Abstract
Motivation: The elucidation of biological concepts enriched with differentially expressed genes has become an integral part of the analysis and interpretation of genomic data. Of additional importance is the ability to explore networks of relationships among previously defined biological concepts from diverse information sources, and to explore results visually from multiple perspectives. Accomplishing these tasks requires a unified framework for agglomeration of data from various genomic resources, novel visualizations, and user functionality.
Results: We have developed ConceptGen, a web-based gene set enrichment and gene set relation mapping tool that is streamlined and simple to use. ConceptGen offers over 20 000 concepts comprising 14 different types of biological knowledge, including data not currently available in any other gene set enrichment or gene set relation mapping tool. We demonstrate the functionalities of ConceptGen using gene expression data modeling TGF-beta-induced epithelial-mesenchymal transition and metabolomics data comparing metastatic versus localized prostate cancers.
Availability: ConceptGen is part of the NIH's National Center for Integrative Biomedical Informatics (NCIBI) and is freely available at http://conceptgen.ncibi.org. For terms of use, visit http://portal.ncibi.org/gateway/pdf/Terms%20of%20use-web.pdf
Contact: conceptgen@umich.edu; sartorma@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
An important step in the analysis and interpretation of gene expression, proteomic, metabolomic or transcription factor binding data is answering the question, ‘What biologically related sets of genes are enriched with the interesting genes/proteins/compounds identified in my experiment?’ Such analysis applied to gene expression data is often referred to as gene set, or functional, enrichment testing. Gene sets defined by Gene Ontology (GO) (Ashburner et al., 2000; Harris et al., 2004) or KEGG pathways (Kanehisa et al., 2006) are often employed, and the statistical significance of enrichment can be established using the Fisher's exact test and the hypergeometric distribution. Several web-based or downloadable tools performing this or a similar test have been developed, such as Onto-Express (Draghici et al., 2007; Khatri et al., 2005), David/EASE (Dennis et al., 2003; Hosack et al., 2003), the Gostats package of Bioconductor (Gentleman, 2007), GOMiner (Zeeberg et al., 2003, 2005) and FuncAssociate (Berriz et al., 2003). A second research question, often viewed as separate from the first, is based on testing hypotheses of correlated signatures between disparate sources of biological knowledge. For example, are genes targeted by a specific microRNA more likely to be involved in a disease progression process than expected by chance? These two common types of research questions can be answered within the same analysis framework of gene set relation mapping.
While there is a plethora of tools for enrichment testing, few offer the level of visualization and interactivity desired by many biomedical researchers to explore results. Gene set relation mapping is a technique that extends beyond enrichment testing and can enable wide-spanning exploratory analysis and hypothesis generation by visualizing relationships among concepts. In addition to testing the overlap between an experimental gene list and predefined gene sets (concepts), the significant overlap among all predefined gene sets (concepts) is assessed. Two concepts are related when they have significantly more genes in common than expected by chance, and these relationships can form a network. Testing among concepts allows one to visualize the networked relationships among concepts enriched with genes in an experimental dataset. For example, one may observe that the concepts enriched with their data cluster into three distinct groups each having previously unsuspected relationships between concepts from diverse concept types. Concept types represent data from different sources of biological knowledge, such as biological processes, microRNA target lists, chromosomal regions, or drug target lists. Another approach to visualizing gene set relations is by clustering genes versus enriched concepts in a heatmap view. This allows one to see in a glance which subset of genes is responsible for the enrichment of which concepts, in addition to visualizing which concepts are closely related (see Section 2.7). Together, the network and heatmap enable a more biologically comprehensible understanding of functional enrichment results.
One type of gene set relation mapping was implemented and incorporated in the software Oncomine (Rhodes et al., 2007) (referred to as molecular concept mapping). It allows investigators to easily navigate the complex and diverse public domain gene expression knowledge relating to cancers through the use of data integration, manual curation, statistical analyses and visualization tools. This approach has led to important discoveries, particularly in research related to the progression of prostate cancer (Morris et al., 2007). However, this initial gene set relation mapping lacked some key sources of biological information included here in ConceptGen, such as protein–protein interactions other than from HPRD and metabolite information; it also relied on basic statistics for analysis of gene expression data, and restricted gene expression signatures to those related to cancer due to the program's cancer-related focus (see Table 2 for comparison). DAVID/EASE (Dennis et al., 2003) offers a different type of gene set relation mapping in which clusters of related gene sets are formed using kappa statistics; however no visualization is offered, and one cannot observe connections between gene sets in different clusters.
Table 2.
Comparison of selected functional enrichment testing software
| Feature | Concept-Gen | DAVID/EASE | GSEA/MSigDB | Oncomine concept mapping | GeneDecks |
|---|---|---|---|---|---|
| Performs concept mapping? | Yes | No | No | Yes | No |
| Contains experimental data? | Yes | No | Yes | Yes, cancer-related | Yes |
| Uses modified Fisher's exact test? | Yes | Yes | N/A | No | No |
| Private account? | Yes | No, but down load | No, but down load | Yes | No |
| Freely available? | Yes | Yes | Yes | Limited academic version | Yes |
| Heatmap view of network? | Yes | No | No | No | No |
| Drug targets? | Yes | No | No | Yes | PharmGKB |
| Metabolite? | Yes | No | No | No | Yes |
| Protein interactions? | MiMI | Several | No | HPRD | No |
| MeSH or other literature-based concepts? | Yes | No | No | Yes | Disorders and compounds |
| Phenotypes? | No | No | No | No | Yes |
| Accepts several gene ID types? | No | Yes | No | No | No |
| Does not require cut-off? | No | No | Yes | No | No |
| In depth visualizations among experimental datasets? | No | No | No | Yes | No |
Here we present a new web-based software application, ConceptGen, that may be used as a gene set enrichment and gene set relation mapping tool. It contains several sources of biological knowledge, offers multiple visualizations, and has a convenient user-interface. In addition, we have performed gene-to-gene enrichment testing which identifies closely related genes based on significance of co-occurring concepts. This provides an additional viewpoint and database for addressing further questions. A similar approach to identifying related genes is used in the new Paralog Hunter tool in GeneDecks, which uses a variety of concept types from GeneCards to identify functional paralogs (Safran et al., 2003; Stelzer et al., 2009).
NCBI's Gene Expression Omnibus (GEO) data repository (Edgar et al., 2002) offers a wealth of experimental data. As one concept type in ConceptGen, we have downloaded, processed and analyzed human raw Affymetrix data from GEO to create gene expression-based concepts covering a wide variety of expression signatures from reactions to treatments, diseases, exposures, genotype, development and injury/infection. Taking such an unbiased approach to data inclusion allows one to identify previously unsuspected relationships among diverse biological perturbations, and generating novel hypotheses. The datasets can be expanded to utilize epigenomics, proteomics, metabolomics and microRNA inputs.
2 METHODS
2.1 Concept building
Concepts (gene sets) were defined based on a wide variety of types of biological knowledge (concept types) with the goal of being able to identify novel relationships among diverse sources. The types of biological knowledge are: biological processes, molecular functions, cellular components, protein-interactions, medical literature-derived concepts, human diseases, drug targets, chromosomal location, molecular pathways, transcription factor targets, protein families, microRNA targets, metabolite-centered concepts and gene expression signatures (Table 1). Concepts and concept types were downloaded automatically or manually from various genomic resource centers, entries were converted to NCBI Entrez Gene IDs, and concepts were uploaded and are stored in an Oracle database. In order to avoid non-informative or overly-broad concepts, we limit the scope to between 5 and 1000 genes per concept; when necessary, manual curation was performed. GO, KEGG pathway, Biocarta Pathway, Panther Pathway and Pfam information was downloaded from their respective sources. Chromosomal location was determined by NCBI cytoband assignment, and gene expression signatures were defined as detailed in Methods section 2.2 and Supplementary Methods section. Other concept types (literature-derived concepts, human diseases, drug targets, transcription factor targets, protein-interactions, microRNA targets and metabolite-centered targets) were built as detailed in Supplementary Methods section.
Table 1.
Biological knowledge types represented in ConceptGen and their Concept Type(s)
| Biological knowledge type | Concept type(s) | Number of concepts |
|---|---|---|
| Biological processes | GO biological process | 2477 |
| Molecular functions | GO molecular function | 1075 |
| Cellular components | GO cellular component | 446 |
| Protein-centered interactions | MiMI | 6823 |
| Medical literature derived concepts | MeSH | 5214 |
| Human diseases | OMIM | 52 |
| Drug targets | Drug Bank | 256 |
| Chromosomal location | cytoband | 1178 |
| Molecular pathways | KEGG pathway; | 195 |
| Panther pathway; | 86 | |
| Biocarta pathway | 245 | |
| Transcription factor targets | Transfac | 119 |
| Protein families | Pfam | 770 |
| microRNA targets | MiRBase | 587 |
| Metabolic interactions | Metabolite | 960 |
| Differential expression profiles | Gene expression | 603 |
The total number of concepts is 21 086.
2.2 Gene expression analysis
In order to define expression-based concepts, we developed a gene expression analysis pipeline that uses a carefully chosen, statistical method for each step. The gene expression concept type is populated with human Affymetrix experiments in GEO. Details of the analysis pipeline are provided in the Supplementary Methods section, and outlined briefly here. The pipeline downloads the raw data, pre-processes it with a Entrez ID centered CDF package (Dai et al., 2005) and normalizes it, outputs quality control data, and tests for differentially expressed genes using an empirical Bayes method (Sartor et al., 2006). The tests are set up manually through an interface, and following testing, gene sets (concepts) are defined by the top ranked genes.
2.3 Enrichment testing
For public concepts, all pairs of concepts from all concept types were tested for whether there exists a larger number of overlapping genes than is expected by chance. We use a slightly modified Fisher's exact test, identical to the ‘Ease score’, which helps to stabilize results by penalizing tests with small numbers of overlap (Hosack et al., 2003). P-values are adjusted for multiple testing by calculating q-values using the FDR method (Benjamini and Hochberg, 1995). The default display is those concepts with q-value <0.05, but the user may choose a different q- or p-value cutoff.
Gene lists uploaded by the user are considered concepts in their private, Experimental concept type. The gene lists are converted to human Entrez Gene IDs, if necessary, and stored and tested in a private concept type. Users have the option of converting from a list of mouse or rat genes using NCBI Homologene homolog families with our conversion tool, or from a set of compounds/metabolites. Users also have the option to upload a background gene set consisting of all the genes that were interrogated in their study (e.g. all genes on a microarray platform). If no background set is provided, ConceptGen uses all Entrez IDs as default. The modified Fisher's exact test described above is then implemented, and q-values are calculated for the experimental list within each concept type separately. Users can filter results based on concept type(s) and/or significance levels (p- or q-values) for export in spreadsheet format or visualizations.
A background set is defined as all genes (Entrez IDs) that were interrogated in creating the concept type. For example, for GO concept types, the background set is all genes that are assigned to at least one ontology term. It is important to use the correct background gene set for each enrichment test, and for that we use the intersection of the background gene sets for the two concept types of the concepts being tested. Thus, for example, if we are testing a GO term versus a microRNA target list, we use all genes that are in both the GO background set and the microRNA target background set.
2.4 Gene set relation mapping—graph network visualization
Users can explore concepts from any of the public sources, or load their own sets of genes to define private concepts. Once a concept is selected, the concepts that are paired with it (those whose enrichment scores are significant) can be selected by category, significance or individually, to participate in a concept-to-concept graph (see the figs in Section 3.2), with nodes representing concepts, and edges representing significant enrichment of overlapping genes. The concept networks, nodes and edges are displayed in an interactive web application (coded in Adobe® Flex/flash). The graph is laid out on the display using a standard layout from Adobe's open source code, which implements a force directed layout algorithm (Fruchterman and Reingold, 1991). This layout algorithm results in highly interconnected groups of concepts clustering together. Within that layout, the concept type of each concept node is shown by the color, the size of the concept node is based on the number of genes in the concept, and the thickness of the edge lines is based on the number of overlapping genes. These graphical network displays can be further explored to find the genes that concepts have in common, filter the graph based on sets of genes, display the statistics associated with a concept or edge, or explore protein interactions within a node or edge. By moving among the results panel, the graphics display, and the protein interaction networks, the user can narrow in on a set of concepts of interest and conceptualize results.
2.5 Gene set relation mapping—heatmap view
The heatmap view offers an alternative view to the network graph, convenient for visualizing large numbers of concepts. It also allows one to see at a glance which genes are responsible for the enrichment of which concepts, and which gene groups co-occur in the same concepts most often (see Supplementary Figures S8 and S9 for examples). The heatmap plots genes (columns) versus enriched concepts (rows), and values used are 0 when a gene does not belong to the concept, or the number of enriched concepts to which the gene belongs otherwise. Genes and concepts are clustered using the complete linkage hierarchical clustering method with the Euclidean distance measure. The color of columns ranges from black (gene does not belong to the enriched concept) to bright red (genes belonging to the most enriched concepts.) Users can toggle between the heatmap and graph network views, and can use a ‘draw tool’ to choose a cluster or section of the heatmap to explore in the graph network display.
2.6 Gene–gene relations by enriched concepts
To visualize how genes are related by concepts and nominate genes by common annotations, we developed and performed gene-to-gene enrichment testing. Similar to gene set enrichment testing, a series of modified Fisher's exact tests is performed, but with genes replacing concepts, and concept membership replacing genes. For the application, q-values are calculated and a q-value <0.01 cutoff is used as default. This testing provides an alternative to viewing the relationships among genes via known protein-interaction networks (see Section 3.4). It can also be used simply to query all concepts that any specific gene is assigned to in ConceptGen. The gene-to-gene enrichment testing provides a statistical measure of the closeness of any two genes by annotations, and can be reached through a link on the main ConceptGen website. Users have many of the same options and visualizations as for the standard ConceptGen analysis.
3 RESULTS
First, we describe general aspects of ConceptGen and summarize the concept types and their relationships. We then demonstrate the performance and functionality of ConceptGen with a typical use-case scenario: a time course gene expression data set obtained from a cell culture model of TGF-β-induced epithelial-mesenchymal transition (EMT) (Keshamouni et al., 2006). We show how ConceptGen was useful in visualizing the data and contextualizing prior knowledge to generate new hypotheses, thereby contributing to the overall understanding of this complex biological process. The final two results sections demonstrate the use of ConceptGen in more specific applications: a metabolomics study of prostate cancer (Sreekumar et al., 2009) and the use of the gene-to-gene enrichment testing for gene function prediction, respectively.
3.1 Properties of the data in ConceptGen
The general aspects involved in development of a gene set enrichment or gene set relation mapping tool are: (i) the test used to assess significance of enrichment, (ii) the concepts and concept types used, (iii) usability of the software tool, (iv) visualizations offered by the tool, (v) quality of statistical methods used to build concepts from experimental data (if applicable); and (vi) adjusting the significance levels for multiple comparisons. The quality of the software application will be a function of the above aspects. Thus in the development of ConceptGen, we have carefully chosen the implementation of each of these aspects to create an easily accessible, powerful, stream-lined and accurate enrichment testing and gene set relation mapping tool.
Table 1 lists the types of biological knowledge incorporated in ConceptGen and the names of the concept type(s) used to represent each of them. These concept types encompass pathways, molecular processes, chromosome location, biological targets, protein–protein interactions, diseases, and experimental and literature-derived data. Each biological knowledge type provides the ability to generate a different type of hypothesis; which concept types are of greatest interest will depend on the context of the application. For example, testing the protein interactions concept type will identify proteins that interact with a significant number of user-supplied gene products, possibly generating a hypothesis for what other proteins may be activated or play a central role in the process under study. For protein interactions, we used the Michigan molecular interactions (MiMI) database, which deep-merges several sources of interactions, resulting in a comprehensive database of human protein-protein interactions and thus greater power to detect significant enrichments than using any one data source alone. A notable member of our concept types is MeSH [derived from Medical Subject Headings (MeSH®)]. Medical subject headings is the National Library of Medicine's controlled vocabulary built in a hierarchical structure, which covers subjects ranging from anatomy, drugs, and diseases to social phenomena, geographical locations and proteins. Therefore, the MeSH concept type in itself contains concepts relating to various types of biological knowledge. Use of the MeSH concept type will help users identify relationships with concepts in literature that may otherwise be overlooked.
Table 2 compares features of ConceptGen with four related software programs performing gene set enrichment and/or gene set relation mapping: DAVID/EASE (Dennis et al., 2003), GSEA and MSigDB (Subramanian et al., 2005), Oncomine's molecular concept mapping (Rhodes et al., 2007) and GeneDecks (Stelzer et al., 2005). Similar to other programs, such as DAVID, certain concept types tend to have larger or smaller concept sizes (number of genes populating each concept) than others, and different distributions (Supplementary Fig. S1). For example, gene sets based on gene expression experiments and microRNA targets had the largest concepts on average. Others, such as GO and MeSH have a broad range of concept sizes due to their hierarchical nature.
3.1.1 Inter-connectivity among concept types
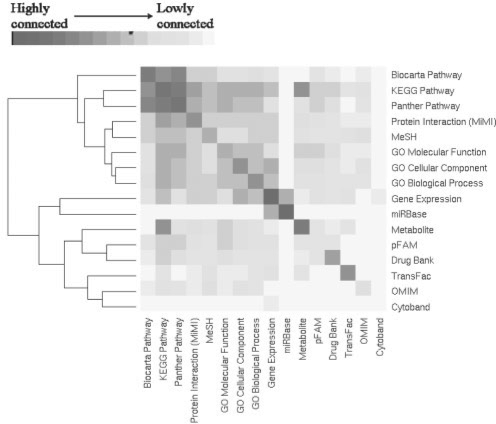
To determine how closely-related the concept types are amongst themselves, we calculated a measure of connectivity. Connectivity is defined as s/n where s = No. of tests between concept types with q-value <0.05 and n = total number of possible connections. As seen in Figure 1, most although not all concept types have highest connectivity with self, and the pathway databases KEGG, Biocarta and Panther form a block of high inter-connectivity. In addition to these expected results, we observe that the pathways have high inter-connectivity with protein interactions and GO. Gene expression data has relatively high connectivity with KEGG Pathway, GO cellular component, GO biological process, MiMI, MeSH, and interestingly, microRNA targets (miRBase). We can speculate that the higher connectivity of gene expression data with KEGG compared with the other pathway databases may be an indicator of how well KEGG approximates the reality of biological systems, although more research is needed to test this prediction. The clustering divides the concept types into two groups, which can be interpreted as (i) those that assess the pathways, molecular processes and cellular networks, and (ii) all others.
Fig. 1.
Inter-connectivity among concept types. Connectivity is defined as s/n where s = No. of tests between concept types with q-value <0.05 and n = total number of possible connections. Values were clustered using complete linkage hierarchical clustering in R.
3.1.2 Properties of Gene Expression data
As part of our creation of the Gene Expression concept type, we developed a database with analysis results from all analyzed experiments, which is a valuable source of information in and of itself, and is available upon request. As an example of its use, we wished to determine which genes are most often and least often differentially expressed. We observed a broad distribution in how often a gene was differentially expressed, ranging from 0 to 53% (108) of the 203 experiments (Supplementary Fig. S2). Genes that were most often differentially expressed were significantly enriched for involvement in cell cycle (q = 3.0 × 10−13) (such as CITED2, RGS2, Il8, PTTG1, UBE2C and several CDC's and CCN's), cell proliferation (q = 4.5 × 10−9), programmed cell death (q = 1.6 × 10−5) (VGEFA, HMOX1, IGF1R, BCL6, GADD45A, NFKBIA and TOP2A), transcription factors (q = 2.8 × 10−5) (JUN, FOS, MYC, CEBPB, EGR1, AHR and DDIT3) and immediate-early proteins (q = 8.2 × 10−5). The genes least often differentially expressed (three or fewer experiments) were enriched for G-protein coupled receptors (q = 3.8 × 10−18) and other sensory receptors, ion channel activity (q = 6.4 × 10−9) and neurotransmitter binding (q = 6.3 × 10−3).
Although this analysis is dependent on what processes are most often studied, the specific genes do provide important and useful information. We thought it may be especially interesting to identify genes involved in apoptosis and cell cycle that very rarely change, and genes involved in receptor and sensory activity that often change. Upon researching this question, we found that the majority of the most often-changed genes involved in receptor and sensory activity were also involved in secondary roles, such as development or transcription factor activity. On the opposite end of the spectrum, we found only two genes involved in mitotic cell cycle that were rarely changed, NEUROG1 and CDC20B. Eight genes playing a role in apoptosis changed least often. One example is CIDEA, known to play a role in the activation of apoptosis. Since this protein activates apoptosis by moving from its sequestered location in the mitochondria to the nucleus (Valouskova et al., 2008), it is not surprising that CIDEA was only found to change twice in its total mRNA level among all the experiments in which apoptosis played a role.
3.1.3 Use of ConceptGen
Users may begin their analyses with ConceptGen from one of two main entry points: with a public concept or a private, user-uploaded concept (gene list). The simplest way to begin is to query or browse the public concepts from the main page (Supplementary Fig. S3). Alternatively, one can register for a free academic account and upload, analyze, and store one or more private concepts. We now demonstrate the use of ConceptGen with an application with microarray data.
3.2 Application to TGFβ-induced EMT in lung adenocarcinoma cells
EMT is a highly conserved embryonic and developmental process that facilitates the dispersion of cells. During EMT, cells lose their epithelial properties, while acquiring mesenchymal properties which enable them to migrate to a predetermined destination in order to generate distinct tissue types (Kalluri and Weinberg, 2009). A similar process is reactivated in cancer cells as an early event during tumor metastasis and confers certain fundamental abilities essential for the process of metastasis. These include the ability to migrate, invade, resist apoptosis and evade immune surveillance (Kalluri and Weinberg, 2009). Multifunctional cytokine TGF-β is a potent inducer of EMT (Thiery and Sleeman, 2006). TGF-β-induced EMT in A549 lung adenocarcinoma cell line provides an excellent in vitro correlate for mechanistic investigations of EMT in the context of tumor progression (Keshamouni et al., 2006, 2009). Here we demonstrate the functionalities of ConceptGen in enabling visualization of data and contextualizing prior knowledge to generate new hypotheses, by utilizing time course gene expression data (Control versus 0.5, 1, 2, 4, 8, 16, 24 and 72 h) obtained from A549 cells undergoing TGF-β-induced EMT.
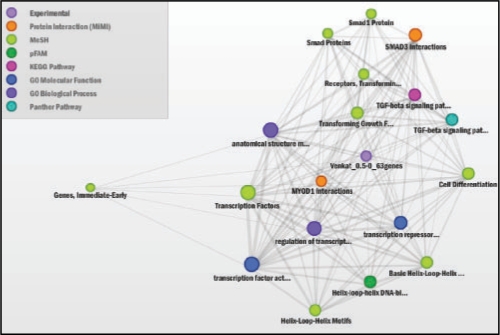
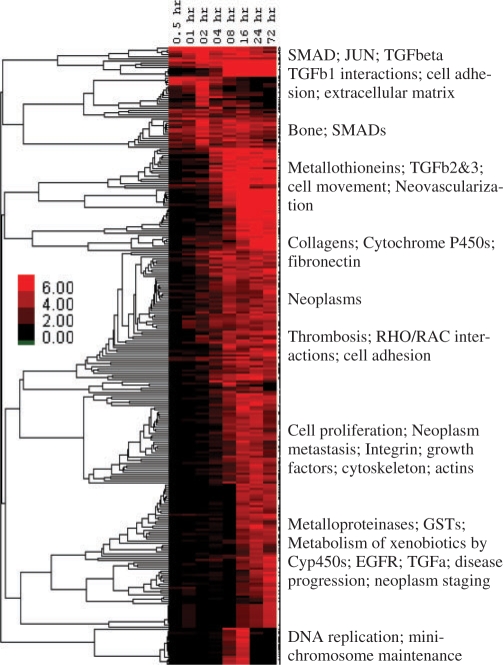
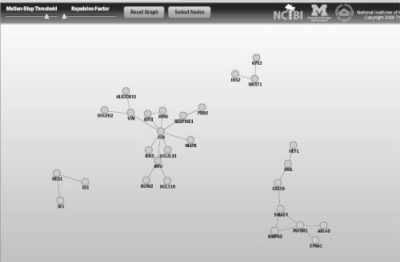
Data were processed and analyzed as previously described (Keshamouni et al., 2009). The data have been deposited in NCBI's GEO (Barrett et al., 2007) and are accessible as GSE17708. The set of differentially expressed genes at each time (defined by p-value <0.001 and >2-fold change) was uploaded to ConceptGen for analysis, and network graphs and heatmaps of results were visualized for each time point. All concept types were used for testing, although in other situations one may wish to limit testing to a subset of concept types. Supplementary Figure 4 illustrates the main results page of ConceptGen and Figure 2 displays the network graph for the 30 min time point. In this figure, colors represent different knowledge types. Results for each time point were then exported from ConceptGen in spreadsheet format and the enrichment significance profiles for concepts with q-value <0.05 for at least one time point were clustered for an overall visualization of enriched concept profiles. This allowed us to easily visualize which processes were being affected throughout the time course. Model-based clustering (Medvedovic and Sivaganesan, 2002) of the −log10(p-values) for enrichment was performed separately for GO terms (Supplementary Fig. S5), Gene expression enrichment (Supplementary Fig. S6), and other concept types (Fig. 3) for ease of interpretation.
Fig. 2.
ConceptGen Network Graph after 30 min TGFβ-induction of A549 cells shows TGF-β receptors, SMAD proteins, Myod1 interactions, and transcription factor activity to be enriched. Shown are all concepts with q-value <0.05 after filtering for Gene Expression.
Fig. 3.
Heatmap of enriched Concept profiles throughout TGF-β-induced EMT transition in the A549 cell line for concept types other than GO and Gene Expression: rows are concepts from KEGG, Biocarta, and Panther Pathways, MiMI protein interactions, MeSH, Pfam and Transfac. All concepts with q-value <0.05 for ≥1 time point were clustered using −log10(p-values) to create a ‘bird's eye view’ of which processes were turning on and off throughout the time course. For presentation, enriched concepts were summarized for each main cluster. Heatmaps for GO and gene expression concepts are presented in Supplementary Material.
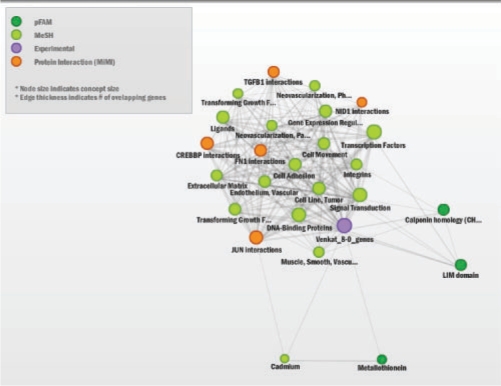
TGF-β initiates signaling by binding to type II receptors on the cell surface, triggering the activation of type I receptors. Activated type I receptors phosphorylate SMAD proteins. Phosphorylated/activated SMADs move into the nucleus where they bind to transcriptional co-activators or co-repressors to regulate target gene expression (Massague, 2000). Enrichment analysis for 30 min to 6 h time points indicated effects of a robust transcriptional reprogramming with the modulation of several transcriptional factors (summarized in Fig. 3 and Supplementary Fig. S5), including SMADs, MyoD and induction of type I TGF-β receptors (Fig. 2), reflecting the primary mode of action of TGF-β. Consistent with growth inhibitory affects of TGF-β (Massague, 2000), we observed enrichment of concepts that are reflective of negative growth regulation in the middle time points (Fig. 3 and Supplementary Fig. S5). Similarly, around the same time points we also observed the enrichment of GO terms reflecting inhibition of apoptosis (Supplementary Fig. S5), consistent with the EMT induced apoptotic resistance demonstrated in earlier studies (Lee et al., 2006). Enrichment of concepts of cell movement, cell adhesion, extracellular matrix and matrix metalloproteases (Figs 3 and 4) are reflective of migratory and invasive phenotypes acquired during EMT (Keshamouni et al., 2006), whereas the concepts such as regulation of cell size, actin binding and cytoskeletal protein binding are consistent with the dramatic change in morphology and robust cytoskeletal reorganization that occurs during EMT (Thiery and Sleeman, 2006). On the whole, functional enrichment analysis of multiple biological knowledge types with ConceptGen coupled with a heatmap viewer provided a reliable overview of the entire data set, accurately representing the underlying biology across time points.
Fig. 4.
ConceptGen Network Graph after 8hrs of TGFβ induction in A549 cells shows extracellular matrix, cell adhesion, cell movement and metallothioneins are enriched, consistent with the migratory and invasive phenotypes acquired during EMT. Shown are the top significant concepts from Pfam, MeSH, and MiMI. GO biological processes indicated that vascular and blood vessel development were also enriched.
ConceptGen also allowed us to contextualize the data in hand with prior knowledge in the public domain which includes previously published gene expression data sets (Supplementary Fig. S5) and protein interaction maps. Enrichment analysis of EMT time course data with publicaly available gene expression data sets has returned interesting results. Similar to the analysis of other concept types, these results were reflective of TGF-β biology and the complexity of EMT process in a time depedent manner, with specific examples provided in supplemental material. Overlap with other expression sets allows identification of common sets of responses that in combination are not well represented in any other knowledge domain. For example, one could detect other conditions that resulted in a certin mix of DNA repair, response to stress and apoptosis. Using the MiMI NetBrowser tool in ConceptGen, we can also visualize a network of protein–protein interactions for the genes belonging to any node or edge, and can identify potential regulatory hubs at each time point. For example, at the 1 h time point, we identified JUN and TGFBR1 as hubs, consistent with the induction and activation of these two proteins respectively in response to TGF-β (Fig. 5).
Fig. 5.
Protein interactions among differentially expressed genes after 1hr of TGFβ induction in A549 cells, visualized by clicking on the gear icon for the uploaded concept in the network browser view, which links to the MiMI NetBrowser view of known protein interactions.
3.3 Application with compounds
A novel feature of ConceptGen is its ability to take one or more compounds as input (in the Upload Gene Set page) and find the genes encoding metabolic enzymes related to those compounds. As an example, we input six metabolites that were found to significantly change in LnCAP cells in the disease progression from benign to localized to metastatic prostate cancer (Sreekumar et al., 2009). This list of metabolites (glycerol-3-phosphate, kynurenine, leucine, proline, sarcosine and uracil) was generated by unbiased metabolomics experiments, and was linked to a total of 37 genes using the methods described for building the Metabolite concept type (see Supplemental Methods section). The first step in characterizing a set of metabolites is identification of the biochemical pathways to which they belong. ConceptGen offers a convenient way to do this, without having to manually map the compounds individually onto KEGG or a similar database.
The conclusions from this simple analysis are consistent with the findings reported in Sreekumar et al. (2009), namely that amino acid metabolism (q-value = 7.4 × 10−12), nitrogen breakdown (q = 2.3 × 10−4) and amino methyltransferase activity (q = 9.6 × 10−3) from GO are enriched (Supplementary Fig. S7). Upon inspection with the heatmap view, we saw that the above-mentioned concepts all clustered with the compound sarcosine (Supplementary Fig. S8), indicating that mainly enzymes related to sarcosine drove the enrichment of these concepts. Additional processes identified as enriched were aminoacyl tRNA biosynthesis KEGG pathway [recently identified as disregulated by androgen in prostate cancer (Vellaichamy et al., 2009)], and procollagen–proline dioxygenase activity from GO molecular function, which is involved in collagen biosynthesis and folding. This is interesting because collagen production is known to be involved in cancer progression, and because the prolyl hydroxylase gene family was recently noted as being a novel class of tumor suppressors, specifically in breast cancer (Shah et al., 2009).
3.4 Gene-to-gene enrichment application
A noteworthy feature of ConceptGen is its gene-to-gene enrichment analysis, which can be used to generate hypotheses about the function or pathway of a gene, and which is based on a significant overlap in concepts to which the genes belong. As an example application of this feature, we queried the gene Chac1 [Cation transport regulator homolog 1 (E.coli)] in the ConceptGen gene-to-gene browser. This gene was identified as the most significantly differentially expressed gene in a recent, unrelated RNA-Seq study, is not annotated to any GO or pathway term, and very little is known of the function of the CHAC1 protein. The results of our ConceptGen query showed several related genes, including Ddit3 (Chop), Ddit4, Atf3, Cebpb, Trib3 and several tRNA synthetases, as being top ranked as most significantly related. The above-mentioned genes are known to be involved in the unfolded protein response and apoptosis. However, to objectively predict the function of Chac1, we uploaded the 100 top ranking genes into our private account in ConceptGen and determined their enriched concepts. This analysis showed, as expected based on simple observation of the gene list, that apoptosis, amino acid transport, tRNA synthetase activity (a known target group of ATF4), and the activity of Cebpb, ATF4 and Chop/Ddit3 are enriched (Supplementary Fig. S9).
One year ago, the involvement of CHAC1 in these processes would have been a novel hypothesis to test. However, recently the connection between CHAC1 and the ATF4/ATF3/ CHOP cascade and apoptosis was identified experimentally (Mungrue et al., 2009), validating our finding with ConceptGen which was based almost solely on the public microarray data concept type, since Chac1 had no functional annotations and only two protein interactions. Testing CHAC1 in GeneDeck's paralog hunter tool did not result in an enrichment of unfolded protein response or apoptosis genes, however it did identify cation/ion channel and transport activity genes. This demonstrates that these two tools are complementary. Based on these findings, we envision that gene–gene enrichment analysis will be useful for predicting the pathways and processes of other unannotated genes.
4 DISCUSSION
Identifying relationships among differentially expressed gene lists and biological concepts is now known to be very helpful in assessing the biological relevance of microarray experiments and in the study of diseases. The ability to visualize such relationships in a variety of ways further enhances this understanding. While there is an abundance of tools for functional enrichment testing, few currently offer the level of visualization and interactivity desired by many biomedical researchers. We have presented a freely-available web-based software application, ConceptGen, that offers such enrichment testing, visualizations and interactivity using gene set relation mapping and protein interactions. The incorporation of several concept types covering a variety of types of biological knowledge provides more opportunities for hypothesis generation than would testing simply against GO and/or KEGG pathways. These features, together with ConceptGen's easy-to-use interface and private account, make it a desirable complement to related tools such as DAVID/EASE, GSEA, and the concept mapping in Oncomine. The concept mapping in Oncomine is supported by a team of scientists who manually curate the concepts. Oncomine is also useful due to the additional features and visualizations it offers for published microarray data which conveniently links directly to concept mapping. Although Oncomine does have more microarray datasets in their database than ConceptGen, they are limited to cancer-related experiments.
We have demonstrated the performance of ConceptGen with a time series gene expression experiment of TGFβ-induced EMT in lung adenocarcinoma cells, and have shown how ConceptGen can also be used for enrichment testing with a list of metabolites/compounds. Furthermore, we showed how ConceptGen's gene-to-gene enrichment testing based on overlapping concepts can be useful. Future work planned for ConceptGen includes increasing the size of the gene expression database by expanding it to include additional microarray platforms and other experimental data, including experimentally identified transcription factor binding sites from ChIP-seq, and differentially expressed proteins identified from proteomics experiments. In order to allow users to visually compare enrichment results across time points or other experimental conditions, we plan to implement a feature to produce heatmaps similar to that created in Figure 3. Finally, in order to aid in the visualization of networks involving a large number of concepts, we will augment the basic network graph with a power graphing technique. This will group highly correlated concepts into larger meta-concepts that will be easier to grasp visually.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all members of the National Center for Integrative Biomedical Informatics (NCIBI) who contributed to database support, gave us valuable input on improving the user interface, or other discussion, particularly Xiaosong Wang. We thank Jyoti Athanikar for a thorough reading of the manuscript.
Funding: This work was funded by the NIGMS (grant U54 DA021519-01A1) (NCIBI), NIH R01 (CA132571), American Cancer Society (CSM-116801), and MTTC GR687.
Conflict of interest: none declared
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- Berriz GF, et al. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr., et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Draghici S, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, et al. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchterman T.MJ, Reingold EM. Graph drawing by force-directed placement. Software: Pract. Exper. 1991;21:1129–1164. [Google Scholar]
- Gentleman RC. Bioconductor package, GOstats vignette. 2007 http://bioconductor.org/packages/2.1/bioc/vignettes/GOstats/inst/doc/GOstatsHyperG.pdf. [Google Scholar]
- Harris MA, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, et al. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshamouni VG, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- Keshamouni VG, et al. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-induced epithelial-mesenchymal transition. J. Proteome Res. 2009;8:35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- Khatri P, et al. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 2005;33:W762–W765. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, et al. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Medvedovic M, Sivaganesan S. Bayesian infinite mixture model based clustering of gene expression profiles. Bioinformatics. 2002;18:1194–1206. doi: 10.1093/bioinformatics/18.9.1194. [DOI] [PubMed] [Google Scholar]
- Morris DS, et al. Integrating biomedical knowledge to model pathways of prostate cancer progression. Cell Cycle. 2007;6:1177–1187. doi: 10.4161/cc.6.10.4247. [DOI] [PubMed] [Google Scholar]
- Mungrue IN, et al. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J. Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, et al. Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor MA, et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:538. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, et al. The prolyl 3-hydroxylases P3H2 and P3H3 are novel targets for epigenetic silencing in breast cancer. Br. J. Cancer. 2009;100:1687–1696. doi: 10.1038/sj.bjc.6605042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stelzer G, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- Valouskova E, et al. Redistribution of cell death-inducing DNA fragmentation factor-like effector-a (CIDEa) from mitochondria to nucleus is associated with apoptosis in HeLa cells. Gen. Physiol. Biophys. 2008;27:92–100. [PubMed] [Google Scholar]
- Vellaichamy A, et al. Proteomic interrogation of androgen action in prostate cancer cells reveals roles of aminoacyl tRNA synthetases. PLOS ONE. 2009;4:e7075. doi: 10.1371/journal.pone.0007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC.Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.