Abstract
Background
TNF inhibitors have revolutionized the treatment of psoriasis vulgaris, as well as psoriatic and rheumatoid arthritis, and Crohns disease. Despite our understanding that these agents block TNF, their complex mechanism of action in disease resolution is still unclear.
Objective
To globally analyze the genomic effects of TNF inhibition in psoriasis patients, and compare genomic profiles of patients who responded or did not respond to treatment.
Methods
In a clinical trial using etanercept TNF inhibitor to treat psoriasis vulgaris (n=15), Affymetrix gene arrays were used to analyze gene profiles in lesional skin at multiple time-points during drug treatment (baseline, and weeks 1, 2, 4 and 12) compared to non-lesional skin. Patients were stratified as responders (n=11) or non-responders (n=4) based on histological disease resolution. Cluster analysis was used to define gene sets that were modulated with similar magnitude and velocity over time.
Results
In responders, 4 clusters of down-regulated genes and 3 clusters of up-regulated genes were identified. Genes down-modulated most rapidly reflected direct inhibition of myeloid lineage immune genes. Up-regulated genes included stable dendritic cell population genes CD1c and CD207 (Langerin). Comparison of responders and non-responders revealed rapid down-modulation of innate IL-1β and IL-8 “sepsis cascade” cytokines in both groups, but only responders down-regulated IL-17 pathway genes to baseline levels.
Conclusion
While both responders and non-responders to etanercept inactivated “sepsis cascade” cytokines, response to etanercept is dependent on inactivation of myeloid dendritic cell genes and inactivation of Th17 immune response.
Capsule Summary
Cutaneous genes regulated during psoriasis treatment by etanercept provide a global view of response in disease tissue. Only responders down-regulated IL-17 pathway genes.
Keywords: TNF, psoriasis, etanercept, gene, Th17, Th1
Introduction
TNF was initially identified in 1976 as the soluble, LPS-dependent serum factor that killed murine fibrosarcoma cells but not normal fibroblasts 1. Later, TNF was characterized as a macrophage-derived product that rapidly induced expression of IL-1, IL-6, and IL-8, leading to recruitment of innate inflammatory leukocytes within hours of an infectious insult 2, 3. This cytokine sequence, which can be termed the “sepsis cascade model,” 4 has largely dominated thinking about the function of TNF in chronic inflammatory diseases. The initial rationale for testing TNF antagonists in rheumatoid arthritis was based on demonstrated increases in all sepsis cascade cytokines in synovial fluid of affected joints, as well as abundant neutrophils producing collagen-destroying matrix metalloproteases 5. Indeed, genomic analysis of peripheral blood during anti-TNF treatment for rheumatoid arthritis showed a down-modulation of several of these acute-response genes during response (CCL4, IL-8, IL-1β) 6.
Extension of these findings led to clinical trials of TNF antagonists in inflammatory bowel disease and psoriasis with subsequent therapeutic success and FDA approval 7, 8. There has been a strong temptation to link pathogenesis of human inflammatory diseases, and especially those successfully treated with TNF antagonists such as psoriasis and rheumatoid arthritis, to direct effects of sepsis cascade cytokines on innate immunity. However, despite the success of TNF blocking agents, psoriasis vulgaris has some elements of pathogenesis that are not well explained by sepsis cascade cytokines and neutrophil activation. First, there is a strong T cell component in psoriasis, including reports of T cell clonality 9-11, and there is considerable clinical benefit provided by putative T cell targeted therapeutics 12, 13. Second, psoriasis has been induced in several transplanted skin models without apparent involvement of neutrophils in converted grafts 14. Third, recent success in treating psoriasis with antibodies to the p40 cytokine subunit shared between IL-12 and IL-23 implies a central role for adaptive immunity, specifically Th1 and/or Th17 T cells, in the pathogenesis of this disease 15-19. We sought to understand how TNF is linked to pathogenic actions of T cells in psoriasis.
In the present study we used Affymetrix arrays to study gene expression in psoriasis lesions at several time points during treatment with etanercept. Responding and non-responding patents show equivalent suppression of innate, sepsis cytokines (IL-1β, IL-8), but only responding patients demonstrate strong modulation of downstream genes linked to IL-17 signaling. Hence, mechanisms for response success and failure appear to be distinct in psoriasis versus rheumatoid arthritis. These studies define a new way to judge the extent of overall disease reversal based on the degree to which broad disease-related gene sets are suppressed by a specific treatment, and may enable the design of even more effective and simple anti-psoriatic therapeutics.
Materials and Methods
Material and Methods, and statistical analysis are described in greater detail in Supplemental Materials and Methods (SMM, Online Repository (OR)).
Patient studies and classification
Twenty adult patients with moderate to severe psoriasis were treated with etanercept 50mg s.c. twice weekly for 12 weeks. Clinical trial number NCT00116181. Clinical and histological response of patients in this trial was previously published 20. Gene array was performed on 15 sequential patients.
Immunohistochemistry and immunofluorescence
Frozen skin sections were stained for in a standard manner (SMM, section 1). A one-tailed students t-test was used to compare baseline and week 2 cell counts.
Tissue gene expression by microarray
RNA was extracted from skin biopsies using the RNeasy Mini Kit (Qiagen). For each Affymetrix gene chip, 2μg total RNA was reverse-transcribed, amplified, and labeled 21. Microarray data has been deposited in Geo-Omnibus (GSE 11903).
Statistical Analysis
The analysis was conducted using R software (www.R-project.org) with bioconductor packages (www.bioconductor.org). Briefly, expression measures were obtained using GCRMA, and probe sets with a SD>0.15, and expression greater than 4 in at least 5 samples, were kept for further analysis (SMM, section 2). Statistical methods used in the analyses of data are summarized in OR Table 1.
Differential expression criteria
A mixed-effect linear model was used to model gene expression in order to account for the time-course design of the clinical trial. To define “etanercept-modulated genes”, a moderated F statistic was used to determine if any of the expression values across time differed from lesional baseline at any time-point. An F test was used to determine genes where the time profile was different between responders and non-responders. The p-values of both tests were adjusted for multiple hypothesis using Benjamini-Hochberg method.
Consensus clustering
To find distinct profiles among etanercept-modulated genes, as well as between responders and non-responders, we performed hierarchical average linkage clustering with distance measure equal to 1-Pearsons’s correlation coefficient. The optimal number of discovered clusters was assessed using Consensus Clustering algorithm available in GenePattern software.
RT-PCR
RT-PCR was conducted for IL-1β and IL-8 as previously described 20.
Response profile of cytokine keratinocyte “pathways”
We recently defined sets of keratinocyte genes induced by specific cytokines, termed IL-17, TNF and IFNγ keratinocyte “pathways”, respectively 22. A linear model was used to define the response profile of these pathways during etanercept treatment . Gene Set Enrichment Analysis (GSEA) approach 23 was used to compare the response profile of the keratinocyte pathways in responders and non-responders (OR Figure 1). GSEA is a method to mine correlations of any gene set (eg keratinocyte “pathways”) with a specific phenotype (eg response) rather than single gene approach.
Results
Patient population
RNA was extracted for gene array from skin collected at each biopsy time point (non-lesional baseline, lesional baseline, weeks 1, 2, 4, and 12). Quantifiers of disease resolution included epidermal thickness (a linear measurement in mm), Ki67 (a marker of cell proliferation), and K16 gene expression (indicating aberrant keratinocyte differentiation). Patients were stratified as “responders” (n=11) or “non-responders” (n=4) based on whether or not they achieved histologic disease resolution by week 12 of etanercept treatment (decreased epidermal thickening, normalization of Ki67 and K16) 20.
Disease genes were temporally modulated by etanercept in a “reverse cascade”
Genes with expression values that changed during etanercept treatment (Fold change [FCH] > 2 for any time-point, false discovery rate [FDR] <0.05) were selected as “etanercept-modulated genes”. We identified 978 probe-sets that were down-regulated during the 12 weeks of treatment with etanercept, and 999 probe-sets that were up-regulated (OR Tables 2, 3, respectively). We used consensus clustering to identify 4 clusters that were down-regulated (OR Figure 2a), and 3 clusters that were up-regulated (OR Figure 2b), with distinct kinetics. This gave a cascade of genes that were modulated by etanercept, and as this was in a reverse fashion as inflammation was quenched, thus we termed this modulation by etanercept a “reverse cascade”.
Genes down-modulated by etanercept
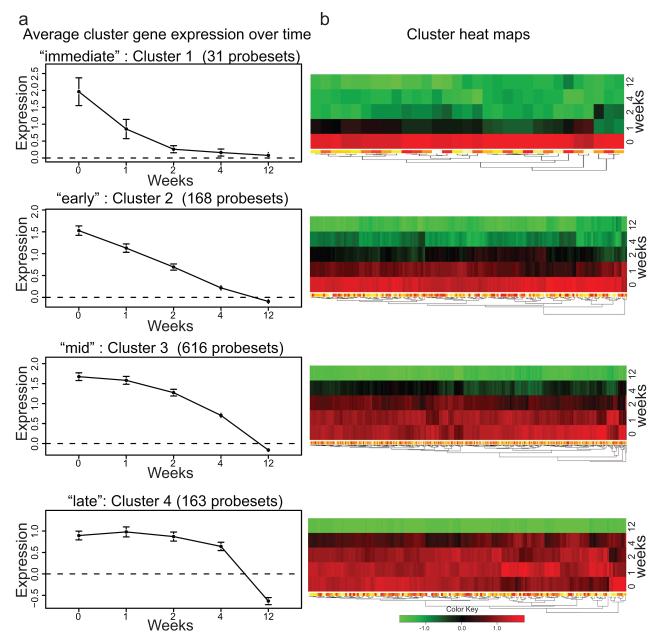
Genes down-modulated by etanercept clustered into 4 groups (Figure 1). Gene expression at each time-point was normalized to non-lesional baseline expression, and average expression of genes within each cluster was plotted over time (Figure 1a) with corresponding heat maps (Figure 1b). The set of genes that were maximally suppressed by 1 week of treatment (Figure 1, cluster 1) are likely to be genes that are directly modulated by TNF. However, with more time, many additional down-regulated genes were identified and these expanded to progressively larger sets of regulated genes over time (Figure 1, clusters 2-4).
Figure 1. Consensus clustering grouped genes down-regulated by etanercept into 4 clusters.
(a) Consensus clustering of genes down-modulated in patients who responded to etanercept treatment (n=11) identified 4 gene clusters that were down-modulated at different time points during the course of etanercept treatment: “immediate”, “early”, “mid”, and “late” clusters. Gene expression in lesional skin at weeks 0, 1, 2, 4 and 12 was normalized to non-lesional gene expression, and shown as average cluster gene expression +/− SEM. (b) Heat map of each down-modulated gene cluster.
Cluster 1 genes (31 probe sets) were down-modulated most rapidly and defined as “immediately” down-modulated genes, cluster 2 with 168 probe sets was “early,” cluster 3 with 616 probe sets was “mid”, and cluster 4 with 163 probe sets was “late”. All down-regulated genes are listed by cluster in OR Table 2. Rapidly down-modulated cluster 1 genes included those involved in leukocyte chemotaxis IL-8, CCL4 (MIP-1β), CCL3 (MIP-1α), FPR1 and plasminogen activator of urokinase receptor (PLAUR). This cluster also contained several genes involved in anti-apoptosis, such as BCL2A1; cell cycle genes AURKA, NCAPG, CDC6, and SPC25; and keratinocyte genes DSC2, SPRR3 and heparin-binding epidermal growth factor-like protein (HBEGF). There were also several genes involved in lipid metabolism, such as LIPG, LDLR, LRP8 and APOBEC3A. Two cytokines included in this cluster were IL-1β and IL-19. IL-1β is an acute-phase cytokine produced by many cell types, particularly monocytes. The regulation of IL-1β by TNF is well appreciated, and hence rapid down-modulation of IL-1β by TNF-inhibition is to be expected. IL-19 is a recently discovered cytokine belonging to the IL-10 family of cytokines, and is produced by monocytes as well as epithelial and endothelial cells during inflammation 24. Although some of these genes are known TNF early response genes, many of them have not been previously identified as TNF early response genes in human skin.
Genes most rapidly down-modulated by etanercept were enriched with myeloid specific genes
In order to determine which leukocyte lineages were most rapidly inhibited by etanercept treatment, we used the expression values of myeloid cells (CD33+ and CD14+), T cells (CD4+ and CD8+), and skin from the Novartis normal tissue compendium 25. OR Figure 3a shows the expression of immediately down-regulated genes (cluster 1) in this data set. Myeloid cells, not T cells, expressed genes that were rapidly down-modulated with etanercept. T cell specific genes were modulated later, in cluster 2 (OR Figure 4), suggesting that TNF inhibition directly modulates myeloid-lineage gene products, which subsequently effect T cells.
There were two particularly interesting myeloid-specific genes contained within the rapidly down-modulated cluster 1 that we confirmed using double label immunofluorescence: HBEGF (OR Figure 3b) and PLAUR (OR Figure 3c). There was a large increase in HBEGF within myeloid CD11c+ dermal DCs (co-expression giving a yellow color) in lesional compared to non-lesional skin. At week 2 of etanercept treatment, HBEGF and CD11c expression were decreased, and by week 12, expression of both markers normalized to non-lesional levels. PLAUR, another down-modulated “immediate” cluster 1 myeloid-specific gene, was also rapidly decreased at the protein level by week 2 of etanercept treatment. PLAUR is a key molecule in the regulation of cell-surface plasminogen activation, and may be involved in inflammation 26.
Resident DC genes were up-regulated during etanercept treatment
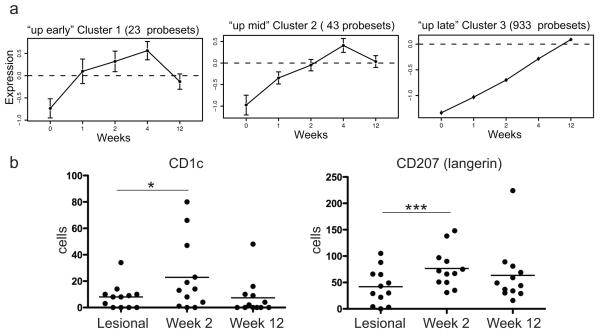
Etanercept up-regulated 999 probe-sets (816 genes) in responding patients, and these were clustered into three groups. The three clusters are shown in Figure 2a (“up early”, “up mid”, and “up late”) and genes in each cluster are listed in OR Table 3. Inspection of genes contained within the most rapidly up-regulated clusters 1 and 2 revealed several dendritic cell (DC) genes: myeloid DC genes CD1c and CD1e were contained within cluster 1, and Langerhans cell genes CD207 (Langerin) and CD1a were both in cluster 2. In normal human skin there are two resident DC populations: dermal DCs that co-express the myeloid lineage marker CD11c and CD1c, and epidermal Langerhans cells (Langerin and CD1a+) 27.Therefore, the genomic up-regulation of CD1c and CD207 could represent a normalization of these stable resident DC populations with treatment. We confirmed this on a cellular level by performing cell counts on sections from biopsies collected at lesional baseline and week 2 of etanercept treatment (Figure 2b). CD1c+ resident myeloid DCs were increased by approximately three-fold (mean 7 cells at baseline and 23 cells at week 2; p=0.03), and CD207+ cells were increased approximately two-fold (mean 39 and 72 cells, respectively; p=0.001). However, week 12 counts of CD1c and CD207+ cells (mean 8 and 64 cells, respectively) were not significantly different from baseline lesional skin. In contrast, there was a large influx of inflammatory CD11c+ cells that do not co-express CD1c in psoriasis lesions (we have termed “inflammatory” myeloid DCs) 28, and these cells were decreased rapidly with etanercept treatment by week 2 20. Therefore, although etanercept decreases inflammatory myeloid DCs after two weeks of treatment, stable resident myeloid DCs and Langerhans cells were temporarily increased.
Figure 2. Consensus clustering grouped genes up-regulated by etanercept into 3 clusters.
(a) Consensus clustering of genes up-regulated in patients who responded to etanercept treatment (n=11) identified 3 clusters, “up early”, up mid”, and up late”. (b) Cell counts for CD1c and CD207 (Langerin) for psoriasis lesional skin, week 2 and at wk 12. Mean cell counts indicated. *p<0.05, ***p<0.001.
Cluster 3 was the largest group of up-regulated genes containing 933 probe-sets. The genes in this group include cutaneous structural proteins such as laminin (α4 and β4), collagen type V and VI, heparin sulfate proteoglycan 2, fibronectin, and keratins such as 15, 18 and 19. Up-regulation of these genes during effective treatment suggests normalization of regenerative hyperplasia.
Both responders and non-responders rapidly down-modulated TNF early response genes IL-1β and IL-8
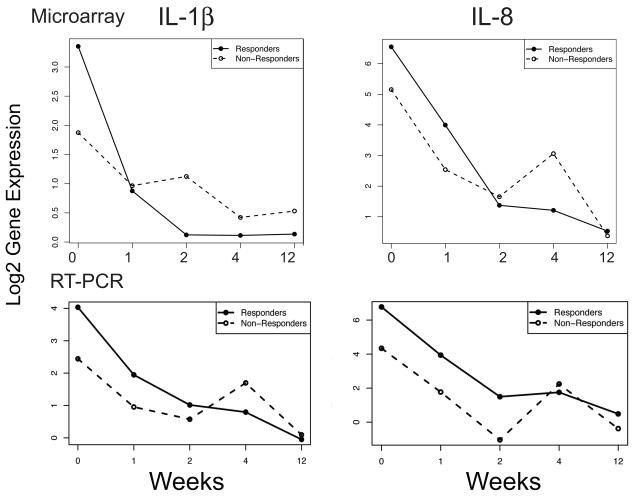
It is possible that some patients do not respond to etanercept treatment because complete TNF-blockade is not achieved. We tested this hypothesis by determining the genomic behavior of TNF immediate response genes IL-1β and IL-8 in both responders and non-responders (Figure 3). We determined that IL-1β and IL-8 are rapidly down-modulated with etanercept treatment, irrespective of the patient’s ultimate phenotypic response, suggesting that non-response cannot be explained by inadequate dosing. PCR analysis of IL-1β and IL-8 confirms that these cytokines are similarly down-regulated in responders and non-responders (Figure 3b). We have previously published RT-PCR data for these two cytokines, for responders (n=11) 20. For the non-responders (n=4), there was also a significant reduction in these two cytokines by week 12. There was no significant difference in the time profiles of either of these cytokines between the responders and non-responders (IL-1β: F-statistic 1.57, p=0.177; IL-8: F-statistic 1.47, p=0.209).
Figure 3. Mean gene expression profile for “sepsis cascade” genes IL-1β and IL-8.
(a) Down-modulation of gene expression for IL-1β and IL-8 in both responders (n=11, filled line) and non-responders (n=4, dashed line) (microarray data). (b) Mean log2 expression normalized to HARP for IL-1β and IL-8 in responders (filled line) and non-responders (dashed line) during etanercept treatment (RT-PCR data). At week 12 there was significant reduction of IL-1β in responders (p<0.0001), and non-responders (p=0.020). Similarly, at week 12 there was significant reduction of IL-8 in responders (p<0.0001), and non-responders (p=0.004).
Differences between responders and non-responders
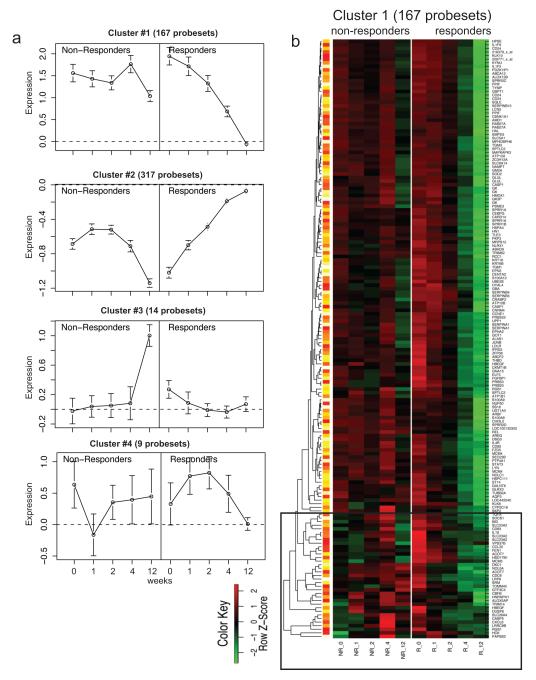
In order to determine the key genomic differences over time between responders and non-responders, we performed consensus clustering on probe-sets where the time-profile differed for responders versus non-responders (FCH >2, FDR<0.2 using the moderated F-statistic). A seven cluster structure was identified, with 4 clusters containing more than 6 probe sets (OR Figure 5; Figure 4a). These genes are listed in OR Table 4.
Figure 4. Consensus clustering grouped genes that were different in responders and non-responders.
(a) Consensus clustering of genes that were different in responders and non-responders grouped genes into 4 clusters. (b) Heat map of cluster 1. There was a group of genes that were down-regulated at baseline in non-responders but not responders (black box).
Cluster 1 contained genes that were strongly down-modulated over time in responders but not in non-responders. Inspection of the cluster 1 heat-map revealed an interesting pattern (Figure 4b). In non-responders, a group of probe-sets (black box) were down-regulated at week 0 compared with non-lesional skin (green), but up-regulated compared to non-lesional skin (red) in responders. Patients who responded to etanercept down-regulated these genes during treatment, while those who did not respond had increased expression of these genes by week four. Most notably, this group contains CCL20, a T cell and DC chemoattractant involved in the trafficking of CCR6+ Th17 T cells into inflamed psoriatic lesions 29. High expression of these “black box” genes in lesional skin may be predictive of response to etanercept. Also contained in cluster 1 was STAT3, a transcription factor that induces Th17 polarization. Thus, key IL-17 pathway genes were strongly down-modulated in responders, but not in non-responders.
Cluster 2 contained genes that were up-regulated in responders, and after a slight increase in the first week, these genes were down-regulated in non-responders. Included in this list are many cell cycle proteins and keratinocyte genes, likely representing the normalization of KC proliferation and differentiation in responders, but not in non-responders.
IL-17 pathway genes are strongly down-modulated in etanercept responders, but not in non-responders
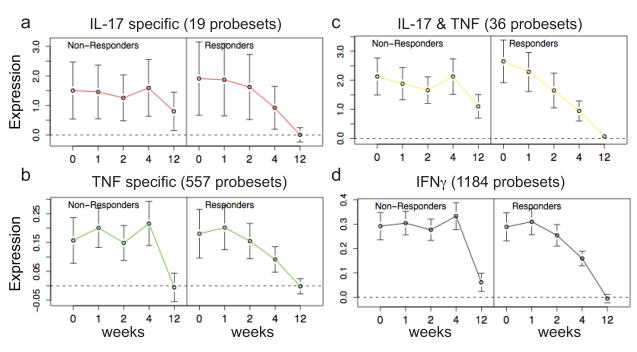
To further evaluate the difference between responders and non-responders in keratinocyte pathways, we performed GSEA analysis to determine how genes contained within each cytokine pathway were modulated over time in responders and non-responders (Figure 5). GSEA analysis demonstrated that the IL-17, TNF, IL-17 plus TNF (genes regulated both by TNF and IL-17), and IFNγ pathways were significantly different in responders and non-responders (OR Table 5). IL-17 pathway genes were highly expressed in both responders and non-responders but returned to baseline non-lesional levels by week 12 only in the responders (Figure 5a). The TNF pathway genes were reduced to baseline in both responders and non-responders (Figure 5b). This supports our interpretation that there is adequate dosing of etanercept. The combination of TNF and IL-17 had a pattern closer to IL-17, again suggesting that resolution of disease is associated with reduction in IL-17 genes (Figure 5c). In fact, the IL-17 plus TNF pathway genes exhibit the most different profile in responders and non-responders. IFNγ pathway genes were down-regulated in both responders and non-responders but with different kinetics (responders were more gradual) and non-responders did not reach baseline non-lesional levels completely (Figure 5d). In summary, IL-17 regulated genes were expressed at higher levels than TNF and IFNγ genes in psoriatic lesional skin, and IL-17 genes were only down-modulated in responders.
Figure 5. Profiles of “cytokine pathways” in etanercept gene sets in responders and non-ersponders, using GSEA analysis.
Keratinocyte cytokine “pathways” were created by culturing primary human keratinocytes with either IL-17, TNF, or IFNγ and defining cytokine pathway gene sets (“IL-17”, “TNF specific”, “IL-17 & TNF”, and “IFNγ”) 22. (a, b, c, d) Genes modulated by etanercept over time were analyzed by GSEA. Average normalized expression (+/− 95% confidence interval) was plotted over time. Responders showed significant down-modulation of IL-17 pathway genes compared to non-responders.
Discussion
The original impetus for testing anti-TNF drugs in rheumatoid arthritis was born from established elevation of TNF regulated “sepsis cascade” cytokines IL-1β and IL-8 in inflamed synovial fluid. The same principle was applied to patients with psoriatic arthritis. Interestingly, however, it was soon noted that the kinetics of disease resolution differed between the joint and the skin component of psoriatic arthritis. In most patients, joint disease improved within 2 weeks while the skin inflammation did not maximally resolve for greater than 12 weeks 30. This in itself presented the possibility that the arthritic and skin disease components had a different mechanism of action, with TNF as a common upstream cytokine.
In rheumatoid arthritis, suppression of the sepsis cascade does indeed govern response to etanercept TNF inhibition: responders down-regulate IL-1β and IL-8 in PBMCs by 72 hours and non-responders do not down-regulate these cytokines 6. Our current study however, suggests that a more complex model is operating in psoriasis skin lesions. Both responders and non-responders rapidly down-regulate IL-1β and IL-8, indicating that down-regulation of sepsis cascade cytokines is not sufficient for psoriasis lesion resolution. Therefore, the rapid response of arthritis to TNF inhibition may be explained by disease directly regulated by TNF and TNF early response genes. Psoriasis however, requires a more complex model of disease that is only indirectly modulated by TNF.
Recent studies in genetics have shown that alleles of genes encoding p19 and p40 subunits of IL-23, and IL-23R confer risk for development of psoriasis 31. This pathway controls the development of Th17 T cells, which may contribute IL-17 and IL-22 to an inflammatory skin environment. The importance of this pathway is also supported by therapeutic response of psoriasis to monoclonal antibodies that inhibit IL-12 and IL-23, producing major improvement in 70-80% of cases 32. Since TNF inhibitors can also confer nearly equivalent therapeutic benefit, there may be some overlap in inflammatory circuits regulated by TNF and p40 cytokines. One possible intersection between TNF and Th17 T cells has been suggested from our prior study of the effect of etanercept effect on synthesis of IL-23, IL-17 and IL-22, namely that TNF may serve as a DC activator for IL-23 production, which then promotes activation of Th17 T cells.
The present study further expands the analysis of the effect of etanercept on inflammatory pathways. Broad sets of genes distinctly regulated by TNF, IL-17, and IFNγ can be measured during a many week period of disease improvement induced by etanercept. Since the set of inflammatory genes regulated by etanercept increases over time, our results suggest additional indirect effects of TNF on more complex cellular or molecular circuits. For example, TNF has been shown to be a “co-stimulus” for T cell activation as well as a critical molecule for DC development and maturation 33.
We considered two hypotheses for the actions of TNF in psoriasis, and therefore the effects of etancercept on gene expression. Under the first model, TNF would act as a direct activator of NFκB (classical TNF transcription factor) and therefore genes primarily regulated by this transcription factor would normalize as the activation stimulus (TNF) was attenuated by etanercept. Under the second hypothesis, we considered that TNF has additional, broader downstream or indirect actions, e.g., as an activator of DCs and as a T cell co-stimulus. In this hypothesis, etanercept might regulate many different transcription factor pathways over time and TNF inhibition might produce different kinetics for normalization of genes driven by diverse transcription factors in multiple cell types, and the involution response might be considered as a reverse “cascade” over time. Hence an effect of TNF blockade may be progressive collapse of cellular elements needed to sustain skin inflammation, a “domino” effect.
The earliest effects of TNF blockade in responders, as we show in this manuscript, are the down-regulation of myeloid lineage cells, and the rapid up-modulation of stable DC lineage genes CD1c and CD207 (Langerin). This suggests that both resident CD1c and Langerhans cells may either not play an active role in TNF-mediated inflammation, or may be “protective.” Previous work by our group has shown rapid down-regulation of CD11c+ inflammatory DC cell counts and their inflammatory products iNOS and IL-23 (p19 and p40 subunit) by RT-PCR 20. Later down-modulation of IL-17 pathway genes, but not TNF or IFNγ pathway genes, are predictive of response to etanercept, thus solidifying the importance of IL-17 in TNF-dependent psoriatic inflammation.
OR Figure 6a outlines how TNF-dependant DC activation might be primarily blocked by etanercept in responding patients. Patients with psoriasis who do not respond to TNF-inhibition must have TNF-independent inflammatory pathways leading to the psoriatic phenotype (OR Figure 6b). Three pathway models could explain why non-responders do not down-regulate the IL-17 pathway. In non-responders, DCs may be activated independently of TNF by either cytokines (for example IFN); direct T cell-induced DC maturation (for example CD40L); or non-responders may have DC-independent activation of Th17 cells by cytokines other than IL-23, such as IL-6, TGFβ and IL-15 34, 35. Clearly, further studies will be required to determine how the IL-17 pathway is sustained in non-responders.
In conclusion, our study characterizes a sequence of gene expression changes in inflamed skin lesions as these resolve with etanercept treatment. Genes that are regulated immediately after starting etanercept are candidates for “direct” TNF gene targets, and expand the roster of potential TNF regulated genes in skin inflammation. The progressively larger set of genes that increase over time suggest that many cellular activation circuits, and especially those linked to DCs and adaptive immunity, are impacted by blocking TNF in a therapeutic context. The resolution of psoriasis by etenercept thus appears to be linked more to modulation of adaptive immune circuits, rather than innate immune circuits, although both pathways are modulated by this TNF antagonist.
Key Messages.
Cluster analysis on psoriasis skin lesions over time during treatment with the TNF inhibiting drug etanercept demonstrated a “domino-effect” model of disease involution.
Many down-modulated TNF early response genes were myeloid lineage genes, while up-regulated genes include resident dendritic cell genes.
In psoriasis, both etanercept responders and non-responders rapidly down-modulated “sepsis cascade” cytokines IL-1β and IL-8. This contrasts with rheumatoid arthritis patients in which only responders down-regulated IL-1β and IL-8.
In psoriasis, only responders down-regulated IL-17 pathway cytokines to baseline values after 12 weeks, suggesting that ultimate Th17 suppression is necessary for response to TNF inhibiting drugs.
Supplementary Material
Acknowledgments
Research was supported by a Clinical and Translational Science Award grant UL1RR024143, LZ is supported by NIH MSTP grant GM07739, and ML by K23 AR052404-01A1 and the Doris Duke Foundation. Amgen supported this study by an unrestricted research grant to Rockefeller University. JGK was a visiting lecturer at Amgen and received a small honorarium.
Abbreviations
- DC
dendritic cell
- FDR
false discovery rate
- FCH
fold change
- GSEA
gene set enrichment analysis
- HBEGF
heparin-binding epidermal growth factor
- PLAUR
plasminogen activator of urokinase receptor
- SMM
Supplemental Materials and Methods
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green S, Dobrjansky A, Carswell EA, Kassel RL, Old LJ, Fiore N, et al. Partial purification of a serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A. 1976;73:381–5. doi: 10.1073/pnas.73.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman MK. The effects of tumor necrosis factor on the production of interleukin-1 by macrophages. Lymphokine Res. 1986;5:255–60. [PubMed] [Google Scholar]
- 3.Clark IA, Virelizier JL, Carswell EA, Wood PR. Possible importance of macrophage-derived mediators in acute malaria. Infect Immun. 1981;32:1058–66. doi: 10.1128/iai.32.3.1058-1066.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nickoloff BJ, Stevens SR. What have we learned in dermatology from the biologic therapies? J Am Acad Dermatol. 2006;54:S143–51. doi: 10.1016/j.jaad.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 5.van Meurs JB, van Lent PL, van de Loo AA, Holthuysen AE, Bayne EK, Singer II, et al. Increased vulnerability of postarthritic cartilage to a second arthritic insult: accelerated MMP activity in a flare up of arthritis. Ann Rheum Dis. 1999;58:350–6. doi: 10.1136/ard.58.6.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koczan D, Drynda S, Hecker M, Drynda A, Guthke R, Kekow J, et al. Molecular discrimination of responders and nonresponders to anti-TNF alpha therapy in rheumatoid arthritis by etanercept. Arthritis Res Ther. 2008;10:R50. doi: 10.1186/ar2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb AB. Etanercept for the treatment of psoriasis and psoriatic arthritis. Dermatol Ther. 2004;17:401–8. doi: 10.1111/j.1396-0296.2004.04043.x. [DOI] [PubMed] [Google Scholar]
- 8.D’Haens G, Daperno M. Advances in biologic therapy for ulcerative colitis and Crohn’s disease. Curr Gastroenterol Rep. 2006;8:506–12. doi: 10.1007/s11894-006-0041-5. [DOI] [PubMed] [Google Scholar]
- 9.Diluvio L, Vollmer S, Besgen P, Ellwart JW, Chimenti S, Prinz JC. Identical TCR beta-chain rearrangements in streptococcal angina and skin lesions of patients with psoriasis vulgaris. J Immunol. 2006;176:7104–11. doi: 10.4049/jimmunol.176.11.7104. [DOI] [PubMed] [Google Scholar]
- 10.Chang JC, Smith LR, Froning KJ, Kurland HH, Schwabe BJ, Blumeyer KK, et al. Persistence of T-cell clones in psoriatic lesions. Arch Dermatol. 1997;133:703–8. [PubMed] [Google Scholar]
- 11.Prinz JC, Grob B, Vollmer S, Trommler P, Strobel I, Meurer M, et al. T cell clones from psoriasis skin lesions can promote keratinocyte proliferation in vitro via secreted products. European Journal of Immunology. 1994;24:593–8. doi: 10.1002/eji.1830240315. [DOI] [PubMed] [Google Scholar]
- 12.Weinshenker BG, Bass BH, Ebers GC, Rice GP. Remission of psoriatic lesions with muromonab-CD3 (orthoclone OKT3) treatment. J Am Acad Dermatol. 1989;20:1132–3. doi: 10.1016/s0190-9622(89)80200-2. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb SL, Gilleaudeau P, Johnson R, Estes L, Woodworth TG, Gottlieb AB, et al. Response of psoriasis to a lymphocyte-selective toxin (DAB389IL-2) suggests a primary immune, but not keratinocyte, pathogenic basis. Nat Med. 1995;1:442–7. doi: 10.1038/nm0595-442. [DOI] [PubMed] [Google Scholar]
- 14.Nestle FO, Nickoloff BJ. From classical mouse models of psoriasis to a spontaneous xenograft model featuring use of AGR mice. Ernst Schering Res Found Workshop. 2005:203–12. doi: 10.1007/3-540-26811-1_11. [DOI] [PubMed] [Google Scholar]
- 15.Krueger GG, Langley RG, Leonardi C, Yeilding N, Guzzo C, Wang Y, et al. A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med. 2007;356:580–92. doi: 10.1056/NEJMoa062382. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb AB, Cooper KD, McCormick TS, Toichi E, Everitt DE, Frederick B, et al. A phase 1, double-blind, placebo-controlled study evaluating single subcutaneous administrations of a human interleukin-12/23 monoclonal antibody in subjects with plaque psoriasis. Curr Med Res Opin. 2007;23:1081–92. doi: 10.1185/030079907x182112. [DOI] [PubMed] [Google Scholar]
- 17.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toichi E, Torres G, McCormick TS, Chang T, Mascelli MA, Kauffman CL, et al. An Anti-IL-12p40 Antibody Down-Regulates Type 1 Cytokines, Chemokines, and IL-12/IL-23 in Psoriasis. J Immunol. 2006;177:4917–26. doi: 10.4049/jimmunol.177.7.4917. [DOI] [PubMed] [Google Scholar]
- 19.McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Zaba LC, Cardinale I, Gilleaudeau P, Sullivan-Whalen M, Farinas M Suarez, Fuentes-Duculan J, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204:3183–94. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haider AS, Lowes MA, Suarez-Farinas M, Zaba LC, Cardinale I, Khatcherian A, et al. Identification of Cellular Pathways of “Type 1,” Th17 T Cells, and TNF- and Inducible Nitric Oxide Synthase-Producing Dendritic Cells in Autoimmune Inflammation through Pharmacogenomic Study of Cyclosporine A in Psoriasis. J Immunol. 2008;180:1913–20. doi: 10.4049/jimmunol.180.3.1913. [DOI] [PubMed] [Google Scholar]
- 22.Nograles K, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Farinas M Suarez, Cardinale I, et al. Th17 cytokines IL-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159(5):1092. doi: 10.1111/j.1365-2133.2008.08769.x. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108–11. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 25.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasperska-Zajac A, Rogala B. Circulating levels of urokinase-type plasminogen activator (uPA) and its soluble receptor (suPAR) in patients with atopic eczema/dermatitis syndrome. Inflammation. 2005;29:90–3. doi: 10.1007/s10753-006-9004-0. [DOI] [PubMed] [Google Scholar]
- 27.Zaba LC, Fuentes-Duculan J, Steinman RM, Krueger JG, Lowes MA. Normal human dermis contains distinct populations of CD11c+BDCA-1+ dendritic cells and CD163+FXIIIA+ macrophages. J Clin Invest. 2007;117:2517–25. doi: 10.1172/JCI32282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC, et al. Psoriasis is characterized by accumulation of immunostimulatory and Th1/Th17 cell-polarizing myeloid dendritic cells. J Invest Dermatol. 2009;129:79–88. doi: 10.1038/jid.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181:8391–401. doi: 10.4049/jimmunol.181.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger G, Callis K. Potential of tumor necrosis factor inhibitors in psoriasis and psoriatic arthritis. Arch Dermatol. 2004;140:218–25. doi: 10.1001/archderm.140.2.218. [DOI] [PubMed] [Google Scholar]
- 31.Nair RP, Ruether A, Stuart PE, Jenisch S, Tejasvi T, Hiremagalore R, et al. Polymorphisms of the IL12B and IL23R genes are associated with psoriasis. J Invest Dermatol. 2008;128:1653–61. doi: 10.1038/sj.jid.5701255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papp KA, Langley RG, Lebwohl M, Krueger GG, Szapary P, Yeilding N, et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 52-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 2) Lancet. 2008;371:1675–84. doi: 10.1016/S0140-6736(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 33.Chomarat P, Dantin C, Bennett L, Banchereau J, Palucka AK. TNF skews monocyte differentiation from macrophages to dendritic cells. J Immunol. 2003;171:2262–9. doi: 10.4049/jimmunol.171.5.2262. [DOI] [PubMed] [Google Scholar]
- 34.Basso AS, Cheroutre H, Mucida D. More stories on Th17 cells. Cell Res. 2009;19:399–411. doi: 10.1038/cr.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega C, Fernandez AS, Carrillo JM, Romero P, Molina IJ, Moreno JC, et al. IL-17-producing CD8+ T lymphocytes from psoriasis skin plaques are cytotoxic effector cells that secrete Th17-related cytokines. J Leukoc Biol. 2009 doi: 10.1189/JLB.0109046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.