 gL sequence (Fig. 6B).
Figure 6.
Pairwise comparison of amino acid and nucleotide differences in selected KSHV reading frames. A) gH; B) gL; C) gB. Numbers above the black boxes are the numbers for pairwise amino acid differences. Numbers below the black boxes are the numbers for pairwise nucleotide differences.
RRV gB
Two distinct phylogenic groupings were also observed for RRV gB sequences (Fig. 1, Fig. 3C, Fig. 7, and Table 1). Seven of the 18 new RRV isolates that we analyzed were 26-95-like in their gB sequence and 11 of the 18 were 17577-like in their gB sequence. Again, sequence divergence was essentially confined to the extracellular domain (Fig. 7). However, the divergence between gBa and gBb was nowhere near as great as between gHa and gHb or between gLa and gLb. Also, gB phylogenetic groupings were unrelated to gH/gL phylogenetic groupings (Fig. 1). Among gBa sequences, median pairwise nucleotide differences were zero (range 0 to 13) for the 2487 nucleotides of gBa coding sequence and median pairwise amino acid differences were zero (range 0 to 4) for the 829 amino acids of gBa. Among gBb sequences, median pairwise nucleotide differences were only 3 (range 0 to 7) for the 2487 nucleotides of gBb coding sequence and median pairwise amino acid differences were zero (range 0 to 1) for the 829 amino acids of gBb.
Figure 7.
Comparison of consensus amino acid sequences of gBa and gBb of RRV. Identical residues are indicated with dots. Gaps in the sequence are indicated by dashes. Shading indicates variable positions. The sequences were aligned using the Mafft program with default parameters. The putative membrane-spanning domain is underlined.
KSHV gB
Nine of the ten KSHV gB sequences that we analyzed were very similar to one another (Fig. 6C). The gB sequence from KSHV VG-1 was an outlier from these other nine, exhibiting 17-19 nucleotide differences and 5-6 amino acid differences. But discrete phylogenetic groupings were not observed and the extent of pairwise differences among the KSHV gB sequences was similar to, or even less than, the pairwise differences within an individual phylogenetic grouping of RRV gB.
RRV R1; KSHV K1
There is an extensive literature on the considerable diversity among K1 gene sequences of KSHV (Meng et al., 2001; Nicholas et al., 1998; Zong et al., 1999; Zong et al., 1997). In fact, K1 is the most diverse of all KSHV proteins. K1 sequences have been placed into four discrete phylogenetic groupings, with as many as 90 amino acid differences (32%) in individual pairwise cross-clade comparisons (Meng et al., 2001; Meng et al., 1999). We thus examined our collection of 20 new RRV isolates for variation at the corresponding R1 locus. Little to no variation was observed at the R1 locus (Fig. 3D, Table 1). The original sequence comparisons of the NEPRC RRV isolate 26-95 and the ONPRC RRV isolate 17577 similarly showed high amino acid identity in the R1 reading frame (Alexander et al., 2000).
gM, gN, R8.1/K8.1, orf68
Minimal variation was observed in the RRV reading frames for gM, gN and orf68 (Table 1). Except for VG-1, which was again a sequence outlier, the KSHV gM, gN, and orf68 reading frames exhibited a level of variation similar to the minimal level observed in the corresponding reading frames of RRV (Table 2). Slightly more variation was observed in the R8.1 reading frame of RRV (Table 1). Again, VG-1 was an outlier when KSHV K8.1 sequences were compared; eight of our nine KSHV K8.1 sequences were virtually identical with one another and identical to K8.1 sequences for KSHV strain BCBL-1 in the database.
Table 2.
Summary of pairwise amino acid differences and non-synonymous to synonymous substitutions (dN/dS) within the individual KSHV genes that were analyzed.
| Glycoprotein | mean1 | median | range | dN/dS | length2 |
|---|---|---|---|---|---|
| gB | 1.4 | 1 | 0–6 | 0.32 | 845 |
| gH | 2.8 | 1 | 0–8 | 0.42 | 730 |
| gL | 0.4 | 0 | 0–2 | 0.15 | 167 |
| gM | 1.1 | 0 | 0–5 | 0.12 | 400 |
| gN | 0.2 | 0 | 0–1 | 0.07 | 110 |
| K8.1 | 1.4 | 0 | 0–7 | 0.34 | 228 |
| orf68 | 0.4 | 0 | 0–2 | 0.12 | 467 |
Mean number of amino acid differences in pair-wise comparisons. n=10 strains.
Number of amino acids.
RRV sequences from other sources
In order to expand the sources of rhesus monkeys used for analysis of RRV glycoprotein sequence variation, we obtained blood samples from rhesus monkeys from the island of Cayo Santiago off Puerto Rico and from rhesus monkeys in the process of importation from China. The history of the rhesus monkeys on Cayo Santiago dates back to the 1930s. Rhesus monkeys were captured near Lucknow India in 1938 and brought to the island of Cayo Santiago in 1939 for the purposes of biomedical research. They have been a closed colony since that time (Rawlins, 1986). Although rhesus monkeys from China and India can inter-breed, they are considered distinct subspecies (Cawthon, Lang KA. 2005). Blood samples were used to recover RRV and recovered RRV was used to amplify gH and gL sequences as above. All twelve new RRV isolates yielded amplified gH and gL products. Again, gHa was always paired with gLa and gHb was always paired with gL b. Among the five RRV isolates from the Cayo Santiago monkeys, two were type a and three were type b. The two gHa and the two gLa amino acid sequences from the Cayo Santiago monkeys were identical to the consensus sequences shown in Figures 2 and 4 and identical to each other. The gHb sequences from the Cayo monkeys differed at 2, 2, and 2 positions from the consensus shown in Fig 2 and gLb sequences differed at 1, 1, and 1 position from the consensus shown in Fig. 4. Among the seven Chinese RRV isolates, three were type a and four were type b. The range of pairwise amino acid differences compared to the consensus sequences shown in Figures 2 and 4 were 2–5, 2–5, 0-0 and 1-1 for gHa, gHb, gLa and gLb respectively. Thus, RRV isolates from the closed Cayo Santiago colony of rhesus monkeys and from Chinese-origin rhesus monkeys showed the same two phylogenetic groupings and the same patterns of gH and gL sequence conservation.
Discussion
The discrete groupings of RRV gH and gL sequences have a number of curious features. The sequences in the external domain of gH and across the full length of gL are markedly different between the phylogenetic groups (58.7% for gH and 54.4% for gL in amino acid identity). In fact, they are barely recognizable at first glance as being related sequences. Despite this considerable divergence in sequence, variation within a phylogenetic grouping is extremely minimal (99.7%, 99.5%, 100% and 99.4% for gHa, gHb, gLa, and gLb in amino acid identity, respectively). Thus, although these glycoproteins are able to tolerate, and in fact have evolved, marked differences in sequences, there is very very little variation within a phylogenetic grouping. The sequence differences between gHa and gHb are confined to the extracellular, external domain of the protein; there are no sequence differences in the C-terminal portion of the extracellular domain, the membrane-spanning domain or the cytoplasmic domain between these phylogenetic groups. And finally, despite the fact that the gH and gL genes are separated by 32,000 base pairs in the viral genome, gHa is always paired with gLa and gHb is always paired with gLb.
How does one explain the origin of these very different phylogenetic groupings and how does one explain this collection of curious features? The lack of significant sequence variation within a phylogenetic grouping, which we assume to have been circulating among rhesus monkeys for thousands of years, is not consistent with continuous sequence pressure from neutralizing antibodies. Evolution of use of a different cellular receptor or cellular binding partner seems to be one explanation that would fit the curious patterns of sequence variation in gH and gL. It is possible that sequence changes in one gene of the gH/gL pair by sequence evolution or by gene capture may have driven sequence change in the other gene of the pair. Since the two glycoproteins form a complex on the virion surface, our findings strongly suggest a need for functional cooperation that has resulted in a linkage of segregated sequence types.
Herpesviruses are DNA viruses whose polymerases for replicating the genetic information are not very error prone, much less error prone than the retroviruses for example. Nonetheless, there is ample evidence to indicate that significant amounts of sequence variation can emerge. Even during infection of a single individual, drug-resistant herpesvirus variants can appear during the course of antiviral drug treatment (Ducancelle et al., 2004; Stranska et al., 2004). Given the short period of time for a single round of viral replication, and the thousands of years that these herpesviruses have presumably been circulating in the population, one would expect selective forces such as neutralizing antibodies to impart specific patterns of sequence variation on the targets of those selective forces. The lack of significant sequence variation for gHa, gHb, gLa, gLb, gM, gN and orf68 of RRV, and for gH, gL, gM, gN, and orf68 of KSHV, suggest that there is little or no selective force to drive sequence change in these genes.
The extreme similarity in sequences for gHa, gHb, gLa, gLb, gM, gN and orf68 is not a peculiar feature of RRV isolates from the New England Primate Research Center. NEPRC has not historically been a closed colony. From its inception in the 1960s, Indian-origin rhesus monkeys have been taken in from a variety of sources and entered into our breeding colony. Nineteen of the 20 rhesus monkeys that were used as the source of new RRV isolates were all born to different parents and raised in different social groupings. Furthermore, gH/gL sequences from one set of these NEPRC RRV isolates match up very closely with the Oregon RRV isolate that was obtained totally independently at the different location. The one monkey (373-03) that came from an external source already infected with RRV yielded RRV sequences closely matched to those from NEPRC-born monkeys. Finally, and most importantly for this point, RRV isolates from the closed Cayo Santiago colony of rhesus monkeys and from Chinese-origin rhesus monkeys showed the exact same patterns of gH and gL sequence conservation. It is important to note that such patterns of glycoprotein sequence conservation are not confined to RRV. Certainly, our KSHV sequences and those in the database for KSHV gH, gL, gM, gN, and orf68 are remarkably conserved. Similar patterns exist for some of the CMV glycoprotein genes as well. For example, Pignatelli et al have demonstrated discrete phylogenetic groupings of gN of CMV, but there is remarkably little variation within each phylogenetic grouping (Pignatelli et al., 2003).
The lack of amino acid variation in the genes specified above suggests little or no selective pressure for change, such as what would be expected from the pressure of neutralizing antibodies over the course of thousands of years of evolutionary history. Even more remarkable is the dearth of third base synonymous changes within these selected glycoprotein genes of independent isolates. This suggests that there is selective pressure to maintain the codon usage within these highly conserved glycoprotein gene clusters. Such pressure to maintain a particular codon usage is not, however, a pressure to maintain the standard cellular optimal codon usage since it has been shown that the codon usage for gH and gL of RRV, and to some extent other herpesviruses as well, is highly suboptimal for expression in uninfected cells (Bilello, Morgan, and Desrosiers, 2008). Thus, there appears to be selective pressure to maintain this highly unusual codon usage.
Materials and Methods
Viruses
Peripheral blood mononuclear cells (PBMCs) from rhesus monkeys were co-cultured with rhesus monkey fibroblasts (RF) in Dulbecco’s modified Eagle’s medium (GIBCO) supplemented with 20% fetal bovine serum (GIBCO), 2 mM L-glutamine, and penicillin-streptomycin (GIBCO, 10 unit and 10 μg/ml, respectively) as described elsewhere (Desrosiers et al., 1997). The KSHV-harboring cells were obtained from the ATCC repository (BC-1, BC-2, BC-3, BCP-1, and JSC-1) or Dr. Jae Jung (BCBL-1, VG-1, 239A, and APK1) (University of Southern California). VG-1 is an African B-subtype genome; the other KSHV-containing PEL lines are of Caucasian origin (Zong et al., 2007). They were cultured according to the suggestions of the provider for subsequent viral DNA isolation.
DNA amplification and sequencing
Viral genomic DNA was partially purified from cell culture supernatants or cell pellets using QIAamp DNA blood mini kit (QIAGEN). Approximately 10 ng of the obtained DNA was used as PCR template. Conserved sequence regions flanking the genes of interest were identified by inspection of the aligned complete genomic sequences of RRV (AF210726 and NC_003401) and KSHV (NC_009333 and NC_003409). Primers directed to these regions were selected manually and checked for the criteria such as dimer formation capability, false annealing, annealing temperature and self-annealing capability using MacVector program. The optimal annealing conditions for each primer set were determined by gradient PCR in order to maximize specificity and DNA yield. The sequence of forward (f) and reverse (r) primers used for the amplification of RRV and KSHV genes are presented in the parenthesis with the optimized annealing temperature.
RRV gH (f-gctacattcaaacgctaacca, r-gttttacgctttattaacagt, 55°C), RRV gL (f-atttaagccatgagtcgctaa, r-agctgggcggatatccggaag, 55°C), RRV R1 (f-tgccatcaacctttgcttgca, r-ataccgggcaaagatacaaac, 55°C), RRV gM (f-tgtacaaacccaaaaccaagcc, r-taagccacttgctgattttactgc, 48.2°C), RRV R8.1 (f-cgtttgnggtttgnccatttcc, r-gcggaatcgctgccagcgcggacg, 56.7°C), RRV orf68 (f-acgccaataaatcgtcaccg, r-cgacntctgggctgttttgg, 53.4°C), RRV gN (f-acgcgtggaagacatggc, r-agaggttctcccggtttgacc, 56.7°C), RRV gB (f-ctccatctccnacctagacg, r-gtgcgcgaatcgattggc, 53.4°C), KSHV K8.1 (f-ccgggagaaccatgccag, r-caccgctaaaccgcctcc, 59.6°C), KSHV orf68 (f-gagtggtcacctgccctgc, r-tgtggctggacactgatttcg, 61.8°C), KSHV gN (f-atctctcggatcggcagtgg, r-ffrcccacarcafrcacraccc, 56.7°C), KSHV gB (f-cccttggtgttggtggat, r-gtctgtatgtggtgcttc, 56.7°C), KSHV gH (f-caggcagatcctgtccaatc, r-ggtgctcggatttcttgc, 58.4°C), KSHV gM (f-cagtatggttttctgtacgtatt, r-gcgataggcagtggcatcag, 58.4°C) KSHV gL (f-taggtgccagtaacagatcc, r-tcattagtcgggactcg, 55.5°C).
The PCR was performed using platinum PCR supermix (Invitrogen) and the cycles consisted initial denaturation (94°C for 5 minutes) followed by 35 cycles of amplification (94°C for 30 seconds, optimized annealing temperature for 1 minute, and 68°C for 1 minute per Kb of extension). The PCR products were purified using QIAquick PCR purification kit (QIAGEN) and sequenced in both directions. The PCR products were purified using QIAquick PCR purification kit (QIAGEN) and sequenced in both directions. All the sequences were submitted to GenBank with the following accession numbers: KSHV gB (GU233080 - GU233088), KSHV gH (GU233089 - GU233097), KSHV gL (GU233098 - GU233106), KSHV gM (GU233107 - GU233115), KSHV gN (GU233116 - GU233124), KSHV K8.1 (GU233125 - GU233133), KSHV ORF68 (GU233134 - GU233142), RRV gB (GU233143 - GU233160), RRV gH (GU233161 - GU233180), RRV gL (GU233181 - GU233200), RRV gM (GU233201 - GU233218), RRV gN (GU233219 - GU233236), RRV ORF68 (GU233237 - GU233254), RRV R1 (GU233255 - GU233274), RRV R8.1 (GU233275 - GU233292). gH and gL sequences of the RRV isolates from Cayo Santiago and Chinese rhesus monkeys have also been submitted to GenBank (GU254256-GU254279).
PCR amplification products were sequenced directly without cloning. 20:80 mixtures would have been readily detected from these direct sequencing reactions. Dual infections were not detected with these methodologies. In every instance, the sequences shown represent the only population detected in the recovered virus and consequently the matched genes must be present in the same genomes. We did not design methodologies to detect small amounts of one genotype in the presence of much larger quantities of the other.
Sequence analysis
The sequences were aligned using the Mafft program (Katoh et al., 2005; Katoh et al., 2002). Amino acid sequences were obtained from the nucleotide sequences using the Genetic Data Environment (GDE) program (Eisen, 1997; Smith et al., 1994). Sequence statistics were calculated from the alignments using macros written in the R Package language (http://www.R-project.org). Pair-wise nucleotide or amino acid differences (D) between a given pair of sequences were obtained by the following formula:
where P is the number of positions in the alignment of the sequences a and b, and
Likewise, the mean number of pair-wise differences between two sequence clusters, x and y, with nx and ny sequences each, were obtained by the formula:
The rates of nonsynonymous to synonymous substitutions, dN/dS (Hurst, 2002; Nei and Gojobori, 1986), were obtained with the Seqinr package (Charif, 2007). Phylogenetic analyses were performed by bootstrapped (n=1000) Neighbor-Joining using the ClustalX program (Thompson, 2002).
Immunoprecipitation
Plasmids encoding gHa and ORF57 of RRV 26-95 have been described previously (Bilello, 2008). The coding regions of gL from RRV 26-95 (gLa) and 17577 (gLb) were cloned in-frame into the pCDNA6-V5/HisA (Invitrogen) after PCR amplification. HEK293T cells, used in transient expression, were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and penicillin (100 units/ml)/streptomycin (100 μg/ml). One day post seeding of HEK293T cells onto 100 mm culture dishes, cells were transfected with different combinations of plasmids as described in Fig. 5 using the Calphos mammalian transfection kit (Clontech) according to the manufacturer’s instructions. At 36 h post transfection, cells were harvested and resuspended with 1 ml of lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Triton X-100] containing protease inhibitors (Sigma) and centrifuged (12,000 × g) for 2 min. The supernatants were transferred to new tubes and their respective protein levels were measured using the bicinchoninic acid (BCA) protein assay kit (Pierce) for normalization purposes.
For sodium dodecyl sulfate-polyacylamide gel electrophoresis (SDS-PAGE), 50 μl of cell lysates were mixed with the equal volume of 2X SDS sample buffer and boiled for 5 min before SDS-PAGE analysis. For immunoprecipitation, the remaining cell lysates were precleared with Sepharose beads for 1 hour at 4°C and anti-myc antibody (Invitrogen) was added into the precleared cell lysates and incubated for 3 hours at 4°C. Protein A/G agarose (Pierce) was added into each tube and mixed for an additional 2 hours. The beads were washed three times with cold lysis buffer. The purified proteins were eluted with 1X SDS sample buffer, separated by SDS-PAGE, and transferred onto polyvinylidene difluoride membrane (Roche). The membrane was subjected to immunoblot assay. Briefly, the membranes were blocked with PBS containing 5% skim milk for 30 min at room temperature and incubated with anti-myc (Invitrogen) or anti-v5 antibody (Invitrogen) for 2 hours followed by 1 hour of incubation with horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Cell Signaling). Specific signals were detected by an enhanced chemiluminescence system.
Acknowledgments
This work was supported by PHS grants AI084698 and AI63928 to R.C.D., RR00168 to NEPRC, and by continuous support to L.R.J. from National Council for Scientific and Technical Research (CONICET) of Argentina. We thank Jae Jung and David Scadden for providing KSHV cell lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. The primary sequence of rhesus monkey rhadinovirus isolate 26–95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74 (7):3388–98. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello J, Morgan JS, Desrosiers RC. Extreme dependence of gH and gL expression on ORF57 and association with highly unusual codon usage in rhesus monkey rhadinovirus. J Virol. 2008;82:7231–7237. doi: 10.1128/JVI.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello JP, Morgan JS, Damania B, Lang SM, Desrosiers RC. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J Virol. 2006;80 (3):1549–62. doi: 10.1128/JVI.80.3.1549-1562.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello JP, Morgan JS, Desrosiers RC. Extreme dependence of gH and gL expression on ORF57 and association with highly unusual codon usage in rhesus monkey rhadinovirus. J Virol. 2008;82 (14):7231–7. doi: 10.1128/JVI.00564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon Lang KA. Primate Factsheets: Rhesus macaque (Macaca mulatta) Taxonomy, Mophology, & Ecology. 2005 July 20; http://pin.primate.wisc.edu/factsheets/entry/rhesus_macaque.
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chandran B, Smith MS, Koelle DM, Corey L, Horvat R, Goldstein E. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology. 1998;243 (1):208–17. doi: 10.1006/viro.1998.9055. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266 (5192):1865–9. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR. SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: Molecules, networks, populations. Springer Verlag; New York: 2007. [Google Scholar]
- Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71 (12):9764–9. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducancelle A, Belloc S, Alain S, Scieux C, Malphettes M, Petit F, Brouet JC, Sanson Le Pors MJ, Mazeron MC. Comparison of sequential cytomegalovirus isolates in a patient with lymphoma and failing antiviral therapy. J Clin Virol. 2004;29 (4):241–7. doi: 10.1016/S1386-6532(03)00163-X. [DOI] [PubMed] [Google Scholar]
- Eisen JA. The Genetic Data Environment. A user modifiable and expandable multiple sequence analysis package. Methods Mol Biol. 1997;70:13–38. [PubMed] [Google Scholar]
- Glenn M, Rainbow L, Aurade F, Davison A, Schulz TF. Identification of a spliced gene from Kaposi’s sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73 (8):6953–63. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YQ, Li JJ, Kaplan MH, Poiesz B, Katabira E, Zhang WC, Feiner D, Friedman-Kien AE. Human herpesvirus-like nucleic acid in various forms of Kaposi’s sarcoma. Lancet. 1995;345 (8952):759–61. doi: 10.1016/s0140-6736(95)90641-x. [DOI] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18(9):486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33 (2):511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30 (14):3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Schoppel K, Amvrossiadis N, Mach M. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J Virol. 1999;73 (2):878–86. doi: 10.1128/jvi.73.2.878-886.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S, Mar EC, Stamey FR, Inoue N. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol. 2003;84 (Pt 6):1485–91. doi: 10.1099/vir.0.18941-0. [DOI] [PubMed] [Google Scholar]
- Lang D, Hinderer W, Rothe M, Sonneborn HH, Neipel F, Raab M, Rabenau H, Masquelier B, Fleury H. Comparison of the immunoglobulin-G-specific seroreactivity of different recombinant antigens of the human herpesvirus 8. Virology. 1999;260(1):47–54. doi: 10.1006/viro.1999.9804. [DOI] [PubMed] [Google Scholar]
- Luna RE, Zhou F, Baghian A, Chouljenko V, Forghani B, Gao SJ, Kousoulas KG. Kaposi’s sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J Virol. 2004;78 (12):6389–98. doi: 10.1128/JVI.78.12.6389-6398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Colaco S, Stevenson PG. Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J Virol. 2005;79 (6):3459–67. doi: 10.1128/JVI.79.6.3459-3467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Smith CM, Gill MB, Stevenson PG. An essential role for the proximal but not the distal cytoplasmic tail of glycoprotein M in murid herpesvirus 4 infection. PLoS One. 2008;3 (5):e2131. doi: 10.1371/journal.pone.0002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ. Molecular evolution of the gamma-Herpesvirinae. Philos Trans R Soc Lond B Biol Sci. 2001;356 (1408):421–35. doi: 10.1098/rstb.2000.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YX, Sata T, Stamey FR, Voevodin A, Katano H, Koizumi H, Deleon M, De Cristofano MA, Galimberti R, Pellett PE. Molecular characterization of strains of Human herpesvirus 8 from Japan, Argentina and Kuwait. J Gen Virol. 2001;82 (Pt 3):499–506. doi: 10.1099/0022-1317-82-3-499. [DOI] [PubMed] [Google Scholar]
- Meng YX, Spira TJ, Bhat GJ, Birch CJ, Druce JD, Edlin BR, Edwards R, Gunthel C, Newton R, Stamey FR, Wood C, Pellett PE. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology. 1999;261 (1):106–19. doi: 10.1006/viro.1999.9853. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332(18):1181–5. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol. 2000;81 (Pt 8):2017–27. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3 (5):418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nicholas J, Zong JC, Alcendor DJ, Ciufo DM, Poole LJ, Sarisky RT, Chiou CJ, Zhang X, Wan X, Guo HG, Reitz MS, Hayward GS. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- Pertel PE. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J Virol. 2002;76 (9):4390–400. doi: 10.1128/JVI.76.9.4390-4400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279 (1):313–24. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- Pignatelli S, Dal Monte P, Landini MP. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J Gen Virol. 2001;82 (Pt 11):2777–84. doi: 10.1099/0022-1317-82-11-2777. [DOI] [PubMed] [Google Scholar]
- Pignatelli S, Dal Monte P, Rossini G, Chou S, Gojobori T, Hanada K, Guo JJ, Rawlinson W, Britt W, Mach M, Landini MP. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J Gen Virol. 2003;84 (Pt 3):647–55. doi: 10.1099/vir.0.18704-0. [DOI] [PubMed] [Google Scholar]
- Rawlins RG, Kessler MJ. The Cayo Santiago Macaques: History, Behavior and Biology. State University of New York Press; New York, NY: 1986. [Google Scholar]
- Searles RP, Bergquam EP, Axthelm MK, Wong SW. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73 (4):3040–53. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM. The genetic data environment an expandable GUI for multiple sequence analysis. Comput Appl Biosci. 1994;10 (6):671–5. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86(4):1276–80. [PubMed] [Google Scholar]
- Stranska R, van Loon AM, Bredius RG, Polman M, Nienhuis E, Beersma MF, Lankester AC, Schuurman R. Sequential switching of DNA polymerase and thymidine kinase-mediated HSV-1 drug resistance in an immunocompromised child. Antivir Ther. 2004;9 (1):97–104. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Curr Protoc Bioinformatics. unit 23. Chapter 2. 2002. Multiple sequence alignment using ClustalW and ClustalX. [DOI] [PubMed] [Google Scholar]
- Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72 (1):873–5. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FZ, Akula SM, Pramod NP, Zeng L, Chandran B. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J Virol. 2001;75 (16):7517–27. doi: 10.1128/JVI.75.16.7517-7527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73 (5):4156–70. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Kajumbula H, Boto W, Hayward GS. Evaluation of global clustering patterns and strain variation over an extended ORF26 gene locus from Kaposi’s sarcoma herpesvirus. J Clin Virol. 2007;40 (1):19–25. doi: 10.1016/j.jcv.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Metroka C, Reitz MS, Nicholas J, Hayward GS. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J Virol. 1997;71 (3):2505–11. doi: 10.1128/jvi.71.3.2505-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]