Abstract
Ionotropic GABA receptors are widely distributed throughout the vertebrate and invertebrate central nervous system (CNS) where they mediate inhibitory neurotransmission. One of the most widely studied insect GABA receptors is constructed from RDL (resistance to dieldrin) subunits from Drosophila melanogaster. The aim of this study was to determine critical features of agonists binding to RDL receptors using in silico and experimental data. Partial atomic charges and dipole separation distances of a range of GABA analogues were calculated, and the potency of the analogues was determined using RDL receptors expressed in Xenopus oocytes. These data revealed functional agonists require an ammonium group and an acidic group with an optimum separation distance of ∼5 Å. To determine how the agonists bind to the receptor, a homology model of the extracellular domain was generated and agonists were docked into the binding site. The docking studies support the requirements for functional agonists and also revealed a range of potential interactions with binding site residues, including hydrogen bonds and cation−π interactions. We conclude that the model and docking procedures yield a good model of the insect GABA receptor binding site and the location of agonists within it.
Ionotropic GABA1 receptors are widely distributed throughout the vertebrate and invertebrate central nervous system (CNS) where they predominantly mediate inhibitory neurotransmission. To date, three ionotropic insect GABA receptor subunits from Drosophila melanogaster have been identified and successfully cloned: resistance to dieldrin (RDL) (1), glycine-like receptor of Drosophila (GRD) (2), and ligand-gated chloride channel homologue 3 (LCCH3) (3). RDL is the gene product of the rdl gene, which was identified in mutant Drosophila strains showing resistance to the insecticide dieldrin (1). Homomeric receptors constituted from RDL subunits (RDL receptors) are similar to vertebrate GABAA receptors in that they are inhibited by picrotoxin but differ in that they are not inhibited by bicuculline; these characteristics are similar to those of GABA receptors of cultured Drosophila neurons (4).
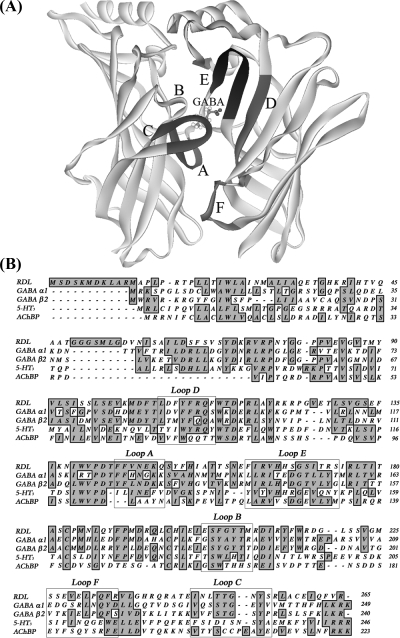
Cys-loop receptors, such as GABAA, GABAC (a subclass of GABAA), glycine, 5-HT3, and nicotinic acetylcholine (nACh) receptors, have homologous regions that form their agonist binding sites. These are located in the extracellular domain and are comprised of six discontinuous loops, named A−F (Figure 1). There are as yet no high-resolution structural data of a complete vertebrate Cys-loop receptor, but the lower-resolution nACh receptor structure and homologous structures, such as those from the related acetylcholine binding protein (AChBP), have been useful for creating homology models (5−8). Homology models have been created for many different vertebrate Cys-loop receptors, including nACh, 5-HT3, GABA, and glycine receptors (9−12), but invertebrate Cys-loop receptors have been largely ignored. The paucity of models for these proteins is surprising, as these proteins are the targets of a number of invertebrate specific ligands, such as insecticides, and a better understanding of critical binding site features could assist the development of novel, more effective compounds (13,14). Some information can be extrapolated from vertebrate models, which support previous mutagenesis and labeling studies in showing that aromatic residues in the binding loops contribute to the formation of an “aromatic box” that is important for ligand binding in all of these receptors (12,15−18). Critical aromatic residues, however, are not necessarily equivalent for different receptors; even in the closely related GABAA and GABAC receptors, for example, GABA has different orientations in the binding pocket. In the former, there is a cation−π interaction between GABA and a tyrosine residue in loop A, while in the latter, this interaction is with a tyrosine in loop B (12,19). Nevertheless, in both these receptors, there is evidence that the carboxylate residue is close to arginine residues in loop D (16,20), suggesting that GABA has broadly similar orientations in both GABA receptor binding sites.
Figure 1.
(A) RDL extracellular domain dimer. Extracellular loops A−F that form the agonist binding site region in related Cys-loop receptors are labeled. (B) Sequence alignment of extracellular domains of RDL and homologous Cys-loop receptor subunits (human GABAA α1, human GABAA β2, mouse 5-HT3A, and AChBP).
There are a range of invertebrate Cys-loop receptors that are activated by GABA, but as mentioned above, we know little about the molecular details of insect GABA receptor binding sites. The aim of this study was to identify the molecular determinants of agonist binding and potential interactions with binding site residues in RDL receptors, one of the best studied classes of insect GABA receptors.
Experimental Procedures
Oocyte Maintenance
Xenopus laevis oocyte-positive females were purchased from NASCO (Fort Atkinson, WI) and maintained according to standard methods (21). Harvested stage V−VI Xenopus oocytes were washed in four changes of ND96 [96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES (pH 7.5)], defolliculated in 1.5 mg/mL collagenase type 1A for approximately 2 h, washed again in four changes of ND96, and stored at 17 °C in ND96 containing 2.5 mM sodium pyruvate, 50 mM gentamycin, and 0.7 mM theophylline.
Receptor Expression
RDL subunit cDNA (GenBnk accession number P25123) was subcloned from pRmHa3-RDL (22) into the oocyte expression vector pGEMHE. cRNA was transcribed in vitro from the linearized plasmid cDNA template using the mMessage mMachine T7 transcription kit (Ambion, Austin, TX). Stage V−VI oocytes were injected with 5 ng of cRNA, and currents were recorded 12−72 h postinjection.
Electrophysiology
Xenopus oocytes were voltage clamped at −60 mV using an OC-725 amplifier (Warner Instruments, Hamden, CT) and Digidata 1322A. Currents were filtered at a frequency of 1 kHz and sampled at 350 Hz. Microelectrodes were made from borosilicate glass (GC120TF-10, Harvard Apparatus, Edenbridge, Kent, U.K.) using a two-stage horizontal pull (P-87, Sutter Instrument Co., Novato, CA) and filled with 3 M KCl. Pipette resistances ranged from 0.5 to 1.5 MΩ. Oocytes were perfused with saline [96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM HEPES (pH 7.4)] at a rate of 15 mL/min. Drug application was via a simple gravity-fed system calibrated to run at the same rate.
Analysis and curve fitting were performed using Prism version 3.02 (GraphPad Software, San Diego, CA). Concentration−response data for each oocyte were normalized to the maximum current for that oocyte. Responses were normalized to a maximal GABA-evoked response. The mean response for a series of oocytes was iteratively fitted to the equation I = Imin + (Imax − Imin)/[1 + 10log(EC50−[A])nH], where Imax is the maximal response, Imin is the minimum response, [A] is the concentration of agonist, and nH is the Hill coefficient. Significance was calculated using a one-way ANOVA or an unpaired t test (Prism version 3.02).
Computational Ligand Analysis
Molecular modeling was conducted with ChemBio3D Ultra 11.0 (CambridgeSoft, Cambridge, U.K.). Ligand structures were generated in the charged zwitterionic state using ChemDraw. Ligand structures were energy minimized using the MM2 force field, and Mulliken charges (partial atomic charges) were calculated using the GAMESS interface. Dipole distances were calculated using the atomic distance tool in ChemBio3D Ultra 11.0.
Homology Modeling and Docking
The protein sequence of the RDL subunit was aligned with the sequence of AChBP (P58154), using FUGUE (23). A three-dimensional homology model was generated using MODELER 6v2 (24) on the basis of the crystal structure of AChBP at 2.7 Å resolution [Protein Data Bank (PDB) entry 1i9b]. The pentamer was generated by superimposition of the RDL extracellular domain onto each protomer of AChBP and was then energy minimized using the force field implemented in MODELER 6v2. The best model was selected after Ramachandran plot analysis of all the generated models using RAMPAGE (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php) (25).
Ligand structures generated using ChemBio3D Ultra 11.0 were energy minimized using the MM2 force field. Docking of the ligands into the RDL receptor homology model was conducted using GOLD 3.0. The binding site was defined using the αC atoms of conserved aromatics Phe206, Tyr254, and Tyr109, with a binding site radius of 10 Å. Ten genetic algorithm runs were performed on each docking exercise, giving a total of 10 solutions for each analogue. The structures were analyzed using the implemented GoldScore fitness function to identify the most accurate simulation. Hydrogen bonding interactions between ligands and binding site residues were identified using the hydrogen bond monitor function in ViewerLite (www.accelrys.com).
Results
Functional Responses
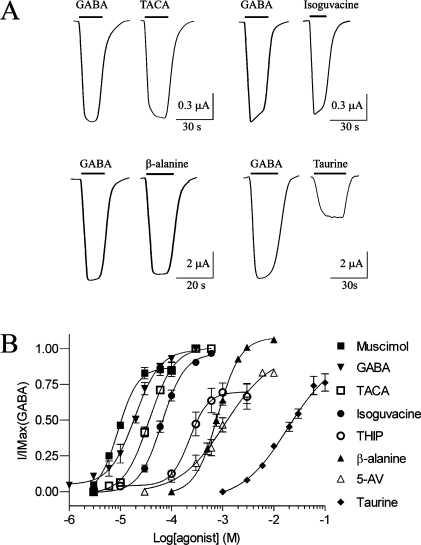
Application of GABA to Xenopus oocytes expressing RDL receptors elicited concentration-dependent inward currents. Plotting current amplitude against a range of GABA concentrations revealed an EC50 of 19 μM (Figure 2) which is similar to previously published results (10−31 μM) (26−30).
Figure 2.
(A) Electrophysiological traces showing maximal currents elicited by agonists tested at RDL receptors: GABA (100 μM), TACA (300 μM), isoguvacine (600 μM), β-alanine (10 mM), and taurine (10 mM). (B) Concentration response curves showing responses elicited by GABA and analogues at wild-type RDL receptors expressed in Xenopus oocytes: muscimol > GABA > TACA > isoguvacine > THIP > 5-AV > β-alanine > taurine. Data are means ± SEM; n ≥ 3.
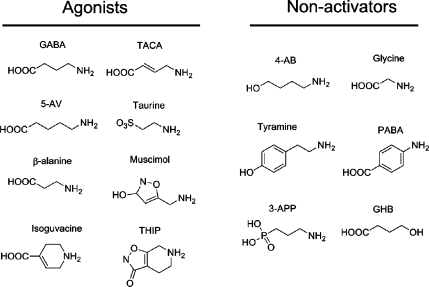
A range of GABA analogues were tested for activity at RDL receptors (Figure 3). The most potent of these were muscimol, TACA, and isoguvacine (Table 1). THIP and β-alanine are weaker agonists with EC50 values of 220 and 800 μM, respectively, while 5-aminovaleric acid (5-AV) and taurine were very weak agonists with EC50 values of 1.1 and >10 mM, respectively. GHB, a GABAB receptor agonist, and 3-APP, a GABAC and GABAB receptor antagonist, had no activating effect at RDL receptors. Glycine, 4-AB, PABA, and tyramine also failed to activate RDL receptors when applied at concentrations up to 10 mM. EC50 values for previously tested analogues (muscimol, β-alanine, TACA, and isoguvacine) are close to published values (26).
Figure 3.
Chemical structures of GABA analogues examined in this study.
Table 1. Parameters Derived from Concentration Response Curvesa.
| agonist | pEC50 (μM) | EC50 (μM) | nHb | nc | Imax/Imax(GABA) |
|---|---|---|---|---|---|
| muscimol | 5.04 ± 0.04 | 9.04 | 2.2 ± 0.6 | 5 | 0.87 ± 0.03 |
| GABA | 4.72 ± 0.03 | 19.3 | 1.8 ± 0.2 | 19 | 1.0 ± 0.06 |
| TACA | 4.44 ± 0.02 | 36.7 | 1.8 ± 0.2 | 3 | 1.0 ± 0.02 |
| isoguvacine | 4.19 ± 0.03 | 64.9 | 1.9 ± 0.2 | 5 | 0.97 ± 0.03 |
| THIP | 3.65 ± 0.10 | 226 | 2.4 ± 1.0 | 5 | 0.57 ± 0.06 |
| β-alanine | 3.09 ± 0.03 | 807 | 2.0 ± 0.3 | 10 | 1.0 ± 0.03 |
| 5-AV | 2.95 ± 0.15 | 1120 | 1.0 ± 0.3 | 4 | 0.85 ± 0.1 |
| taurine | <2.0 | >10000 | − | 3 | − |
| GHB | N/R | − | − | − | − |
| 3-APP | N/R | − | − | − | − |
| glycine | N/R | − | − | − | − |
| 4-AB | N/R | − | − | − | − |
| PABA | N/R | − | − | − | − |
| tyramine | N/R | − | − | − | − |
Data are means ± SEM. N/R indicates no response.
Hill coefficient.
Number of replicates.
Computational Ligand Analysis
The atomic distance between the ammonium nitrogen and the carboxylate oxygen, or its equivalent substituent, was calculated for all ligands tested. Comparing these data to the ligand EC50 values (Table 2) indicates a dipole separation distance for receptor activation of ∼5 Å. Glycine has a dipole separation distance of 2.35 Å and may be too short to activate RDL receptors, while for tyramine, the distance was 7.94 Å; thus, this molecule may be too long to fit into the binding site cleft. GHB and 3-APP, however, which have dipole separation distances of 5.1 and 4.5 Å, respectively, are not agonists, while 5-AV, which has a dipole separation distance of 6.5 Å, can activate receptors. These data suggest that other factors also play a role. One of these may be charge distribution; partial atomic charge calculations suggest there is a dependence on the electrostatic potentials of atomic groups at either end of the ligand. Partial atomic charges of +0.3 at ammonium hydrogen atoms and −0.5 to −0.6 on carboxylate oxygen atoms were common among the most potent ligands (Table 2). However, for 3-APP and 4-amino-1-butanol (4-AB), which do not activate receptors, the charges on phosphonic acid and hydroxyl oxygens are −1.2 and −0.4 respectively. Additionally, 4-AB and GHB are not zwitterions and thus carry no formal charge, so even if their length is close to optimal for receptor activation, their inactivity could be attributed to the absence of charge at either end of the ligand. The hydroxyl of GHB cannot substitute for the ammonium of GABA, confirming the importance of this group for ligand binding at RDL receptors.
Table 2. Dipole Separation Distances of GABA Analoguesa.
| ligand | potency pEC50 | dipole separation (Å) | carboxylate oxygensb | ammonium hydrogens |
|---|---|---|---|---|
| muscimol | 5.04 | 5.22 | −0.26 (isoxazole N), −0.47 | 0.32, 0.32, 0.30 |
| GABA | 4.72 | 4.83 | −0.60, −0.54 | 0.31, 0.31, 0.29 |
| TACA | 4.44 | 4.88 | −0.57, −0.53 | 0.31, 0.31, 0.29 |
| isoguvacine | 4.19 | 4.86 | −0.55, −0.53 | 0.29, 0.31, 0.29 |
| THIP | 3.65 | 5.14 | −0.25 (isoxazole N), −0.46 | 0.29, 0.31, 0.31 |
| β-alanine | 3.09 | 4.37 | −0.58, −0.51 | 0.29, 0.29, 0.29 |
| 5-AV | 3.06 | 6.47 | −0.61, −0.56 | 0.31, 0.31, 0.30 |
| taurine | <2.0 | 4.46 | −1.11, −1.12 | 0.30, 0.30, 0.28 |
| GHB | NRd | 5.10 | −0.61, −0.61 | −0.38 (hydroxyl O) c |
| 3-APP | NR | 4.50 | −1.23, −1.23 (phosphate O) | 0.29, 0.31, 0.31 |
| glycine | NR | 2.35 | −0.47, −0.57 | 0.32, 0.32, 0.27 |
| 4-AB | NR | 6.20 | −0.38 (hydroxyl O) | 0.19, 0.20, 0.24 |
| PABA | NR | 6.39 | −0.54, −0.54 | 0.30, 0.30, 0.30 |
| tyramine | NR | 7.94 | −0.47 (hydroxyl O) | 0.31, 0.30, 0.30 |
Mulliken charges of GABA analogues calculated using the GAMESS interface. Ammonium hydrogens and carboxylate oxygens, or their equivalent substituents for the dipole, are listed.
Ligands without carboxyl groups were assessed by their equivalent groups: isoxazole nitrogens in THIP and muscimol, phosphate oxygen in 3-APP, and hydroxyl oxygen in 4-AB and tyramine.
GHB does not have an ammonium group. The charge on the hydroxyl oxygen is negative.
NR = no response.
Homology Modeling and Docking
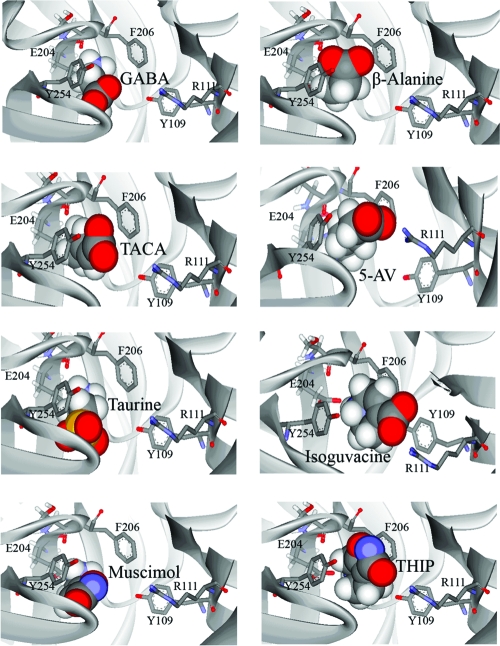
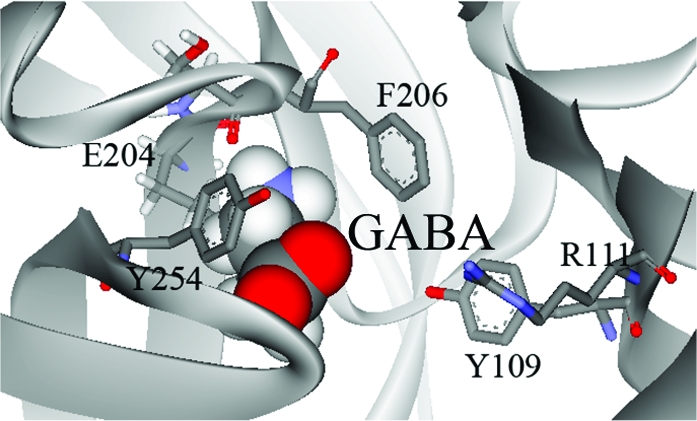
Homology modeling of the extracellular domain of RDL was based on the crystal structure of AChBP, and analogue docking was ranked on the basis of the GOLDscore fitness function, which ranks simulations by comparing interaction energies by consideration of predicted protein−ligand hydrogen bond energy, protein−ligand van der Waals energy, ligand internal van der Waals energy, and ligand torsional strain energy, and has been established as an accurate method for scoring ligand−protein docking (31). The most favored orientation for GABA in the binding site was with the ligand in the cleft among loops B−D (Figure 4). The carboxyl group of GABA was deepest in the cleft, located close to residues Tyr109 and Arg111 in loop D. The ammonium was located between aromatic residues contributed by loop B (Phe206) and loop C (Tyr254), suggesting a cation−π interaction with one or both of these residues. A hydrogen bond was predicted between the ammonium of GABA and the backbone carbonyl of Ser205. 5-AV docked in a similar orientation with a hydrogen bond predicted at the same location.
Figure 4.
Docking of GABA and active analogues (β-alanine, TACA, 5-AV, taurine, isoguvacine, muscimol, and THIP) into the RDL binding site. Analogues docked with the ammonium group located deep in the cleft formed between loop B and loop C aromatic residues. The carboxylate moiety or equivalent substituent was predicted to be located facing toward Arg111.
β-Alanine was found to dock in two almost equally favored orientations. One of these was similar to the orientation of GABA described above, with the carboxyl deep in the cleft and the ammonium sandwiched between Phe206 and Tyr254. In this orientation, there was a hydrogen bond between the ligand ammonium and the Ser205 backbone carbonyl oxygen. In the second orientation, β-alanine was close to Ser131 above loop E, and there was a hydrogen bond between the ligand ammonium and the side chain hydroxyl.
All other agonists docked with the ammonium moiety oriented close to loop B residues Glu204 and Ser205, with hydrogen bonds predicted between these residues and TACA and taurine (Table 3). Muscimol and isoguvacine were predicted to form hydrogen bonds with residues Leu249 and Tyr254, respectively. These orientations also position the carboxylate moieties close to loop D residue Arg111.
Table 3. Hydrogen Bonding Partner Residues Predicted from Docking of Functional GABA Analogues.
| analogue | H-bonding residues predicted | analogue | H-bonding residues predicted |
|---|---|---|---|
| GABA | Ser205 | isoguvacine | Tyr254 |
| β-alanine | Ser205 | TACA | Ser205, Glu204 |
| 5-AV | Ser205 | taurine | Ser205, Glu204 |
| muscimol | Leu249 | THIP | none detected |
Discussion
In this study, we have examined the characteristics of agonist binding in RDL receptors, GABA-gated chloride selective Cys-loop receptors found in insect nervous systems. They are important to understand as insecticidal targets, and knowledge of their structure and function could clarify how these receptors function in other species. Here we have created a homology model of the RDL binding site and demonstrated that GABA and a range of GABA analogues can dock into this site with a range of potential interaction with binding site residues. We have also examined the characteristics of the agonists and, using functional and modeling data, described the critical features required to activate these receptors.
The GABA-gated chloride channels produced when RDL subunits are expressed in Xenopus oocytes have similarities to GABA-activated receptors in vertebrates but have distinct pharmacological characteristics. They are activated by GABA with an EC50 of ∼20 μM, exhibit minimal desensitization, can be inhibited by PTX but not by bicuculline, and can be activated by the GABAA receptor agonist muscimol and the GABAC receptor agonist TACA. Muscimol, which is a full agonist of GABAA receptors and a partial agonist of GABAC receptors, is a full agonist of RDL receptors, while THIP, which is a full agonist of GABAA receptors and an antagonist of GABAC receptors, is a partial agonist of RDL receptors. These characteristics are similar to those in cockroach neurons as previously reported (32,33). Other GABA-gated channels that have been heterologously expressed include those from Musca (MdRdl) and Laodelphax where similar characteristics (e.g., GABA EC50 values) have been observed; some or all of these characteristics have also been observed for GABA-gated chloride channels in lobster, cockroach, and crab neurons and are in contrast to those of GABA-gated cation channels that have been identified in Caenorhabditis elegans, small crab, and Drosophila(34−38).
Our examination of the GABA agonists suggests that GABA activates RDL receptors in the extended conformation, with an optimum length of ∼5 Å. This is similar to the extended conformation of GABA at GABAC receptors, but differs from the partially folded conformation of GABA at GABAA receptors (39, 40). 5-AV, which is one CH2 group longer than GABA, and β-alanine, which is one CH2 group shorter, can also activate the receptors, suggesting that they are both close enough in size to generate essential contacts within the binding site, although the potencies of both these compounds (EC50 ∼ 1 mM) are considerably weaker than that of GABA. Glycine, which is two CH2 groups smaller than GABA, is inactive, as is tyramine, which is considerably larger than GABA. Similarly, a correlation between agonist affinity and agonist length has been previously shown in GABAA receptors (39).
A carboxylate group at one end of the molecule is critical; replacement with a hydroxyl, as in 4-AB, results in an inactive ligand, suggesting the requirement for an anionic group at this point of the ligand. Taurine, which is an analogue of β-alanine with the carboxylate replaced with a sulfonate, can also act as an agonist, although its low potency indicates that sulfonate does not substitute very effectively for carboxylate. Replacement of the carboxylate with a phosphoric acid group, however, as in 3-APP, results in an inactive ligand, demonstrating that the type of acidic group is important. These data are therefore in contrast to GABAC receptor data, where replacement of the carboxylic acid group of GABA with a phosphonic acid, phosphinic acid, or sulfonic group produces potent antagonists (40).
The ammonium group is also critical. GHB, the well-known “date rape” drug, is an analogue of GABA with the ammonium group replaced with a hydroxyl. This compound had no activity at RDL receptors.
The data therefore support the requirement for a charged dipole for receptor activation, suggesting electrostatic interactions with binding site residues are important. Computational ligand calculations reveal a partial negative charge at the carboxylate moiety and a partial positive charge at the ammonium moiety for all active agonists. Thus, since ligands must have a dipole distance close to 5 Å, and both ends of the ligand are thought to interact with the receptor, we can assume that the agonist binding site lies at the cleft between the binding loops with a distance from one another of ∼5 Å. This suggests that loops B−D comprise the part of the binding site with which agonists interact in this receptor, as placement of ligands between the other loops would require a longer ligand. This hypothesis is supported by the model which shows that loops B−D form a clearly defined pocket in which the ligands bind (Figure 1).
Docking of ligands identified several residues with hydrogen bonding potential, in particular in loops B and C, which have been shown to have important contact points for ligands in 5-HT3, nACh, glycine, and GABAC receptors (10,11,18,19). Most of the agonists (GABA, β-alanine, TACA, and taurine) could form a hydrogen bond with Ser205 (loop B), and TACA and taurine could also form H-bonds with Glu204 (also loop B). Muscimol and isoguvacine could form H-bonds with loop C residues Leu249 and Tyr254, respectively.
The ammonium group of all the functional agonists docked between loop B and loop C aromatics (Phe206 and Tyr254, respectively), suggesting a cation−π interaction. All Cys-loop receptors studied to date have been shown to have a cation−π interaction with an aromatic residue in the binding pocket and the ammonium of their natural ligand, although the location of the interacting residue varies from receptor to receptor. Such interactions have mostly been with loop B aromatics (in nACh, 5-HT3, glycine, and GABAC receptors), but interactions with loop C (MOD-1) and loop A (GABAA) aromatics have also been reported (12,41).
In conclusion, we have identified the charge and dipole requirements for agonist recognition in RDL receptors and have identified residues within loop B, loop C, and loop D which could interact with agonists. We have also shown that loop B and/or loop C aromatic residues could contribute to the binding and/or function of the receptor via a cation−π interaction. The data therefore provide a model of the agonist binding site, which can be used for further structure−activity studies and rational drug design.
This work was supported by the Wellcome Trust (S.C.R.L. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science) and MRC UK (I.M. is a holder of an MRC graduate studentship).
Footnotes
Abbreviations: nACh, nicotinic acetylcholine; AChBP, acetylcholine binding protein; GABA, γ-aminobutyric acid; 4-AB, 4-amino-1-butanol; 5-AV, 5-aminovaleric acid; THIP, 4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-ol; GHB, γ-hydroxybutyric acid; 3-APP, 3-aminopropylphosphonic acid; PABA, p-aminobenzoic acid; TACA, 4-amino-2-butenoic acid; LGIC, ligand-gated ion channel.
References
- Ffrench-Constant R. H.; Mortlock D. P.; Shaffer C. D.; MacIntyre R. J.; Roush R. T. (1991) Molecular cloning and transformation of cyclodiene resistance in Drosophila: An invertebrate γ-aminobutyric acid subtype A receptor locus. Proc. Natl. Acad. Sci. U.S.A. 88, 7209–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. J.; Schmitt B.; Hermans-Borgmeyer I.; Gundelfinger E. D.; Betz H.; Darlison M. G. (1994) Sequence of a Drosophila ligand-gated ion-channel polypeptide with an unusual amino-terminal extracellular domain. J. Neurochem. 62, 2480–2483. [DOI] [PubMed] [Google Scholar]
- Henderson J. E.; Soderlund D. M.; Knipple D. C. (1993) Characterization of a putative γ-aminobutyric acid (GABA) receptor β subunit gene from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 193, 474–482. [DOI] [PubMed] [Google Scholar]
- Zhang H. G.; ffrench-Constant R. H.; Jackson M. B. (1994) A unique amino acid of the Drosophila GABA receptor with influence on drug sensitivity by two mechanisms. J. Physiol. 479 (Part 1), 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma T. K.; Smit A. B. (2003) Acetylcholine binding protein (AChBP): A secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Annu. Rev. Biophys. Biomol. Struct. 32, 311–334. [DOI] [PubMed] [Google Scholar]
- Brejc K.; van Dijk W. J.; Klaassen R. V.; Schuurmans M.; van Der Oost J.; Smit A. B.; Sixma T. K. (2001) Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276. [DOI] [PubMed] [Google Scholar]
- Miyazawa A.; Fujiyoshi Y.; Unwin N. (2003) Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955. [DOI] [PubMed] [Google Scholar]
- Unwin N.; Miyazawa A.; Li J.; Fujiyoshi Y. (2002) Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the α subunits. J. Mol. Biol. 319, 1165–1176. [DOI] [PubMed] [Google Scholar]
- Bartos M.; Price K. L.; Lummis S. C.; Bouzat C. (2009) Glutamine 57 at the complementary binding site face is a key determinant of morantel selectivity for α7 nicotinic receptors. J. Biol. Chem. 284, 21478–21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J.; Lochner M.; Lummis S. C. (2008) Loop B is a major structural component of the 5-HT3 receptor. Biophys. J. 95, 5728–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless S. A.; Millen K. S.; Hanek A. P.; Lynch J. W.; Lester H. A.; Lummis S. C.; Dougherty D. A. (2008) A cation-π interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J. Neurosci. 28, 10937–10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett C. L.; Hanek A. P.; Lester H. A.; Dougherty D. A.; Lummis S. C. (2007) Unnatural amino acid mutagenesis of the GABAA receptor binding site residues reveals a novel cation-π interaction between GABA and β2Tyr97. J. Neurosci. 27, 886–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida J. E. (2009) Pest toxicology: The primary mechanisms of pesticide action. Chem. Res. Toxicol. 22, 609–619. [DOI] [PubMed] [Google Scholar]
- Casida J. E. (1993) Insecticide action at the GABA-gated chloride channel: Recognition, progress, and prospects. Arch. Insect Biochem. Physiol. 22, 13–23. [DOI] [PubMed] [Google Scholar]
- Pless S. A.; Dibas M. I.; Lester H. A.; Lynch J. W. (2007) Conformational variability of the glycine receptor M2 domain in response to activation by different agonists. J. Biol. Chem. 282, 36057–36067. [DOI] [PubMed] [Google Scholar]
- Harrison N. J.; Lummis S. C. (2006) Locating the carboxylate group of GABA in the homomeric ρ GABA(A) receptor ligand-binding pocket. J. Biol. Chem. 281, 24455–24461. [DOI] [PubMed] [Google Scholar]
- Beene D. L.; Price K. L.; Lester H. A.; Dougherty D. A.; Lummis S. C. (2004) Tyrosine residues that control binding and gating in the 5-hydroxytryptamine 3 receptor revealed by unnatural amino acid mutagenesis. J. Neurosci. 24, 9097–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beene D. L.; Brandt G. S.; Zhong W.; Zacharias N. M.; Lester H. A.; Dougherty D. A. (2002) Cation-π interactions in ligand recognition by serotonergic (5-HT3A) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry 41, 10262–10269. [DOI] [PubMed] [Google Scholar]
- Lummis S. C.; L Beene D.; Harrison N. J.; Lester H. A.; Dougherty D. A. (2005) A cation-π binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem. Biol. 12, 993–997. [DOI] [PubMed] [Google Scholar]
- Wagner D. A.; Czajkowski C.; Jones M. V. (2004) An arginine involved in GABA binding and unbinding but not gating of the GABAA receptor. J. Neurosci. 24, 2733–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin A. L. (1992) Maintenance of Xenopus laevis and oocyte injection. Methods Enzymol. 207, 266–279. [DOI] [PubMed] [Google Scholar]
- Millar N. S.; Buckingham S. D.; Sattelle D. B. (1994) Stable expression of a functional homo-oligomeric Drosophila GABA receptor in a Drosophila cell line. Proc. Biol. Sci. 258, 307–314. [DOI] [PubMed] [Google Scholar]
- Shi J.; Blundell T. L.; Mizuguchi K. (2001) FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257. [DOI] [PubMed] [Google Scholar]
- Sali A.; Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Lovell S. C.; Davis I. W.; Arendall W. B. III; de Bakker P. I.; Word J. M.; Prisant M. G.; Richardson J. S.; Richardson D. C. (2003) Structure validation by Cα geometry: φ,φ and Cβ deviation. Proteins 50, 437–450. [DOI] [PubMed] [Google Scholar]
- Hosie A. M.; Sattelle D. B. (1996) Agonist pharmacology of two Drosophila GABA receptor splice variants. Br. J. Pharmacol. 119, 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie A. M.; Sattelle D. B. (1996) Allosteric modulation of an expressed homo-oligomeric GABA-gated chloride channel of Drosophila melanogaster. Br. J. Pharmacol. 117, 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie A. M.; Buckingham S. D.; Presnail J. K.; Sattelle D. B. (2001) Alternative splicing of a Drosophila GABA receptor subunit gene identifies determinants of agonist potency. Neuroscience 102, 709–714. [DOI] [PubMed] [Google Scholar]
- McGurk K. A.; Pistis M.; Belelli D.; Hope A. G.; Lambert J. J. (1998) The effect of a transmembrane amino acid on etomidate sensitivity of an invertebrate GABA receptor. Br. J. Pharmacol. 124, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belelli D.; Callachan H.; Hill-Venning C.; Peters J. A.; Lambert J. J. (1996) Interaction of positive allosteric modulators with human and Drosophila recombinant GABA receptors expressed in Xenopus laevis oocytes. Br. J. Pharmacol. 118, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk M. L.; Cole J. C.; Hartshorn M. J.; Murray C. W.; Taylor R. D. (2003) Improved protein-ligand docking using GOLD. Proteins 52, 609–623. [DOI] [PubMed] [Google Scholar]
- Sattelle D. B.; Pinnock R. D.; Wafford K. A.; David J. A. (1988) GABA receptors on the cell-body membrane of an identified insect motor neuron. Proc. R. Soc. London, Ser. B 232, 443–456. [DOI] [PubMed] [Google Scholar]
- Schnee M.; Rauh J.; Buckingham S. D.; Sattelle D. B. (1997) Pharmacology of skeletal muscle GABA-gated chloride channels in the cockroach Periplaneta americana. J. Exp. Biol. 200, 2947–2955. [DOI] [PubMed] [Google Scholar]
- Eguchi Y.; Ihara M.; Ochi E.; Shibata Y.; Matsuda K.; Fushiki S.; Sugama H.; Hamasaki Y.; Niwa H.; Wada M.; Ozoe F.; Ozoe Y. (2006) Functional characterization of Musca glutamate- and GABA-gated chloride channels expressed independently and coexpressed in Xenopus oocytes. Insect Mol. Biol. 15, 773–783. [DOI] [PubMed] [Google Scholar]
- Duan S.; Cooke I. M. (2000) Glutamate and GABA activate different receptors and Cl− conductances in crab peptide-secretory neurons. J. Neurophysiol. 83, 31–37. [DOI] [PubMed] [Google Scholar]
- Beg A. A.; Jorgensen E. M. (2003) EXP-1 is an excitatory GABA-gated cation channel. Nat. Neurosci. 6, 1145–1152. [DOI] [PubMed] [Google Scholar]
- Swensen A. M.; Golowasch J.; Christie A. E.; Coleman M. J.; Nusbaum M. P.; Marder E. (2000) GABA and responses to GABA in the stomatogastric ganglion of the crab Cancer borealis. J. Exp. Biol. 203, 2075–2092. [DOI] [PubMed] [Google Scholar]
- Gisselmann G.; Plonka J.; Pusch H.; Hatt H. (2004) Drosophila melanogaster GRD and LCCH3 subunits form heteromultimeric GABA-gated cation channels. Br. J. Pharmacol. 142, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. V.; Sahara Y.; Dzubay J. A.; Westbrook G. L. (1998) Defining affinity with the GABAA receptor. J. Neurosci. 18, 8590–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward R. M.; Polenzani L.; Miledi R. (1993) Characterization of bicuculline/baclofen-insensitive (ρ-like) γ-aminobutyric acid receptors expressed in Xenopus oocytes. II. Pharmacology of γ-aminobutyric acidA and γ-aminobutyric acidB receptor agonists and antagonists. Mol. Pharmacol. 43, 609–625. [PubMed] [Google Scholar]
- Mu T. W.; Lester H. A.; Dougherty D. A. (2003) Different binding orientations for the same agonist at homologous receptors: A lock and key or a simple wedge?. J. Am. Chem. Soc. 125, 6850–6851. [DOI] [PubMed] [Google Scholar]