Abstract
Often considered benign, meningiomas represent 32% of intracranial tumors with 3 grades of malignancy defined by the WHO histology based classification. Malignant meningiomas are associated with less than 2 years median survival. The inability to predict recurrence and progression of meningiomas induces significant anxiety for patients and limits physicians in implementing prophylactic treatment approaches. This report presents an analytical approach to tissue characterization based on MALDI TOF mass spectrometry imaging (MSI) which is introduced in an attempt to develop a reference database for predictive classification of brain tumors. This pilot study was designed to evaluate the potential of such approach and to begin to address limitations of the current methodology. Five recurrent and progressive meningiomas for which surgical specimens were available from the original and progressed grades were selected and tested against non-progressive high-grade meningiomas, high-grade gliomas, and non-tumor brain specimens. The common profiling approach of data acquisition was compared to imaging and revealed significant benefits in spatially resolved acquisition for improved spectral definition. A preliminary classifier based on support vector machine showed the ability to distinguish meningioma image spectra from non-tumor brain and from gliomas, a different type of brain tumors, and to enable class imaging of surgical tissue. Although the development of classifiers was shown to be sensitive to data preparation parameters such as recalibration and peak picking criteria, it also suggested the potential for maturing into a predictive algorithm if provided with larger series of well-defined cases.
Keywords: Meningioma, neurosurgery, mass spectrometry, classification
INTRODUCTION
Meningiomas account for approximately one-third of all primary brain tumors 1. They develop from the meninges, the tissue covering the brain and spinal cord, and more specifically, are thought to originate from meningothelial cells of the arachnoid membrane, the middle membrane layer of the meninges. The current World Health Organization (WHO) grading system recognizes 15 subtypes of meningiomas based on histological features 2; however, these can be classified into three major grades. According to a 10-year retrospective classification of 314 meningiomas, 78% were benign (grade I), 20% atypical (grade II), and 2% malignant (grade III) 3. Although meningiomas are often considered benign neoplasms, their recurrence rates vary from 7-25% for benign, 29-52% for atypical, and 50-94% for anaplastic 2, the latter being associated with a dismal 1.5 years median survival 4. In an attempt to correlate selected immunohistochemical markers with tumor grade and clinical outcome, results indicated that grade and Topollα index were the only two independent predictive factors of meningioma recurrence 5.
Since prognosis is intimately related to the appropriateness of treatment modality, it is of paramount importance to identify tumors with high sensitivity and accuracy to maximize treatment efficiency. Unfortunately, some brain tumors are heterogeneous and consist of populations of cells with different degrees of tumor initiating potential 6, 7, and different susceptibility to treatment 8, 9. These distinct cell sub-populations of tumors can have different proteomes in response to microenvironment conditions and stimuli, or as a result of further genetic aberration. Because proteins are dynamic cellular effectors, assessment of their relative concentrations provides valuable information on cellular identity, activity, and state. Proteomic approaches in the search for diagnostic biomarkers involve extensive discovery and validation phases before considering usage approval, and the quest for single marker identification is opening the way to multivariate/multiparametric approaches in response to the complexity of human disease. In addition to the inherent complexity of the disease, the source and complexity of the material selected to identify new diagnostic content in the discovery phase determine the potential of isolating disease markers. The choice of affected tissue for characterization over serum increases detection ability by both an increased concentration of markers to be discovered at the disease site in comparison to dilute circulating markers, and sequesters the markers from more abundant constituents of serum which interfere with detection of the less abundant constituents in mass spectrometry 10.
Mass spectrometry (MS) is a well-established analytical technique used to identify and characterize molecules based upon their molecular weight. Matrix Assisted Laser Desorption Ionization (MALDI) is the ionization method of choice for the MS analysis of large and/or labile biomolecules in complex biological samples 11, 12. MALDI Time-of-Flight (TOF) mass spectrometry has developed as a method to evaluate biomolecule expression profiles directly from tissue samples, including brain tumor samples 13, 14. In mass spectrometry imaging (MSI), hundreds of closely spaced spectra are taken in a grid pattern where each spectrum is analogous to an image pixel 15-17. Each pixel contains information on the mass and intensity of hundreds of biomolecules, which can be translated into a spatial map of molecular distribution and abundance 18-21. This map can be co-registered to microscopic images to correlate molecular signatures with specific histological features 14, 22-24. Previous reports of mass spectrometry profiling and imaging of brain tumor tissues have shown differentiation of tumor types, grades, and sub-classes of gliomas from large sample sets, and the ability to predict response to treatment by early detection of drug-induced proteomic changes in a breast cancer mouse model 13, 16, 25.
Here, to further adapt mass spectrometry imaging for clinical applications, and with the ultimate goal to deliver an objective and accurate evaluation of brain tumor specimens at the time of surgery, human brain tumor samples were analyzed and used to initiate a reference database. This report examines a set of recurrent and progressive meningiomas, and presents an approach for building classifiers and addresses potential caveats of data preparation. The clinical concept of mass spectrometry imaging of surgical specimens for prediction of disease progression is illustrated in Scheme 1.
Scheme 1.
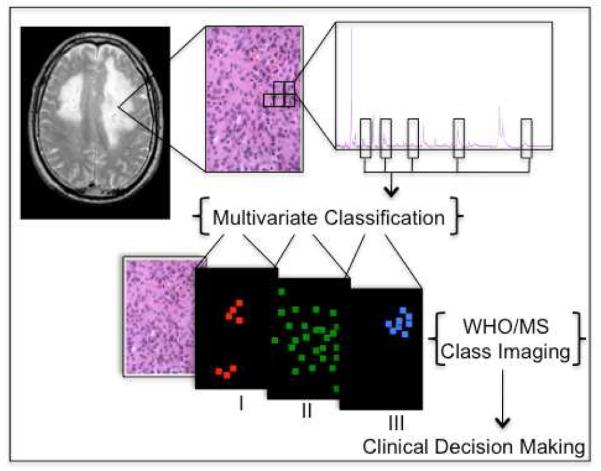
After a tissue biopsy is sectioned and stained, individual mass spectra are obtained uniformly across a sister tissue section surface. Spectra from tissues of different WHO grades are used to perform multivariate classification based on expert histopathology diagnosis. Classification models are then used to grade individual spectra from a tumor specimen, and the molecular images are rendered as class images. From these images, molecular indication of progression could be detected while still not observable by microscopic evaluation, and potentially contribute to clinical decision making.
EXPERIMENTAL SECTION
Specimen Selection
Tumor samples were obtained from the Brain Tumor Bank in the Department of Neurosurgery, Brigham and Women’s Hospital and analyzed under approved Institutional Review Board (IRB) protocol, with informed written consent obtained by licensed neurosurgeons at BWH. Included in this study are eight patients with histologically proven meningiomas, from which five patients progressed over several years. Many of the patients underwent several surgical tumor resections, each time providing a separate tissue sample for a total of 15 unique meningioma specimens. As control sets, six separate patients provided glioma tumor samples and six separate patients provided non-tumor samples (two surgical epilepsy specimens, two post-mortem sporadic amyotrophic lateral sclerosis specimens, and two blood samples from healthy individuals).
Materials
“Protein calibration standard 1” from Bruker Daltonics (Billerica, MA, USA) was used for external calibration of the mass spectrometer. Sinapinic acid was from Sigma. ITO coated coverslips with busbars were 18 × 18 mm with resistivity from 8-12 ohms (thickness 1: 0.13-0.17 mm) from SPI Supplies (West Chester, PA, USA). ImmEdge PEN was from Vector Laboratories, Inc. (Burlingame, CA, USA).
Sample Preparation for MALDI Mass Spectrometry Imaging
Specimens were sectioned using a Microm HM525 cryostat from Mikron Instruments Inc. (San Marcos, CA, USA) at 16 μm thickness. Specimens were prepared by matrix solution fixation 26. Briefly, matrix solution fixation is performed by adding matrix (30 mg/ml final concentration) in a 2:2:1:1 ethanol:methanol:acetonitrile:water 0.2% TFA solution directly to the tissue. 20-40 μl of matrix solution was deposited, and was scaled depending upon the surface area of the sample. After 10 minutes of fixation at −21°C, samples were allowed to dry at room temperature.
Mass Spectrometry Profiling and Imaging
A MALDI-TOF mass spectrometer Microflex interfaced with FlexImaging Software version 2.1 from Bruker Daltonics (Billerica, MA, USA) was operated in linear mode for m/z greater than 3000, and using a custom made coverslip holder 26. Standard instrument parameters were used, although delayed extraction times were optimized to between 500 ns for the mass range of interest, 4000-20000 m/z. The coverslips were held on the target by minimal conductive adhesive tape. Spectral images were acquired at a uniform grid resolution of 100-150 μm using 300 laser shots at each grid position. Each profile spectrum was generated from 5000 laser shots spread randomly over the tissue section and summed to create a single spectrum.
Data Analysis
Data were internally calibrated using FlexAnalysis (Bruker Daltonics). Every spectrum underwent manual internal calibration using a two point linear fit to reference masses for singly and double charged hemoglobin. Normalization and classification were performed using ClinProTools version 2.2 (Bruker Daltonics). The resolution was set at 500 for a mass range of 4000-20000 m/z with a top-hat baseline correction of 10%. Unless otherwise noted, peak calculation was driven by a signal-to-noise ratio of 2.0. Peak areas were calculated with the default zero-level integration.
RESULTS AND DISCUSSION
Specimen Selection and Pathology
The selection of specimens as presented in Figure 1A was designed to perform a pilot study of the mass spectrometry-based classification of human meningiomas, which had clinically progressed. Specifically, specimens were selected from patients who presented for multiple surgeries and with a clear progression from lower to higher grade, thus providing internal controls for comparison. The clinical motivation to perform such a study is to develop a mass spectrometry-based platform to objectively diagnose and potentially provide predictive indicators of tumor recurrence and progression for meningioma patients. Two neuropathologists independently graded the specimens with no inter-observer variability, and the concept of using matching specimens from one patient should minimize variability arising from individuals’ molecular profiles within the series.
Figure 1.
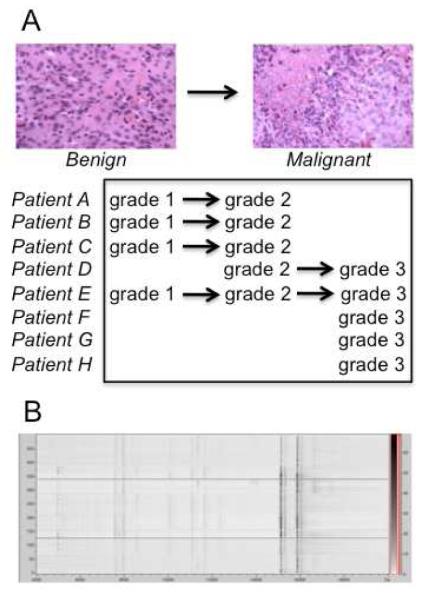
(A) Several cases of meningioma recurred and showed progression in grade; (B) Pseudo-gel view of internally calibrated spectra from MS images used for classification.
Increasing the Resolution of Molecular Profile Mass Spectra with Imaging
This study analyzes and classifies samples at two different levels. First, a single high signal/noise “profile” spectrum representing the entire tissue is generated by signal averaging thousands of spectra acquired across the surface of the tissue, without preserving spatial distribution information. This experiment is analogous to the widely used “molecular profile” experiments performed on various tissue samples using MALDI- and SELDI-TOF platforms 14, 27. Second, a mass spectral image was generated using hundreds to thousands of lower signal/noise spectra (because only ~300 spectra could be acquired at a given location before exhausting signal production) acquired in a grid pattern across the surface of the tissue.
In the course of optimizing instrument parameters, location-dependent peak shifting was observed (e.g. hemoglobin mass measured as 15119 m/z at one position and 15245 m/z at another). Thus, a given peak from a profile spectrum created by averaging spectra from different locations is the convolution of peaks with different maxima. Such profile spectra, generated by randomly collecting spectra from a tissue’s surface were characterized by peak broadening, decreased resolution, and appearance of possible artifact peaks. These problems were overcome by averaging internally calibrated spectra from mass spectral images, which resulted in resolution (FWHH) increases of 1.2 +/− 0.3 fold for evaluated peaks ranging from 5000-15200 m/z. The effect was also more significant (e.g. 1.6) on peaks of higher intensities, whereas low intensity peaks showed a slight increase in resolution from profiling. Such internal calibration using the singly and doubly charged forms of hemoglobin had the added benefit of minimizing data classification errors, since the maximum allowed peak shift for recalibration in ClinProTools (2000 ppm) was significantly less than the experimental variance in molecular mass (~5000 ppm). Thus, spectra that were not internally calibrated resulted in the improper binning of peaks (features) such that the peak that was actually hemoglobin was mistaken for two different molecules during classification, creating artifactual differences between two samples. The pseudo-gel view in Figure 1B shows the resulting quality of alignment from manual recalibration of 500 image spectra.
Classification and Prediction of Progression
The realigned spectra from 15 images were subjected to Principal Component Analysis (PCA) resulting in the apparent unsupervised segregation of each grade in accordance to histopathological grade (Figure 2). Support Vector Machine (SVM) classification models constructed using profile and image spectra suggested to be capable of recognizing image spectra of meningiomas using classifiers built from: 1) a single brain tumor class that compared both glioma and meningioma with non-tumor tissue (including blood); 2) a meningioma brain tumor class and non-tumor brain class; and 3) a meningioma class, a glioma class, and a non-tumor brain class (Figure 3). Including blood specimens in the control group minimizes its contribution to signals derived from surgical tumor specimens, which can be infiltrated and/or retain blood from bleeding in the surgical cavity. Training sets were built using the current gold standard of WHO histopathology classification of tumors of the central nervous system. Although sampling needs to be significantly increased to propose a predictive multivariate indicator of progression, the concept begins to emerge from the results presented in Table 1. The percentage distributions between the 3 grades were more pronounced in samples A through E, while F-H (which did not present with a history of recurrence and progression) showed more segregation to their histological grade.
Figure 2.
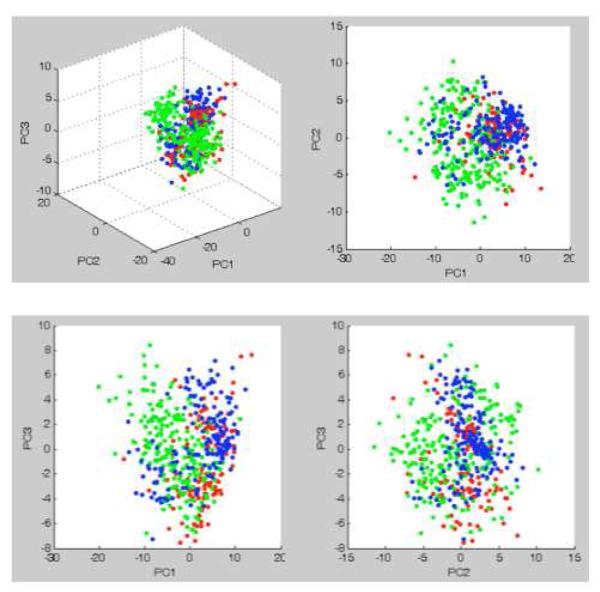
Principal component analysis using peak intensities indicates the potential for automatic grading via cluster analysis: Grade I (red), Grade II (green), Grade III (blue).
Figure 3.
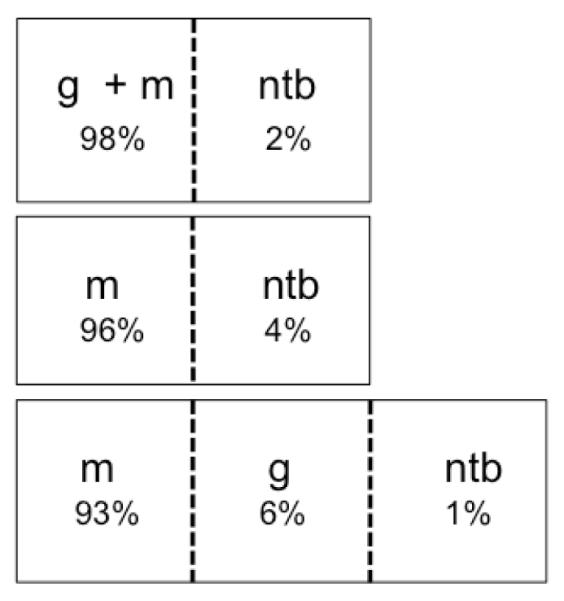
General classification of meningioma images using a support vector machine trained on graded profiles. Combined with a non-tumor training set, several tumor sets were examined: meningiomas combined with gliomas, only meningiomas, meningiomas and gliomas separately. The ability to generally distinguish tumor from non-tumor under each scenario indicates the potential of this approach in a clinical setting. Labels indicate non-tumor brain (ntb), meningioma (m), and glioma (g), and percentages represent the distribution of total meningioma image spectra recognized by a given class.
Table 1.
Classification of imaged biopsies
| WHO Grade | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Histological | I | II | III | |||||||
| MSI | I | II | III | I | II | III | I | II | III | |
| % spectra | ||||||||||
| Patients | A | 83 | 17 | - | 26 | 9 | 65 | |||
| B | 42 | 50 | 8 | 43 | 29 | 28 | ||||
| B* | - | 30 | 70 | |||||||
| C | - | 50 | 50 | 10 | 18 | 72 | ||||
| D | 12 | 18 | 70 | 2 | 37 | 58 | ||||
| E | - | 10 | 90 | - | 4 | 92 | 2 | - | 98 | |
| F | - | - | 100 | |||||||
| G | - | - | 100 | |||||||
| H | 15 | 11 | 74 | |||||||
|
|
||||||||||
| Total | 4 | 24 | 72 | 26 | 20 | 54 | 8 | 10 | 82 | |
Imaged biopsies were manually and automatically graded. Each biopsy is reported within the column indicating its histological WHO classification. Within this group, the MSI classifier reports the percent of image classified according to each grade. B* represents a specimen from patient B with non-progressive recurrence.
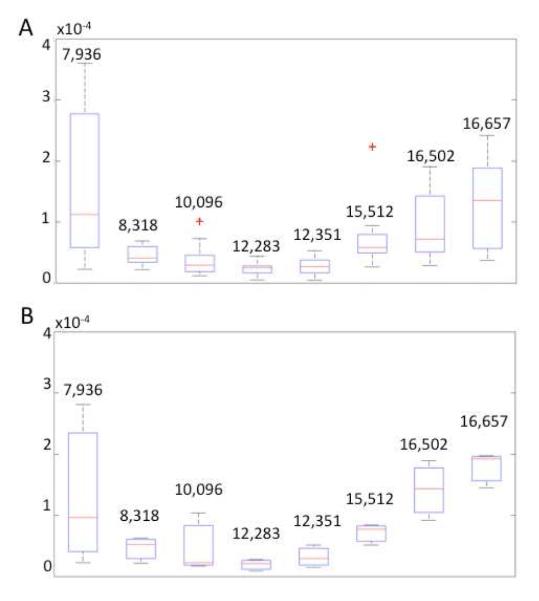
It is also of interest to understand the contribution of inter-individual variance in discriminating between different tumor grades, since the data set does not include all 3 grades imaged for each subject. Towards this goal, the 7 peaks used by the SVM for classification were plotted as a box-and-whisker plot from all grades over all the subjects (Figure 4A), and compared to the equivalent plot for the same 7 classifying peaks for one subject (Figure 4B). The discrimination ability of the peaks is comparable between the two plots, suggesting an inter-grade variance rather than an inter-individual variance. While future increased sampling should increase the robustness of these findings, the box-and-whisker plots indicate a classification dominated by inter-grade variance, with minimal inter-individual variance contribution, justifying further development of the framework for molecular diagnosis. In the identification of a novel serum tumor marker for colorectal cancer using a classical proteomic approach, a group from Roche Diagnostics reported the use of 16-matched colorectal cancer spanning the 4 Dukes stages and adjacent normal tissue samples for the initial discovery phase 10, 28. While it is not possible to retain matched-healthy tissue from the brain, the use of matched recurrence specimens minimizes inter-individual contribution. When combined with control groups constituted of distinct types of brain tumor and independent non-tumor tissues, this approach provides with a valid initial reference frame.
Figure 4.
Box-and-whisker plot statistical representation of peaks used by the SVM classifier. (A) Discrimination ability of the SVM peaks from the complete dataset including all available subjects, for all tumor grades; (B) Box-and-whisker plot of spectra from dataset for one randomly selected subject, for all tumor grades. Discrimination ability of the SVM peaks for a selected subject’s dataset including all tumor grades. The lower and upper quartiles in each box are delineated by the median represented by a red line; the smallest and largest observations are indicated by corresponding whiskers; red crosses outside the whiskers delimited regions report outliers.
This phenomenon and perceived advantage of imaging the specimens in comparison to profiling is represented in Figure 5. This presents the class imaging results of a grade II specimen (selected region of analysis) from subject E, who presented with tumor recurrence and a progression to grade III. The advantages of preserving spatial resolution in the mass spectrometry analysis of surgical specimens include not only higher resolution spectra, but also the ability to detect these spectra with an underlying character of higher grade otherwise not recognizable by standard histopathological evaluation. Taken together with clinical presentation, radiology findings, and standard histopathology evaluation, MSI, once validated with larger sample sets, could contribute to clinical decision-making. Unraveling the grade of a tumor by its molecular characterization could provide access to treatment protocols otherwise not available for a standard histopathology grade, and potentially decrease recurrence probability.
Figure 5.
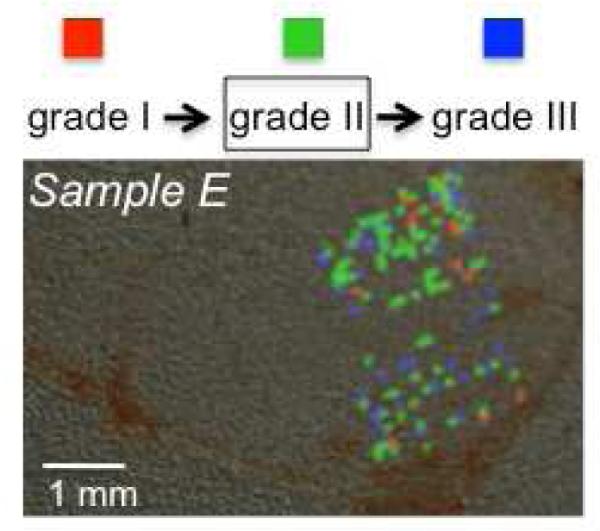
Class imaging of a tissue specimen of histological grade II from a patient with progressive recurrence from grade I to II and to III shows the presence of regions of distinct grades. While many image spectra were discarded as null or noisy, the admissible spectra suggest spatial clustering of the different grade signals. Colored pixels represent Grade I (red), Grade II (green), Grade III (blue) regions.
Although such results are encouraging in developing approaches to assist in the management of neurosurgical diseases, the preparation of the data to build classifiers has a profound effect upon the results of classification and thus one must be very careful in the interpretation of the results. The sensitivity of the overall analysis to data preparation implies that further development of more robust data preprocessing and classification is required. Classification results for the total number of image spectra for each grade is presented in Table 2 for S/N criteria ranging from 1-6, and justify the further development and automation of appropriate data pre-processing applications to streamline the preparation of large numbers of spectra for classification. Further, while the Support Vector Machine technique seemed to work well for our purposes, a Neural Network approach for the data classification will be tested as well as the relevance vector machine (RVM) technique. The RVM classification method is a Bayesian extension of SVM, which achieves comparable performance to SVM while providing posterior probabilities for class memberships and a sparser model. Clustering may also be improved using a kernel PCA methodology in which one does not need to assume linear models.
Table 2.
Effect of peak selection signal to noise ratio (S/N) on classification
| I | II | III | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S/N | Peaks | I | II | III | I | II | III | I | II | III |
| % spectra | ||||||||||
| 1 | 6 | 2 | 37 | 60 | 19 | 23 | 58 | 6 | 15 | 79 |
| 2 | 5 | 4 | 23 | 73 | 27 | 20 | 53 | 8 | 10 | 82 |
| 4 | 8 | 3 | 13 | 84 | 25 | 7 | 68 | 9 | 8 | 83 |
| 6 | 2 | 2 | 2 | 95 | 12 | 12 | 76 | 6 | 6 | 89 |
Classification is sensitive to the signal-to-noise ratio used in selecting peaks. The experiment in Table 1 was repeated under various ratios to illustrate the effect on number of peaks used in the classifier and subsequent totaled classification represented in percentages of total image spectra classified within each MSI class.
CONCLUSIONS
A comprehensive molecular diagnosis obtained during surgery would enable physicians to tailor treatment by accurate evaluation of tumor type, grade, and to select the best suited treatment regimen on an individual basis. MALDI MSI is an approach with potential in image-guided therapy, and its incremental validation will be driven by the inclusion of large and diversified arrays of tumor specimens. Moreover, the diversity of a well-documented database including detailed clinical information will later allow more refined predictive models for personalized cancer management.
ACKNOWLEDGMENT
We thank Dr. Jennifer Chan for pathology assessment. We thank Bruker Daltonics and its employees for continued collaboration. We thank Jane-Marie Kowalski and Paul Kowalski for their continued technical assistance. This research was funded by grants from the Brain Science Foundation and the American Brain Tumor Association (ABTA), and support from the Daniel E. Ponton Fund for the Neurosciences to N.Y.R.A., and supported in part by an award from the DOD (contract W81XWH-04-0158) to J.N.A. This work was also supported in part by NIH Grants (NAC P41 RR-13218) and (NAMIC U54 EB005149) through Brigham and Women’s Hospital.
REFERENCES
- (1).CBTRUS 2008.
- (2).Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Willis J, Smith C, Ironside JW, Erridge S, Whittle IR, Everington D. Neuropathol Appl Neurobiol. 2005;31:141–149. doi: 10.1111/j.1365-2990.2004.00621.x. [DOI] [PubMed] [Google Scholar]
- (4).Perry A, Scheithauer BW, Stafford SL, Lohse CM, Wollan PC. Cancer. 1999;85:2046–2056. doi: 10.1002/(sici)1097-0142(19990501)85:9<2046::aid-cncr23>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- (5).Korshunov A, Shishkina L, Golanov A. Int J Cancer. 2003;104:728–734. doi: 10.1002/ijc.11013. [DOI] [PubMed] [Google Scholar]
- (6).Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- (7).Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- (8).Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- (9).Bao S, Wu Q, Sathornsumetee S, Hao Y, Li Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD, Rich JN. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- (10).Zolg W. Mol Cell Proteomics. 2006;5:1720–1726. doi: 10.1074/mcp.R600001-MCP200. [DOI] [PubMed] [Google Scholar]
- (11).Spengler B, Kaufmann R. Analusis. 1992;20:91–101. [Google Scholar]
- (12).Karas M, Bachmann D, Bahr U, Hillenkamp F. Int. J. Mass. Spectrom. Ion Processes. 1987;78:53–68. [Google Scholar]
- (13).Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Clin Cancer Res. 2004;10:981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- (14).Chaurand P, Sanders ME, Jensen RA, Caprioli RM. Am J Pathol. 2004;165:1057–1068. doi: 10.1016/S0002-9440(10)63367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Nat Med. 2001;7:493–496. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- (16).Schwartz SA, Weil RJ, Thompson RC, Shyr Y, Moore JH, Toms SA, Johnson MD, Caprioli RM. Cancer Res. 2005;65:7674–7681. doi: 10.1158/0008-5472.CAN-04-3016. [DOI] [PubMed] [Google Scholar]
- (17).Seeley EH, Caprioli RM. Proc Natl Acad Sci U S A. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gusev AI, Vasseur OJ, Proctor A, Sharkey AG, Hercules DM. Analytical Chemistry. 1995;67:4565–4570. [Google Scholar]
- (19).Castaing R, Slodzian GJ. Microscopie. 1962;1:395–410. [Google Scholar]
- (20).Stoeckli M, Farmer TB, Caprioli RM. J Am Soc Mass Spectrom. 1999;10:67–71. doi: 10.1016/S1044-0305(98)00126-3. [DOI] [PubMed] [Google Scholar]
- (21).Caprioli RM, Farmer TB, Gile J. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- (22).Chaurand P, Schwartz SA, Billheimer D, Xu BJ, Crecelius A, Caprioli RM. Anal Chem. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- (23).Chaurand P, Schwartz SA, Caprioli RM. J Proteome Res. 2004;3:245–252. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- (24).Caldwell RL, Caprioli RM. Mol Cell Proteomics. 2005;4:394–401. doi: 10.1074/mcp.R500006-MCP200. [DOI] [PubMed] [Google Scholar]
- (25).Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL, Caprioli RM. Cancer Res. 2004;64:9093–9100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- (26).Agar NY, Yang HW, Carroll RS, Black PM, Agar JN. Anal Chem. 2007;79:7416–7423. doi: 10.1021/ac071460e. [DOI] [PubMed] [Google Scholar]
- (27).Whelan LC, Power KA, McDowell DT, Kennedy J, Gallagher WM. J Cell Mol Med. 2008;12:1535–1547. doi: 10.1111/j.1582-4934.2008.00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Roessler M, Rollinger W, Palme S, Hagmann ML, Berndt P, Engel AM, Schneidinger B, Pfeffer M, Andres H, Karl J, Bodenmuller H, Ruschoff J, Henkel T, Rohr G, Rossol S, Rosch W, Langen H, Zolg W, Tacke M. Clin Cancer Res. 2005;11:6550–6557. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]