Abstract
Deletion of the Saccharomyces gene, UTH1, a founding member of the SUN family of fungal genes, has pleiotropic effects. Several phenotypes of Δuth1 cells including their decreased levels of mitochondrial proteins, their impaired autophagic degradation of mitochondria, and their increased viability in the presence of mammalian BAX, a pro-apoptotic regulator localized to the mitochondria, have prompted others to propose that the Uth1p functions primarily at the mitochondria. In this report, we show that cells lacking UTH1 have more robust cell walls with higher levels of β-d-glucan that allows them to grow in the presence of calcofluor white (CFW) or sodium dodecyl sulfate (SDS), two reagents known to perturb the yeast cell wall. Moreover, these Δuth1 cells are also significantly more resistant to spheroplast formation induced by zymolyase treatment than their wildtype counterparts. Surprisingly, our data suggests that several of the enhanced growth phenotypes of Δuth1 cells, including their resistance to BAX-mediated toxicity, arise from a strengthened cell wall. Therefore, we propose that Uth1p's role at the cell wall and not at the mitochondria may better explain many of its effects on yeast physiology and programmed cell death.
Keywords: UTH1, PKC1, SUN genes, cell wall biogenesis, β-d-glucan, chitin
Introduction
A founding member of the SUN (SIM1, UTH1, and NCA3) family of fungal genes, UTH1 was originally identified in a genetic screen for S. cerevisiae mutants that increased the stress resistance and the replicative lifespan of yeast cells (Kennedy et al., 1995; Austriaco, 1996). Mutant cells lacking UTH1 have longer replicative lifespans (Kennedy et al., 1995; Austriaco, 1996); are capable of growing at elevated temperatures (Austriaco, 1996; Camougrand et al., 2003); are resistant to hydrogen peroxide (Austriaco, 1996; Bandara et al., 1998) and rapamycin (Camougrand et al., 2003); and are sensitive to copper (Jo et al., 2008) and to paraquat (Austriaco, 1996; Bandara et al., 1998). Finally, Δuth1 cells are also able to grow in the presence of overexpressed pro-apoptotic mammalian BAX suggesting that UTH1 may be involved in the regulation of yeast programmed cell death (Camougrand et al., 2003).
What does Uth1p do? Several lines of evidence have prompted Camougrand and her colleagues to propose that the Uth1p protein functions primarily at the mitochondria. First, they showed that the inactivation of UTH1 lowers the levels of mitochondrial proteins including cytochrome aa3, c, and b, and citrate synthase (Camougrand et al., 2000). Next, they localized Uth1p to the outer mitochondrial membrane and to the cell wall (Velours et al., 2002). Third, as we have already noted above, they observed that deleting UTH1 allows yeast cells to survive the overexpression of mammalian BAX, a pro-apoptotic protein known to act at yeast mitochondria (Camougrand et al., 2003). Finally, they discovered that the autophagic degradation of mitochondria is impaired in Δuth1 cells (Kissova et al., 2004; Kissova et al., 2007). In light of this data, Camougrand et al. have suggested that Uth1p is a regulator of mitochondrial function and that this role may explain its diverse effects on yeast apoptosis and cell physiology (Camougrand et al., 2004).
In this paper, we report that UTH1 is involved in cell wall biogenesis: Cells lacking UTH1 have more robust cell walls that are relatively more resistant to enzymatic attack by zymolyase than their wildtype counterparts, probably because they contain higher levels of β-d-glucan. Surprisingly, our data also suggests that several of the enhanced growth phenotypes of Δuth1 cells, including their resistance to BAX-mediated toxicity, arise from a strengthened cell wall. Therefore, we propose that Uth1p's role at the cell wall and not at the mitochondria may better explain many of its effects on yeast physiology and programmed cell death.
Materials and Methods
Yeast Strains, Plasmids, and Growth Conditions
All experiments were done with isogenic strains in the W303-1A background (MATa ade2, his3, leu2, trp1, ura3, ssd1-d2). The Δuth1 mutant was created by disrupting the ORF with the kanR marker using a PCR-based knock-out strategy (Brachmann et al., 1998) and verified both by PCR and by phenotypic analysis. To overexpress either human BAX or yeast PKC1 in our strains, we transformed either plasmid pCM189/Bax (Camougrand et al., 2003) or plasmid pFR22 (Roelants et al., 2004) into our cells and plated them on selective media. Doxycycline supplementation was used to regulate BAX expression as previously described (Camougrand et al., 2003). For all the experiments described in this paper, cells were cultured using standard protocols (Amberg et al., 2005), and transformations were accomplished using the lithium acetate method (Gietz and Schiestl, 2007). Unless noted otherwise, all drugs were purchased from SIGMA-Aldrich.
Spot Assays
Cells were grown overnight in either rich or selective media at 30°C and then diluted to a final concentration (an approximate OD660 value of 0.2). For each strain, a series of 10-fold dilutions was then prepared in water over a range of concentrations from 10-1 to 10-5 relative to the initial culture. Spots of 5 μl from each dilution series were then plated on the indicated media and cultured at either 30°C or 39°C for 2, 3 or 5 days, depending upon the particular plate. Plates supplemented with drugs were poured and used on the same day. All spot assays were repeated at least three times and a representative experiment is shown.
Spheroplast Rate Assay
Enzyme preparation and cell wall lysis assay were based on the method described previously (Ovalle et al., 1998). Briefly, cells were grown overnight in rich media at 30°C, harvested and washed three times with deionized water, and resuspended to an OD660 of 0.5 in TE buffer, pH 7.5 (50 mM Tris/HCl, 5 mM EDTA). Zymolyase (5 U/μl; Zymo Research, Orange, CA) was then added to the cells to a final concentration of 12 μg/ml. Cell suspensions were incubated at 23°C and their optical density was recorded at four-minute intervals for the indicated time period.
Analysis of Cell Wall Sugar Composition
Chitin levels were quantified according to a method described previously (Lesage et al., 2005). Yeast cultures were first grown to stationary phase in liquid YPD medium then diluted 1:100 in fresh YPD and incubated at 30°C with shaking for 18-22 h. Typically, one ml of culture was spun in a microfuge tube at 14K for 2 min and the media removed. The cell pellets were then air dried at 37°C for 2-3 days. Next, cell pellets were resuspended in 1 ml 6% KOH and heated at 80°C for 90 min with occasional mixing. Alkaline insoluble material was pelleted (20,000 × g, 20 min) and neutralized with phosphate-buffered saline for 10–20 min with occasional mixing. After centrifugation (20,000 × g, 20 min), 200 μl of McIlvaine's Buffer (0.2 M Na2HPO4/0.1 M citric acid, pH 6.0) was added to pellets. Extracts were then stored at -20°C until processed for chitin measurements. Samples were thawed and subjected to digestion with 5 μl of 5 mg/ml chitinase from Trichoderma viride (SIGMA-Aldrich) at 37°C for 36–40 h and then for 20–24 h. The amounts of chitin were then determined by using a modified Morgan-Elson procedure as described previously (Bulik et al., 2003). The levels of chitin, expressed as GlcNAc concentration, were then normalized to the dry weight of the sample.
Next, β-d-glucan levels were quantified according to the methods of Boone et al. (1990) and Yiannikouris et al (2004). Yeast cells were grown as 10 ml cultures of YPD until stationary phase. Cells were harvested, washed once with distilled water, and then extracted three times with 0.5 ml of 3% NaOH at 75 °C (1 h per extraction). After alkali extraction, the alkali-insoluble material was washed once with 1 ml of 100 mM Tris-HCl (pH 7.5), and once with 1 ml of 10 mM Tris-HCl (pH 7.5). The washed residue was then digested for 16 h at 37 °C with 1 mg of Zymolyase 100T (United States Biological, Swampscott, MA), in 1 ml of 10 mM Tris-HCl (pH 7.5). The insoluble pellet that remains after zymolyase digestion was removed by centrifugation, and the supernatant was dialyzed against distilled water, using Spectra/por tubing with a 6,000-8,000-D pore size (Spectrum Laboratories, Rancho Dominguez, CA), for 16 h. Carbohydrate content prior to dialysis [(1, 3) plus (1, 6) β–glucan] and post dialysis [(1, 6) β–glucan alone] was measured as hexose by the phenol-sulfuric acid method (Dubois et al., 1956). The levels of β-d-glucan, were then normalized to the dry weight of the sample. Finally, supernatants obtained from the alkali extraction containing alkali soluble β–glucans and mannoproteins from yeast cell walls were dialyzed (1:100, v/v) with 0.02 M Tris-HCl buffer (pH 7.4) for at least 16 h at 4 °C with 0.02 M Tris-HCl buffer (ph 7.4). Mannans and β–glucans were separated on a concanavalin A Sepharose column (Pharmacia) at 4°C as previously described (Yiannikouris et al., 2004). Alkali-soluble β–glucans were eluted with 0.02 M Tris-HCl buffer (pH 7.4)/ 0.5 M NaCl and stored at -20 °C until carbohydrate content was measured as above, using the phenol-sulfuric acid method.
Results and Discussion
Deleting UTH1 has pleiotropic effects including phenotypes associated with the endoplasmic reticulum
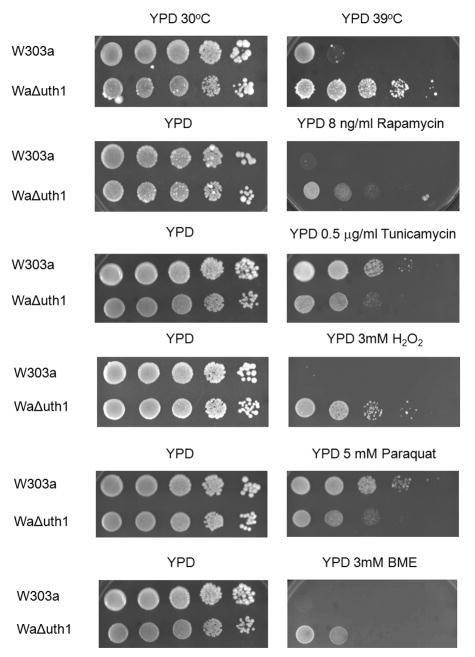
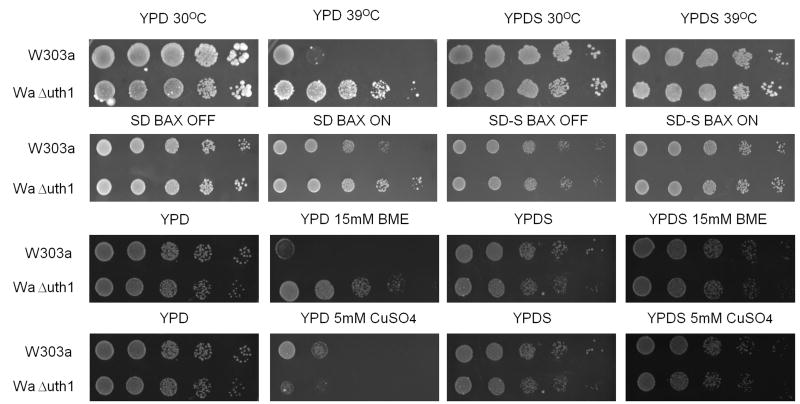
Deletion of the gene, UTH1, has pleiotropic effects. As we and others have previously shown, mutant cells lacking UTH1 are capable of growing at elevated temperatures; are resistant to hydrogen peroxide; and are sensitive to copper and to paraquat (Figure 1). In addition, we have also discovered that they are resistant to β-mercaptoethanol (Figure 1) and to dithiothreitol (data not shown), drugs known to induce ER stress (Cox and Walter, 1996). All of these phenotypes suggest that UTH1 function involves numerous physiological processes in the cell.
Figure 1.
UTH1 is involved in the yeast cell's response to different stresses. 5 μl aliquots of 10-fold serial dilutions of wildtype and Δuth1 mutant cells in the W303-1A strain background were plated onto the designated media and cultured at either 30°C or 39°C for 2, 3 or 5 days, depending upon the particular plate. All spot assays were repeated at least three times and a representative experiment is shown.
UTH1 is involved in maintaining the integrity of the yeast cell wall
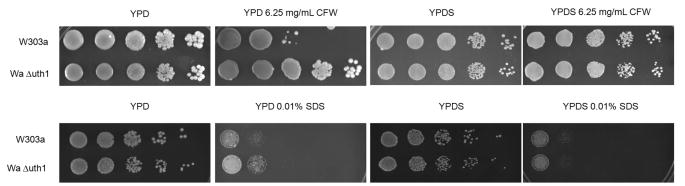
UTH1 is a founding member of the SUN family of fungal genes (Austriaco, 1996). Four other fungal SUN genes, SUN4 in S. cerevisiae (Mouassite et al., 2000), psu1 in S. pombe (Omi et al., 2005), and SUN41 and SIM1/SUN42 in C. albicans (Hiller et al., 2007; Firon et al., 2007; Sosinska et al., 2008), have been implicated in the regulation of the integrity of the yeast cell wall. To test if UTH1 has a similar function, we plated wildtype and Δuth1 cells on media containing either calcofluor white (CFW) or sodium dodecyl sulfate (SDS), two reagents known to perturb the yeast cell wall (Kaeberlein and Guarente, 2002), and observed that the mutant is more resistant to these agents than the wildtype strain (Figure 2A). This suggested that deleting UTH1 strengthens the yeast cell wall.
Figure 2.
UTH1 is involved in maintaining the integrity of the yeast cell wall. (A) Deleting UTH1 enhances cell growth on media supplemented with calcofluor white (CFW) or sodium dodecyl sulfate (SDS). 5 μl aliquots of 10-fold serial dilutions of wildtype and Δuth1 mutant cells in the W303-1A strain background were plated onto the designated media and cultured at 30°C for 2 or 4 days, depending upon the particular plate. YPDS denotes YPD plates supplemented with 1 M sorbitol respectively. (B) Overexpression of PKC1 enhances cell wall integrity mimicking a deletion of UTH1. 5 μl aliquots of 10-fold serial dilutions of wildtype and Δuth1 mutant cells transformed with plasmid pFR22 to overexpress PKC1 were plated onto the designated selective SD plates and cultured at 30°C for 2 or 5 days. SD-S denotes SD plates supplemented with 1 M sorbitol. All spot assays were repeated at least three times and a representative experiment is shown.
To further characterize this phenotype, we plated wildtype and Δuth1 cells on CFW and on SDS plates supplemented with 1 M sorbitol, which provides osmotic stabilization and prevents lysis caused by a weakened cell wall (Kaeberlein and Guarente, 2002). Under these conditions, there is no difference in the growth of our wildtype and mutant strains suggesting that the deletion of UTH1 phenocopies the supplementation of growth media with an osmotic stabilizer (Figure 2A).
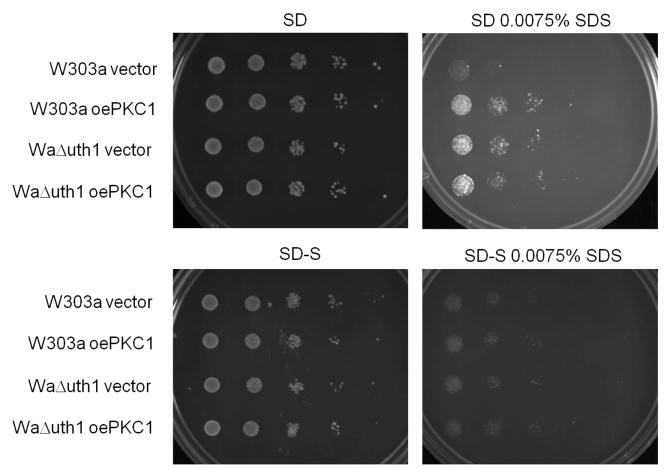
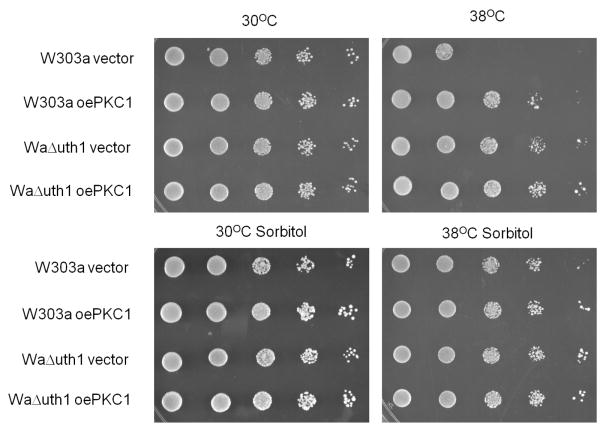
To confirm our findings, we overexpressed PKC1 in wildtype and Δuth1 cells using a high copy plasmid and plated them on SDS plates with or without osmotic stabilization. Pkc1p is a central integrator of cell integrity that acts to promote transcription of cell wall biosynthetic genes (Heinisch et al., 1999). We observed that the growth rates of wildtype cells, wildtype cells overexpressing PKC1, and Δuth1 cells, are indistinguishable on SDS plates supplemented with sorbitol, suggesting that deleting UTH1 and overexpressing PKC1 have the same effect of enhancing cell wall integrity (Figure 2B). Since we see no difference in growth between wildtype cells overexpressing PKC1, wildtype cells on sorbitol, and wildtype cells overexpressing PKC1 on sorbitol, on SDS plates, our data suggests that the phenotypes we observe from either of these manipulations – either the overexpression of PKC1 or the supplementation of media with 1 M sorbitol – can be attributed directly to their enhancement of cell wall integrity rather than to an indirect effect caused either by a non-cell wall-related function of overexpressed PKC1 (Fairn et al., 2007) or by the sorbitol-mediated activation of the osmoregulatory HOG pathway (Saito and Tatebayashi, 2004). Otherwise, we would have seen a synergistic effect on the growth of wildtype cells overexpressing PKC1 on SDS plates supplemented with sorbitol.
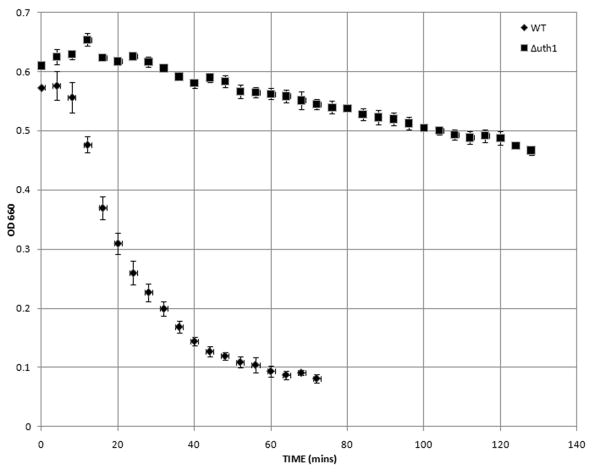
Finally, to more directly assay the structural integrity of the wildtype and Δuth1 yeast cell walls, we compared the rates of formation of spheroplasts of wildtype and mutant Δuth1 yeast cells cultured in a hypotonic solution in the presence of zymolyase, a mixture of cell wall-digesting enzymes (Ovalle et al., 1998). This spheroplast rate assay has been used by others to show that Acb1p (Gaigg et al., 2001), Pho85p (Huang et al., 2002), and Bet1p (Kipnis et al., 2004) are all involved in maintaining the integrity of the yeast cell wall. As shown in Figure 3, mutant cells lacking UTH1 were significantly more resistant to zymolyase treatment than wildtype cells. More specifically, for wildtype cells, the mean lag time (LT) in the presence of zymolyase for three independent samples – where lag time (LT) has been estimated by interpolation of the lysis curve as the time in which the OD660 dropped by 0.05 from its initial value (Ovalle et al., 1998) – was 12 min and the mean maximal lysis rate (MLR) was 0.230 – where MLR has been defined as the absolute value of the slope of the least-squares fit line for the portion of the lysis curve with the steepest log-linear decline (Ovalle et al., 1998). In contrast, for the Δuth1 mutants, the mean lag time (LT) for three independent samples was 18.6 min and the MLR was 0.074. The rate indices (RI) of the wildtype and Δuth1 cells, where RI has been defined as MLR/LT (Ovalle et al., 1998), were 0.0192 and 0.0041 respectively (p<0.002). These results demonstrate that the walls of Δuth1 cells are more resistant to enzymatic attack than those of wildtype controls confirming the findings from the genetic analysis that had suggested that the cell walls of Δuth1 cells are more robust.
Figure 3.
Deleting UTH1 decreases the rate of spheroplast formation of yeast cells. Cells of the indicated genotypes were cultured overnight in rich YPD media at 30°C, harvested and washed three times with deionized water, and then resuspended to an OD660 of 0.5 in TE buffer, pH 7.5 (50 mM Tris/HCl, 5 mM EDTA). Zymolyase (5 U/μl; Zymo Research, Orange, CA) was then added to the cells to a final concentration of 12 μg/ml. Cell suspensions were incubated at 23°C and their optical density (OD660) was recorded at four-minute intervals for the indicated time period. Assays were done in triplicate and mean values are shown along with the standard deviation.
Taken together, both our genetic and biochemical data suggests that UTH1's function parallels the role of four other fungal SUN genes: psu1, which is involved in cell wall synthesis in S. pombe (Omi et al., 2005); SUN4, which is involved in cell septation and which, significantly, appears to act in concert with UTH1 (Mouassite et al., 2000), and SUN41 and SIM1/SUN42, which have been implicated in cell wall remodeling in C. albicans (Firon et al., 2007). In some way, UTH1 is involved in the biogenesis of the cell wall in Saccharomyces cerevisiae.
Deleting UTH1 alters the polysaccharide composition of the yeast cell wall
How does UTH1 regulate the integrity of the yeast cell wall? Firon et al. have shown that UTH1 is able to complement a sun41Δ Δ sun42 Δ double mutant in C. albicans (Firon et al., 2007). Given that SUN41 and SIM1/SUN42 mutants manifest specific cell wall defects at the septa in Candida and that these Candida mutants are sensitive only to cell wall-perturbing agents that are specific to chitin synthesis, these authors have proposed that UTH1 is involved in chitin biosynthesis in S. cerevisiae. To test this hypothesis, we determined the chitin levels in the cell walls of Δuth1 cells and showed that they indeed have decreased amounts of chitin as compared to wildtype controls (Table 1).
Table 1. Chitin and Glucan Composition of the Cell Wall.
Deleting UTH1 alters the polysaccharide content of the yeast cell wall. Chitin and β-d-glucan levels were quantified as described in Materials and Methods. Assays were done in triplicate and mean values are shown along with the standard deviation in brackets. Statistical significance was calculated with the Student's unpaired t-test.
| CHITIN | GLUCAN | |||
| Alkaline Insoluble | Alkaline Soluble | |||
| Total chitin (nmole GlcNAc/mg dry weight) |
Total β-glucan (μg/g dry weight) |
1,6 β–glucan (μg/g dry weight) |
Total β–glucan (μg/g dry weight) |
|
| W303-1A | 21.3 (1.75) | 190 (28) | 51 (8) | 21 (44) |
| W303-1A Δuth1 | 15.9 (1.34) [p<0.002] |
488 (52) [p<0.001] |
90 (22) [p<0.05] |
247 (109) [p<0.05] |
This finding – that Δuth1 cells have lower chitin levels – was unexpected. As shown in Figure 2 and Figure 3, Saccharomyces cells lacking UTH1 are not only more resistant to calcofluor white (CFW) and to sodium dodecyl sulfate (SDS), two reagents that are known to destabilize the cell wall (Kaeberlein and Guarente, 2002), but are also significantly more resistant to zymolyase, a mixture of cell wall-digesting enzymes (Kitamura and Yamamoto, 1972). This would not be expected if deleting UTH1 only lowers the chitin content of the cell wall, an effect expected to weaken and not to strengthen the cell wall. Interestingly, one previous report has shown that mutant cells with increased resistance to CFW have lower chitin levels than their wildtype counterparts (Roncero et al., 1988).
How could lower levels of chitin lead to the strengthening of the yeast cell wall? Some have suggested that a compensatory mechanism exists in yeast in response to cell wall damage whereby decreases in β-d-glucan levels leads to a compensatory increase in chitin levels (Kapteyn et al., 1997; Popolo et al., 1997; Ram et al., 1998; Valdivieso et al., 2000). To explain our data, therefore, we predicted that the reverse mechanism may also exist: The lower levels of chitin in Δuth1 cells may be accompanied by compensatory higher levels of β-d-glucan. To test this hypothesis, we determined the β-d-glucan levels of Δuth1 cells. Indeed, as predicted, these mutant Δuth1 cells had cell walls with significantly higher levels both of total alkaline soluble and total alkaline insoluble β-d-glucan and of alkaline insoluble (1,6) β-d-glucan more specifically (Table 1). This would explain why Δuth1 cells are more resistant to zymolyase, a mixture of cell wall-digesting enzymes composed primarily of β–1, 3–glucan laminaripentaohydrolase and β–1, 3–glucanase (Kitamura and Yamamoto, 1972). The higher levels of β-d-glucan would also explain our genetic data that had suggested that the cell walls of Δuth1 cells are more robust than their wildtype counterparts.
In sum, our data suggests that UTH1 is involved in the biogenesis of the yeast cell wall. The precise molecular mechanism behind this role, however, remains unknown: Uth1p could be involved in any of the regulatory pathways that have been linked to the complex process of cell wall assembly in Saccharomyces cerevisiae (Lesage and Bussey, 2006). It is significant that the Uth1p homolog in Candida albicans, Sim1p, has been identified as a covalently linked cell wall protein by mass spectrometry (Sosinska et al., 2008). Another group has also shown that Uth1p appears to act in concert with another SUN protein, Sun4p, to contribute to cell wall morphogenesis and septation strongly supporting our proposal that the protein is involved in cell wall biogenesis (Mouassite et al., 2000). Finally, it is striking that, like UTH1, many genes involved in the assembly of the yeast cell wall – for example, FKS1 or RHO1 – are pleiotrophic (Lesage and Bussey, 2006).
Deleting UTH1 improves growth under diverse stress conditions by enhancing cell wall integrity
Finally, to determine if UTH1's role in regulating the integrity of the yeast cell wall could explain any of the pleiotrophic phenotypes of the Δuth1 mutant, we took wildtype and Δuth1 cells and stressed them on media with or without sorbitol supplementation. We discovered that there is no difference in growth between these two strains either at 39°C, on media containing either β-mercaptoethanol or copper, or with overexpressed mammalian BAX, as long as they are cultured on media with an osmotic stabilizer (Figure 4A). This suggested that the growth enhancement attributed to the deletion of UTH1 under these conditions could be explained by the gene's role in cell wall biogenesis rather than on mitochondrial function. This was true regardless of whether the BAX cell death assays were done on sorbitol-supplemented media containing either glucose or glycerol as a carbon source (data not shown). In both cases, the viability of wildtype and Δuth1 cells overexpressing mammalian BAX were indistinguishable.
Figure 4.
Deleting UTH1 enhances the cell's response to diverse stresses by strengthening its cell wall. (A) Osmotic stabilization with 1M sorbitol phenocopies a deletion of UTH1. 5 μl aliquots of 10-fold serial dilutions of wildtype and Δuth1 mutant cells were plated onto the designated media and cultured at either 30°C or 39°C for 2, 3 or 5 days. YPDS and SD-S denotes YPD and SD plates supplemented with 1 M sorbitol respectively. To overexpress human BAX in our strains, we transformed plasmid pCM189/Bax into our cells and plated them on selective SD media with or without doxycycline supplementation as previously described (Camougrand et al., 2003). (B) Overexpression of PKC1 enhances cell growth at elevated temperatures mimicking a deletion of UTH1. 5 μl aliquots of 10-fold serial dilutions of wildtype and Δuth1 mutant cells transformed with plasmid pFR22 to overexpress PKC1 were plated onto the designated selective SD plates and cultured at 30°C or 38°C for 2 or 5 days. All spot assays were repeated at least three times and a representative experiment is shown.
To confirm this result, we repeated the assay with wildtype and Δuth1 cells both overexpressing PKC1 on a high copy plasmid and observed that the growth rates of wildtype cells, wildtype cells overexpressing PKC1, and Δuth1 cells, at 38°C are indistinguishable on plates supplemented with sorbitol, suggesting once again that the enhanced growth phenotype of Δuth1 cells at elevated temperatures is linked to the enhancement of cell wall integrity (Figure 4B). As before, the absence of a synergistic effect on the growth at 38°C of wildtype cells overexpressing PKC1 on plates supplemented with sorbitol suggested that the growth enhancement phenotype could be attributed directly to a strengthened cell wall. Finally, parallel results were obtained on copper supplemented plates suggesting that the sensitivity of Δuth1 cells to copper is also mediated by the gene's effect on the integrity of the cell wall (data not shown). It would be interesting to determine if any of the other phenotypes of Δuth1 cells, especially those associated with Uth1p's putative mitochondrial function, can also be linked to its role in regulating the integrity of the yeast cell wall. Finally, in light of our findings it is intriguing to note that one group has been unable to find the reported link between Uth1p function and mitochondrial autophagy (Kanki et al., 2009). However, it is still not clear if this was due to differences in strain background. If so, we speculate that Uth1p's differential effects on mitophagy could be linked to the differences in cell wall composition commonly seen among wildtype yeast strains.
In conclusion, we show that cells lacking UTH1 have more robust cell walls that are resistant to zymolyase treatment because they contain higher levels of β-d-glucan. Surprisingly, our data also suggests that several of the enhanced growth phenotypes of Δuth1 cells, including their resistance to BAX-mediated toxicity, arise from a strengthened cell wall. Thus, we propose that Uth1p's role at the cell wall and not at the mitochondria may better explain many of its effects on yeast physiology and programmed cell death.
Acknowledgments
We thank Mary Blasik, Gina Contuzzi, Kathleen Cornelly, David J. Laprade, Kevin R. Murphy, and Charles Specht (University of Massachusetts School of Medicine, Worchester, MA) for technical assistance; and Brian K. Kennedy (University of Washington, Seattle), Stephen Manon (University of Bordeaux), and Jeremy Thorner (University of California, Berkeley) for strains and/or plasmids. We also thank both Gregory Liszt and Leonard Guarente (M.I.T.) and Charles Specht (University of Massachusetts School of Medicine, Worchester, MA) for helpful discussion. Our laboratory is supported by Rhode Island INBRE Grant P20RR016457 from the National Center for Research Resources (NCRR) of the National Institutes of Health (NIH) and a CAFR grant from Providence College.
Literature Cited
- Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Austriaco N. Ph D Dissertation. Massachusetts Institute of Technology; 1996. UTH1 and the Genetic Control of Aging in the Yeast, Saccharomyces. [Google Scholar]
- Bandara PD, Flattery-O'brien JA, Grant CM, Dawes IW. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr Genet. 1998;34:259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- Boone C, Sommer SS, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Camougrand NM, Mouassite M, Velours GM, Guerin MG. The “SUN” family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;375:154–160. doi: 10.1006/abbi.1999.1655. [DOI] [PubMed] [Google Scholar]
- Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol Microbiol. 2003;47:495–506. doi: 10.1046/j.1365-2958.2003.03311.x. [DOI] [PubMed] [Google Scholar]
- Camougrand N, Kissova I, Velours G, Manon S. Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res. 2004;5:133–140. doi: 10.1016/j.femsyr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcriptional factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric Method for Determination of Sugars and Related Substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Fairn GD, Macdonald K, McMaster CR. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J Biol Chem. 2007;282:4868–4874. doi: 10.1074/jbc.M610575200. [DOI] [PubMed] [Google Scholar]
- Firon A, Aubert S, Iraqui I, Guadagnini S, Goyard S, Prevost MC, Janbon G, d'Enfert C. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol Microbiol. 2007;66:1256–1275. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- Gaigg B, Neergaard TB, Schneiter R, Hansen JK, Faergeman NJ, Jensen NA, Andersen JR, Friis J, Sandhoff R, Schrøder HD, Knudsen J. Depletion of acyl-coenzyme A-binding protein affects sphingolipid synthesis and causes vesicle accumulation and membrane defects in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:1147–1160. doi: 10.1091/mbc.12.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- Heinisch JJ, Lorberg A, Schmitz HP, Jacoby JJ. The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol Microbiol. 1999;32:671–680. doi: 10.1046/j.1365-2958.1999.01375.x. [DOI] [PubMed] [Google Scholar]
- Hiller E, Heine S, Brunner H, Rupp S. Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot Cell. 2007;6:2056–2065. doi: 10.1128/EC.00285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Moffat J, Andrews B. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol Cell Biol. 2002;22:5076–5088. doi: 10.1128/MCB.22.14.5076-5088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Loguinov A, Chang M, Wintz H, et al. Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants. Toxicol Sci. 2008;101:140–151. doi: 10.1093/toxsci/kfm226. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, Ram AF, Groos EM, Kollar R, Montijn RC, Van Den Ende H, Llobell A, Cabib E, Klis FM. Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Khoury CM, Greenwood MT. The pleiotropic effects of heterologous Bax expression in yeast. Biochim Biophys Acta. 2008;1783:1449–1465. doi: 10.1016/j.bbamcr.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Kipnis P, Thomas N, Ovalle R, Lipke PN. The ER-Golgi v-SNARE Bet1p is required for cross-linking alpha-agglutinin to the cell wall in yeast. Microbiology. 2004;150:3219–3228. doi: 10.1099/mic.0.27189-0. [DOI] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yamamoto Y. Purification and properties of an enzyme, zymolyase, which lyses viable yeast cells. Arch Biochem Biophys. 1972;153:403–406. doi: 10.1016/0003-9861(72)90461-4. [DOI] [PubMed] [Google Scholar]
- Lesage G, Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G, Shapiro J, Specht CA, Sdicu AM, et al. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005;6:8. doi: 10.1186/1471-2156-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, et al. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouassite M, Camougrand N, Schwob E, Demaison G, Laclau M, Guerin M. The ‘SUN’ family: Yeast SUN4/SCW3 is involved in cell septation. Yeast. 2000;16:905–919. doi: 10.1002/1097-0061(200007)16:10<905::AID-YEA584>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Omi K, Sonoda H, Nagata K, Sugita K. Cloning and characterization of psu1(+), a new essential fission yeast gene involved in cell wall synthesis. Biochem Biophys Res Commun. 2005;262:368–374. doi: 10.1006/bbrc.1999.1209. [DOI] [PubMed] [Google Scholar]
- Ovalle R, Lim ST, Holder B, Jue CK, Moore CW, Lipke PN. A spheroplast rate assay for determination of cell wall integrity in yeast. Yeast. 1998;14:1159–1166. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1159::AID-YEA317>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AF, Kapteyn JC, Montijn RC, Caro LH, Douwes JE, Baginsky W, Mazur P, van den Ende H, Klis FM. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of beta 1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J Bacteriol. 1998;180:1418–1424. doi: 10.1128/jb.180.6.1418-1424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants FM, Torrance PD, Thorner J. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1, and Sch9. Microbiology. 2004;150:3289–3304. doi: 10.1099/mic.0.27286-0. [DOI] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol. 1988;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Tatebayashi K. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem. 2004;136:267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- Sosinska GJ, de Groot PW, Teixeira de Mattos MJ, Dekker HL, de Koster CG, Hellingwerf KJ, Klis FM. Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology. 2008;154:510–520. doi: 10.1099/mic.0.2007/012617-0. [DOI] [PubMed] [Google Scholar]
- Valdivieso MH, Ferrario L, Vai M, Duran A, Popolo L. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J Bacteriol. 2000;182:4752–4757. doi: 10.1128/jb.182.17.4752-4757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velours G, Boucheron C, Manon S, Camougrand N. Dual cell wall/mitochondria localization of the ‘SUN’ family proteins. FEMS Microbiol Lett. 2002;207:165–172. doi: 10.1111/j.1574-6968.2002.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Wissing S, Ludovico P, Herker E, Buttner S, et al. An AIF orthologue regulates apoptosis in yeast. J Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiannikouris A, Francois J, Poughon L, Dussap C, et al. Alkali Extraction of b-D-Glucans from Saccharomyces cerevisiae Cell Wall and Study of Their Adsorptive Properties toward Zearalenone. J Agric Food Chem. 2004;52:3666–3673. doi: 10.1021/jf035127x. [DOI] [PubMed] [Google Scholar]