Summary
The lineage restriction of prospectively isolated hematopoietic progenitors has been traditionally assessed by bulk in vitro culture and transplantation of large number of cells in vivo. These methods, however, cannot distinguish between homogenous multipotent or heterogeneous lineage-restricted populations. Using clonal assays of 1 or 5 cells in vitro, single cell quantitative gene expression analyses, and transplantation of mice with low numbers of cells, we show that the common myeloid progenitor (CMP) is Sca-1lolin-c-Kit+CD27+Flk-2- (SL-CMP; Sca-1lo CMP) and a granulocyte/macrophage progenitor (GMP) is Sca-1lolin-c-Kit+CD27+Flk-2+CD150-/lo (SL-GMP; Sca-1lo GMP). We found that mast cell progenitor potential is present in the SL-CMP fraction but not in the more differentiated SL-GMP population and is more closely related to megakaryocyte/erythrocyte specification. Our data provide criteria for the prospective isolation of SL-CMP and SL-GMP and support the conclusion that mast cells are specified during hematopoiesis earlier than and independently from granulocytes.
Introduction
The hematopoietic hierarchy that maps the differentiation of the self-renewing hematopoietic stem cells (HSCs) into mature blood and immune cells has been further developed and refined ever since the initial identification of HSCs by surface phenotype (Spangrude, 1988). However, the accurate positioning of mast cells in the hematopoietic cell differentiation scheme had remained undetermined. Mast cells are important effector and immunoregulatory cells derived from HSCs (Galli et al., 2005; Kitamura, 1989; Kitamura et al., 1977). Although they were initially studied for their role in the pathogenesis of allergic and other inflammatory diseases, mast cells are increasingly viewed as being able to perform a spectrum of positive and negative immunoregulatory functions that can either exacerbate disease or help to sustain health (Bischoff, 2007; Galli et al., 2005; Kalesnikoff and Galli, 2008; Rao and Brown, 2008). Consequently, a clear understanding of the developmental pathway from HSCs to mature mast cells may facilitate the design of new approaches to alter mast cell development or function, in order to treat disease or promote health.
Still, there is considerable controversy regarding the developmental pathway of mast cells. Some findings suggest relationships among erythroid, megakaryocytic and mast cell lineages (Martin et al., 1990; Ogawa, 1989; Suda et al., 1983). Other evidence supports the existence of progenitors that can give rise only to mast cells and macrophages (Suda et al., 1983), to mast cells, neutrophils and macrophages (Suda et al., 1983), or to mast cells and basophils (Arinobu et al., 2005), observations which are consistent with the conclusion that mast cells are derived from GMP. By contrast, we identified a clonogenic mast cell progenitor (MCP) in adult mouse bone marrow and provided evidence that MCPs are derived from the early multipotent progenitor (MPP) and possibly from the CMP, but not from the GMP or the megakaryocyte/erythroid (Meg/E) progenitor (MEP) (Chen et al., 2005).
In the present study, we pursued the idea that a more coherent picture of the origin of mast cells might be achieved by increasing the resolution of progenitor-progeny lineage determination assays. We sorted 1 or 5 hematopoietic progenitor cells and examined their lineage readout in vitro, tested the lineage readout of 20-30 cells in vivo, and carried out single cell multigene quantitative real-time PCR (qRT-PCR). We report evidence that mast cells are derived from a Sca-1lo-CMP (SL-CMP) but not from the SL-GMP. Our findings suggest that mast cell development is most closely associated with the Meg/E lineage. Our results also illustrate the importance of analyzing the in vitro differentiation potentials of single progenitors, rather than populations of progenitors that may contain heterogeneous cell types.
Results
Mast cells can be derived in vitro from single Sca-1lo CMP and single Sca-1- β7+ cells, but not from single Sca-1lo GMP
We investigated the lineage potential of Sca-1-lin-c-Kit+ (SN; “Sca-1-”) cells after subdividing them into four fractions based on the expression of CD27, Flk-2, CD150 and β7-integrin and plating them at 1 or 5 cells per well in a cytokine cocktail which promotes full erythromyeloid potential (Figures 1A,B and Table S1). Using two plating densities allowed assessment of the in vitro differentiation potential and heterogeneity of each population. We analyzed individual wells by flow cytometry and cytospin at day 7 (D7) (Figure S1) and D14 (not shown). Consistent with previous findings (Pronk et al., 2007), 1 or 5 SN-CD150+ cells (designated “MEP” in Figure 1B) almost exclusively generated Meg/E lineage cells (Figure 1B). By D14, SN Flk-2-CD150-β7-integrin+ (SN-β7+) cells gave rise only to Meg/E or mast cells. Cultures of 5 SN-β7+ cells also exhibited a few c-Kit-FcεRIα+ colonies at D7, but such cells were not detectable at D14.
Figure 1. Evidence that Sca-1-lin-c-Kit+ (SN, Sca-1 negative) cells have already committed to the GM, Meg/E, or mast cell lineage.
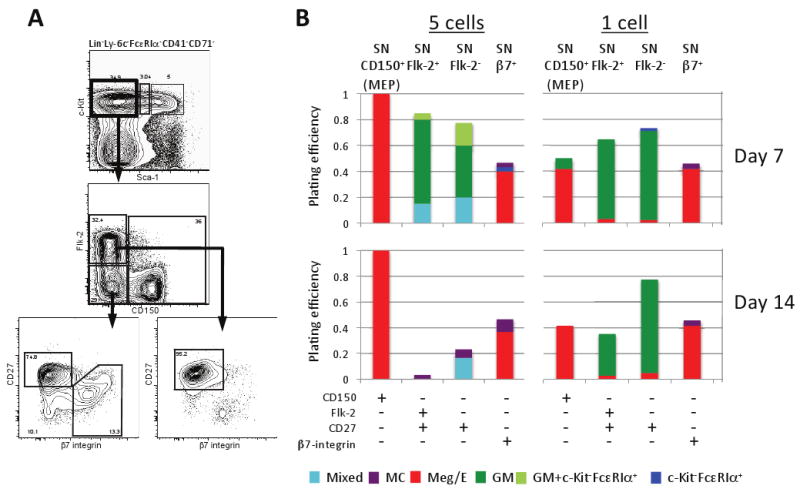
(A) SN cells were sorted into four populations based on surface expression of Flk-2, CD150, CD27, and β7-integrin. (B) Of the four populations that were able to expand in vitro, SN CD150+ (MEP) cells were committed to the Meg/E lineage whereas the two SN CD27+ populations were mostly restricted to GM potential. Mast cells were primarily derived from the SN β7+ population, which contains the previously reported mast cell progenitor (MCP). The results of Chi-square analyses comparing the colony output results (i.e., proportion of various types of colonies derived from 1 or 5 cell assays at day 7 or 14) for each of the types of SN progenitor cells tested are summarized in Table S1. See legend of Figure S1 for definitions of: Mixed, MC, Meg/E, GM and GM + c-Kit-/FcεRIα+ populations; “c-Kit/FcεRIα+” colonies contained only that population.
The fact that SN-β7+ cells (which contains the previously defined MCP [Chen et al., 2005]) gave rise only to Meg/E or mast cells, but not GM lineage cells, supports the idea that the mast cell and GM lineages are committing independently and is consistent with other evidence suggesting that mast cell potential more closely associates with the megakaryocyte and erythrocyte pathway (Martin et al., 1990; Ogawa, 1989). Of the remaining fractions, SN Flk-2-CD150-CD27+ (SN-Flk-2-) and SN Flk-2+CD150-CD27+ (SN-Flk-2+) cells revealed a strong GM bias even in 5-cell D7 wells; SN-Flk-2+ and SN-Flk-2- cells had a GM efficiency of 70% and 57.5%, respectively (Figure 1B).
SN cells generated mixed potential colonies less efficiently than SL cells, particularly at D14 or from a single cell (see below, and compare Figures 1B and 2B). Notably, the single SN cells that did expand were already committed to one lineage, predominately GM or Meg/E. When single cells were tested, the SN populations did not generate mixed lineage colonies at day 7, yet SN subsets were mainly, but not absolutely, biased towards Meg/E or GM outcomes; it is conceivable some CMPs that commit early to one or more potential are in the SN subset, or that these represent GM or Meg/E committed cells within the SN gates. While we can't exclude the possibility that some CMP-like activity resides in the SN populations that were analyzed, we favor the interpretation that any mixed potential observed in wells derived from 5 SN cells reflects the inability of our panel of surface markers to isolate pure populations, but not that SN cells are truly oligopotent.
Figure 2. Single cell in vitro analysis reveals that lineage commitment is already initiated in the Sca-1lolin-c-Kit+ (SL) bone marrow fraction.
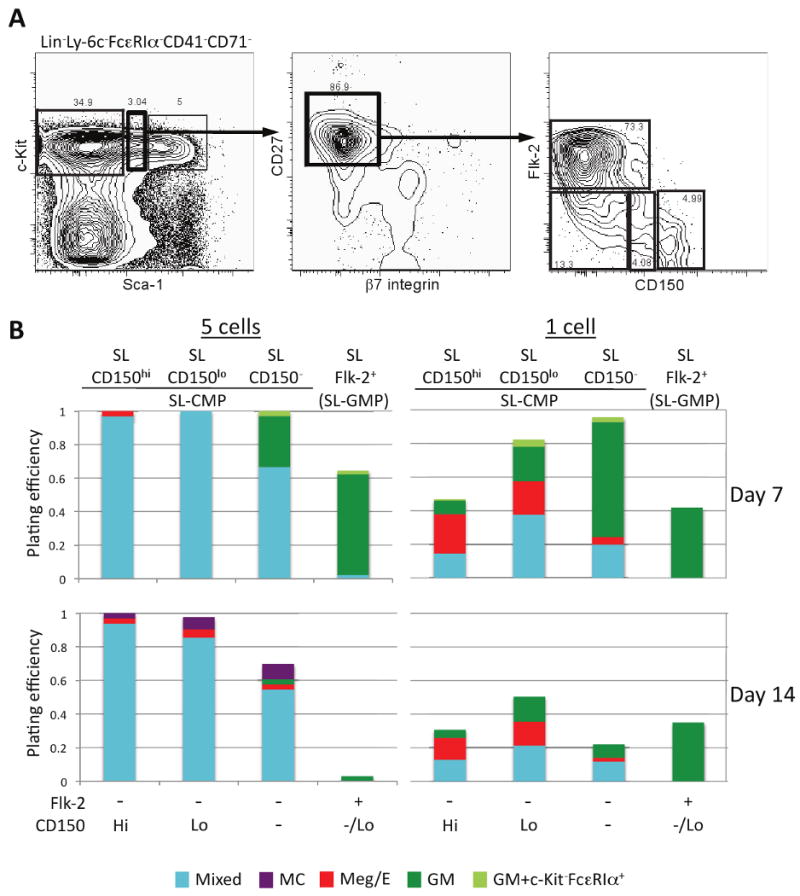
(A) SL bone marrow progenitor cells were sorted into four populations based on surface expression of CD27, β7-integrin, Flk-2, and CD150. (B) The four CD27+ fractions were able to expand in vitro at both a 5 cell and 1 cell per well density. The SL CD150lo fraction retained the most mixed lineage potential at each plating density, suggesting that this is the most immature population. However, SL Flk-2+ cells were already restricted to the GM lineage, notably without any detectable mast cell potential. The data in B summarize all the experiments we performed. See Table S2 for the total number of wells analyzed and the numbers of individual sorts performed for each progenitor population. For the initial 3-4 sorts, we always analyzed wells plated with single cells and 5 cells; for the remaining 2-3 sorts, we analyzed only wells seeded with single cells. See Table S3 for the results of Chi-square analyses comparing colony output (i.e., proportion of various types of colonies derived from 1 or 5 cell assays at day 7 or 14) for the various SL progenitor cells.
CMPs were originally defined as within the Sca-1-lin-c-Kit+ fraction of mouse bone marrow cells (Akashi et al., 2000), but the application of improved antibody labeling and flow cytometric separation technologies, especially using the Sca-1 monoclonal antibody (mAb), has questioned this definition (Pronk et al., 2007; Arinobu et al., 2007). Hypothesizing that some lineage restriction might occur within the Sca-1lolin-c-Kit+ fraction (Sca-1lo cells were formerly contained within the Sca-1- gate; new stains reveal Sca-1lo and Sca-1- subsets), we plated four fractions of Sca-1lolin-c-Kit+ cells, based on expression of CD27, β7-integrin, CD150 and Flk-2 (Figure 2).
Among the Sca-1lolin-c-Kit+CD27+ (SL; “Sca-1lo”) cells analyzed, SL Flk-2-CD150lo cells (SL-CD150lo in Figure 2B) had the greatest mixed lineage potential at D7; there also was substantial mixed lineage potential in SL Flk-2-CD150hi and SL Flk-2-CD150- cells (SL-CD150 hi and SL-CD150- in Figure 2B) but with greater bias towards Meg/E or GM lineages, respectively, suggesting that these could be transitional populations (Figure 2B and Tables S2 and S3). Mast cells were present at D14 in most wells seeded with Flk-2- SL-CD150lo, SL-CD150hi or SL-CD150- cells that were scored on D7 or D14 as having mixed lineage potential, including those derived from 1 or 5 cells (Table S4), and wells plated with 5 cells included some which, at D14, contained only mast cells or only cells with Meg/E-restricted potential (Figure 2B). By contrast, SL-Flk-2+ (“SL-GMP” in Figure 2B) cells yielded almost exclusively GM colonies, and no mast cells (Table S4), at D7 or D14. These data support the conclusions that: (1) lineage specification is initiated in the SL fraction, (2) Flk-2- SL-CD150lo cells may be the least committed but all 3 SL-Flk-2- populations (SL-CMP in Figure 2B) contain CMP-like activity, and (3) expression of Flk-2 in SL cells is associated with a more GM-restricted potential.
In experiments requiring live cells, mast cells are commonly identified by flow cytometry as c-Kit+FcεRIα+ and basophils as c-Kit-FcεRIα+ (Arinobu et al., 2005; Chen et al., 2005). However, there is evidence that some cells in the mast cell lineage can be c-Kit-FcεRIα+ (Baroni et al., 2007). SN-β7+ cells, which contain the MCP (Chen et al., 2005), were the only cells that generated mast cells at the clonal level by D7 (Figure 1B). By contrast, of the more immature SL populations, only SL-Flk-2+ cells were unable to generate mast cells by D14 (Figure 2B). Intriguingly, other SL fractions produced c-Kit-FcεRIα+ cells at low efficiency by D7, but such cells were undetectable at D14 (Figures 2B, S2A,B). Morphologically, c-Kit-FcεRIα+ cells resembled immature myeloblasts with few to no metachromatic granules (not shown). Most of the wells that contained c-Kit-FcεRIα+ cells at D7 generated c-Kit+FcεRIα+ mast cells by D14 (all of these D14 populations lacked c-Kit-FcεRIα+ cells) (Figure S2B). Moreover, the great majority of wells that had generated c-Kit+FcεRIα+ mast cells by D14 had contained c-Kit-FcεRIα+ cells at D7 (Figure S2C). Because mast cells but not basophils can thrive in culture for long periods of time (Dvorak et al., 1994), it is possible that the few wells with c-Kit-FcεRIα+ cells at D7, which did not generate mast cells by D14 (Figure S2C), truly had contained basophils.
We think that the simplest interpretation of our data (particularly those in Figure S2D), is that at least some mast cells go through a c-Kit-FcεRIα+ stage of development in vitro but, if cultured long enough, such populations will upregulate c-Kit expression. Moreover, the occurrence of a c-Kit-FcεRIα+ stage of mast cell development could explain the report of a bipotent basophil-mast cell progenitor in cultured mouse spleen cells, in which basophils were identified as cells that were c-Kit-FcεRIα+ (Arinobu et al., 2005).
Confirmation of the lineage potential of Sca-1lo CMP and Sca-1lo GMP in vivo
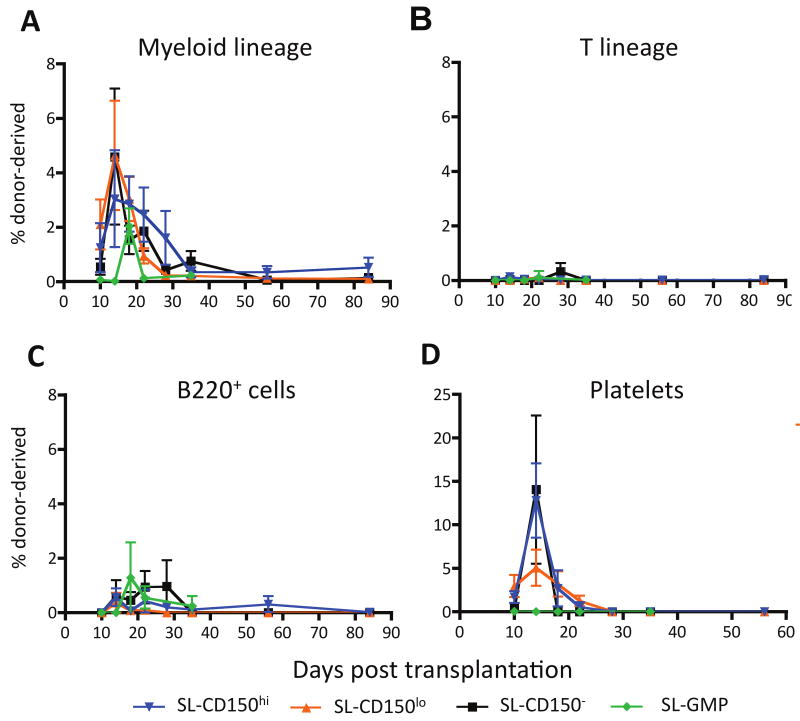
We competitively transplanted 20-30 congenic IL-7Rα- SL cells with 3×105 whole bone marrow cells intravenously into lethally-irradiated mice. Each of the four SL subsets gave robust, yet transient, myeloid engraftment (Figure 3A). SL-CD150lo and SL-CD150- (GM-biased) cells had the most efficient myeloid engraftment, peaking with 4.6% of the total myeloid contribution at D14, but myeloid cells were undetectable by D56 (Figure 3A). SL-CD150hi cells, which have more Meg/E than GM potential in vitro (Figure 2B), also produced transient engraftment that peaked at D14, but with a lower engraftment efficiency of 3%. SL-Flk-2+ (SL-GMP) cells peaked later (D18) with an efficiency of 2%.
Figure 3. Very low cell number in vivo transplantation shows that the Sca-1lolin-c-Kit+ (SL) population contains the common myeloid progenitor (SLCMP) and the granulocyte/macrophage progenitor (SL-GMP) but no long-term engraftment ability.
(A) All four IL-7Rα- SL subsets gave robust short-term myeloid engraftment with SL CD150lo and SL CD150- populations contributing the most. (B) None of the four populations contained detectable T cell potential. (C) However, a modest transient B220+ cell potential was observed from Flk-2+ SL-GMP and Flk-2- SL CD150- cells (B220+ cell output was observed in 3 out of 10 and 4 out of 18 of the mice injected with these cells, respectively, and from 3 out of 4 batches of progenitors tested). By contrast, for mice injected with SL CD150lo cells, such potential was observed in 2 out of 25 mice, from 1 out of 5 batches of progenitors tested). (D) All three Flk-2- populations had strong platelet potential in vivo, but Flk-2+ SL-GMP cells had none.
None of these IL-7Rα- SL populations exhibited lymphoid potential when cultured in vitro on OP9 or OP9-Dl1 stromal cells (Nakano et al., 1994; Schmitt et al., 2004) (Figure S3A). In agreement with our in vitro results, none of the SL subsets had in vivo T cell potential by D84 (Figure 3B), and intrathymic transplantation of isolated populations of SL-CD150lo cells, MPP and CLP into unconditioned hosts revealed substantial lymphoid output from most of the injected populations of MPP and CLP, but little or no lymphoid potential from the SL-CD150lo cells (Figure S3B). However, some B220+ cells (representing B cells and/or B220+ dendritic cells) appeared transiently in mice injected intravenously with SL cells, particularly the SL-CD150- and SL-Flk-2+ fractions (Figure 3C), perhaps reflecting imperfect exclusion of IL7Rα+ cells.
To track the in vivo platelet potential of each population, we transplanted 20-30 GFP+ IL7Rα- SL cells into lethally irradiated hosts (Figure 3D). At their D14 peak (Figure 3D), we observed platelet engraftment from all three Flk-2- SL fractions. No platelet contribution from any of the four SL fractions was detectable by D28, and SL-GMP cells had no Meg/E potential.
Notably, the SL fractions did not give long-term reconstitution, supporting the conclusion that SL populations contain progenitors but not HSCs. The engraftment data thus validate the findings of our in vitro culture system regarding the lymphoid and myeloid potential of the analyzed populations. Most significantly, both the in vitro and in vivo data support the conclusion that SL-Flk-2- cells (especially those that are CD150lo) and SL-Flk-2+ cells constitute newly defined SL-CMP and SL-GMP populations, respectively.
Analysis of lineage-related gene expression in single hematopoietic cells
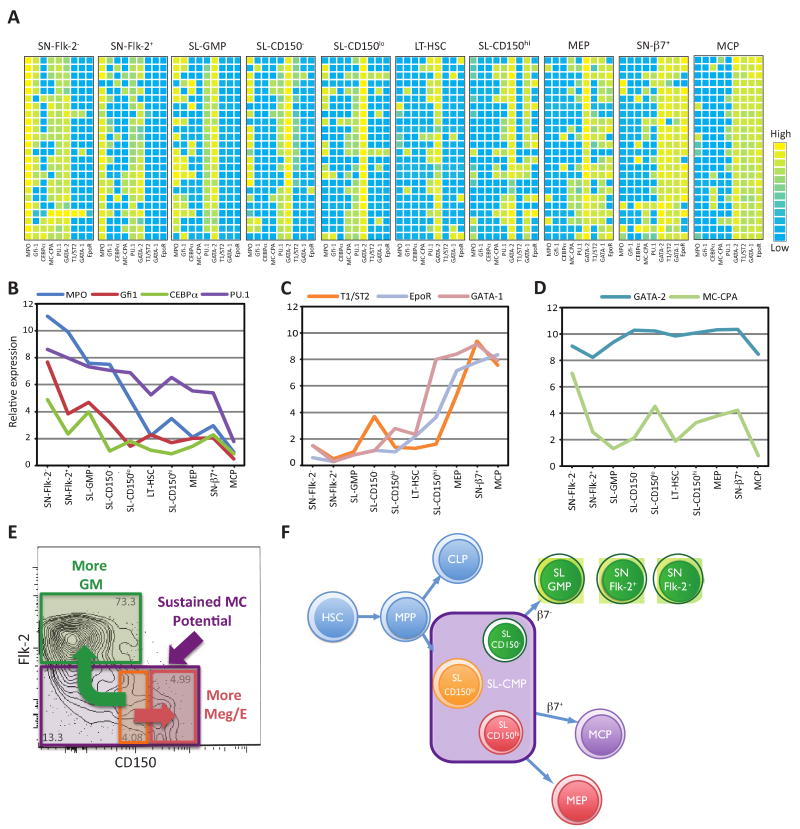
Even in progenitor populations that have been highly purified (Figure S4A), single cell analysis may be required to dissect progenitor lineage potentials (Figure S4B). We therefore devised a method for single cell quantitative real time PCR (qRT-PCR) to characterize the expression of nine lineage-related genes in single SL and SN progenitors, long-term HSCs (LT-HSC, Sca-1hilin-c-Kit+Flk-2-CD150+) and MCPs (Chen et al., 2005) (Figure 4A). Previous reports using conventional assays have suggested that HSCs and other early progenitors “promiscuously” express genes of committed lineages at low levels (Miyamoto et al., 2002). Our data indicate that most LT-HSCs have low to undetectable levels of lineage-related genes while a small fraction of cells express them highly, findings that could explain low-level detection of such genes in bulk assays.
Figure 4. Quantitative single cell gene expression analyses align with results of in vitro culture and in vivo transplantation studies to define the SL-CMP and the SL-GMP and to indicate that mast cells are generated independently of granulocytes and macrophages during hematopoiesis.
(A) qRT-PCR of 24 single cells of each SL or SN progenitor population shows that expression of lineage-related genes correlates with observed lineage potential in vitro and in vivo. Each row within a given population represents expression in an individual cell. Analysis using paired two-way ANOVA revealed that the results for each population were significantly different from those for any other population at the p < 0.0001 level except for LT-HSC versus SL-CD150lo cells (p = 0.0049). (B, C, D) Average expression levels of lineage-related genes within each population. The formula used to calculate average gene expression levels takes into account expression levels of GAPDH as a control. The calculated average expression level of MC-CPA in MCP (in which the transcript was detected in 2/24 cells tested) is slightly lower than the value calculated for SL-GMP (in which the transcript was detected in none of the 24 cells tested) because GAPDH levels were higher in MCP than SL-GMP. (E) Mast cell potential exists in all Flk-2- SL-CMP cells, including SL-CD150lo cells, the Meg/E-biased SL-CD150hi cells, and GM-biased SL-CD150- cells. The SL-GMP is defined as SL-Flk-2+. (F) SL-CMP cells have full myeloerythroid potential but lack the ability to generate lymphoid cells. The SL-GMP, SN-Flk-2+ and SN-Flk-2− populations can give rise to granulocytes and macrophages, but not mast cells. No Meg/E lineage potential was observed in the SL-GMP, and the minimal Meg/E lineage potential observed in the SN-Flk-2+ and SN-Flk-2− populations (Figure 1B) may have reflected minor contamination of those populations. The mast cell progenitor (MCP) and Meg/E progenitor (MEP) are Sca-1-lin-c-Kit+ (SN) cells derived from the SL-CMP.
Our findings are consistent with the possibility that lineage restriction begins in SL progenitors and progresses in SN progenitors (Figure 4A), results mirroring our in vitro and in vivo assessments of lineage potential. However, other explanations of the data have not been ruled out, such as transient expression of various phenotypes by cells with GM potential. Gata-2, the most widely expressed of the genes analyzed, is necessary for survival and cell cycle regulation of progenitor populations (Ling et al., 2004; Rodrigues et al., 2005; Tipping et al., 2009). PU.1 was commonly detected in LT-HSCs, and myeloperoxidase (Mpo) was expressed moderately in SL-CD150lo cells. Expression of both genes increased with greater GM potential and decreased with Meg/E or MC potential. Levels of Mpo, C/EBPα, PU.1, and Gfi-1 expression rose with increased GM potential, with SL-CD150- SL-GMP, SN-Flk-2+ and SN-Flk-2- populations having progressively higher expression (Figures 4A,B). SN-Flk-2- cells expressed the highest levels of Mpo, C/EBPα, and Gfi-1, suggesting that these cells have a lower macrophage potential than SN-Flk-2+ cells (Arinobu et al., 2005; Laslo et al., 2006).
Mast cell carboxypeptidase A3 (MC-CPA) was first reported as a mast cell-associated gene (Reynolds et al., 1989) and is expressed in a mast cell-committed precursor identified in the blood of fetal mice (Rodewald et al., 1996), but this gene is also expressed in thymocytes (Feyerabend et al., 2009). We detected little MC-CPA expression in bone marrow-derived MCPs, but substantial expression in progenitors with either Meg/E or GM potential (Figures 4A, D). Thus, our single cell analyses support newer evidence that MC-CPA is not a mast cell-specific gene, and indicate that there is little or no expression of MC-CPA in bone marrow MCPs.
Gata-1 is thought to influence Meg/E lineage differentiation through its interactions with FOG-1 (Iwasaki et al., 2003). Both Gata-1 and the erythropoietin receptor (EpoR), were detected in SL-CD150hi cells and further elevated in MEPs and SN-β7+ cells (Figures 4A, C). T1/ST2, a marker of MCP (Chen et al., 2005), was also detected in MEPs and SN-β7+ cells, but not in SL-CD150hi cells. MCPs expressed T1/ST2, Gata-1, and EpoR, but not the GM-related genes.
Discussion
In this study we show that the identities of our newly defined SL-CMPs (Sca-1lolin-c-Kit+CD27+Flk-2-) and SL-GMPs (Sca-1lolin-c-Kit+CD27+Flk-2+CD150-/lo) are supported by three independent approaches—in vitro cultures, in vivo transplantations, and quantitative single cell gene expression analyses. The SL populations that were analyzed had little or no detectable lymphoid potential when IL7Rα+ cells were depleted. Our data indicate that hematopoietic lineage commitment is initiated in the Sca-1lolin-c-Kit+ bone marrow fraction, in which Meg/E potential is positively correlated with CD150 expression, while GM potential increases with a loss of CD150 and gain of Flk-2 expression (Figures 4E, F). SL-CD150hi and SL-CD150- cells may be Meg/E- or GM-biased transitional populations, respectively, but each still can generate mast cells (Figures 2B and 4).
Our single cell assays show that the purity of a population based on a given surface profile does not necessarily imply that it has homogeneous lineage potential. Such “purity” is solely based on the arbitrary choice of markers and flow cytometry gates used by the researcher and not necessarily on the developmental potential of the cells. This issue was obvious even in the extremely rare SL Flk-2- fractions (representing approximately 0.001-0.002 % of the total bone marrow cell population), which had a high heterogeneity of lineage potential when plated at 5 cells as compared to 1 cell per well. We hope that the approaches described herein will allow for a much finer resolution of many rare cell populations, not just hematopoietic progenitors, including the isolation of tissue-specific stem cells.
The striking finding that some individual HSCs express lineage-related transcription factors at high levels (as opposed to all of these cells expressing these factors at low levels) would not be possible without using multigene qRT-PCR measurements in single cells. The HSCs analyzed appear to express either GM or Meg/E gene profiles to the exclusion of the other. It remains to be determined whether this finding represents an indication of lineage bias within the LT-HSC compartment. Because we can not analyze the lineage potential of a single cell and still lyse it for qRT-PCR, it is unclear whether such a pattern of gene expression is reflective of commitment bias initiated earlier than previously thought. However, it is intriguing to consider the possibility that lineage commitment mechanisms could be initiated prior to the loss of self-renewal potential.
The development of a method to quantify the expression of multiple genes in a single cell permits us to describe in more detail the phenotype of cells undergoing lineage determination, and may also facilitate a better understanding of the mechanisms which contribute to cell fate determination. However, the analysis of gene expression in various progenitor cells does not permit us to characterize the contributions of the individual genes in the lineage commitment process. Detailed functional studies are needed to address this point. In the present study, we found that our single cell gene expression measurements strongly supported the notion emerging from the SN β7+ cell culture data of a possible mast cell-Meg/E lineage relationship. We do not know whether these findings reflect the existence of a common mast cell and Meg/E progenitor, or the usage of similar molecular mechanisms of differentiation by two independent lineages. However, our results do support the notion, first proposed years ago (Suda et al. 1983, Ogawa 1989, Martin et al. 1990), that mast cell and Meg/E potential track closely during hematopoiesis. Our results also strongly indicate that mast cells are generated independently of the granulocyte and macrophage lineages.
Experimental Procedures
Mice
C57BL/Ka-Thy1.1 (CD45.2), C57BL/Ka-Thy1.1-Ly5.2 (CD45.1), and β-actin GFP transgenic (Wagers et al., 2002) mice (4-10 weeks old) were used for the isolation of progenitors. All animals were maintained in the Stanford University Laboratory Animal Facility in accordance with Stanford Animal Care and Use Committee and National Institutes of Health guidelines.
Flow Cytometry and Cell Sorting
Bone marrow was harvested from 8-10 week old mice and homogenized with a 20-guage needle to a single cell suspension. The suspension was subjected to red blood cell lysis in a hypotonic solution. The remaining bone marrow cells were depleted for the lineage markers CD3, CD4, CD5, CD8, B220, Gr-1, Mac-1/CD11b, and Ter119 by MACS LD columns (Miltenyi Biotec). LT-HSC, SL and SN progenitors were sorted on a FACS Aria (Becton Dickinson) using the labeled monoclonal antibodies CD3-Pacific Blue, CD4-Pacific Blue, CD8-Pacific Blue, CD8-Pacific Blue, Mac-1/CD11b-Pacific Blue, B220-Pacific Blue, Ter119-Pacific Blue, Gr-1-Pacific Blue, Sca-1-PE/Cy5.5, β7-integrin-PE, c-Kit-Alexa750, CD150-PE/Cy5, Ly6c-FITC, FcεRIα-FITC, CD71-FITC, CD41-FITC, CD27-APC, IL-7Rα-Alexa700, CD135-Biotin, SA-PE/Cy7. The Sca-1lo gate was set such that the lower end had a fluorescence intensity of approximately 1.0 × 103 units and the upper end extended to approximately 1.1 × 103. The appropriateness of the Sca-1lo gate was verified by establishing that it contained approximately 2-5% of β7-integrin+ cells. This gating method is very easy to set up in FACSDiva software. By comparing in vitro culture and single cell qPCR results from different sorts, we find that our gating methods produce highly consistent and reproducible results. The CD150lo gate for SL-CMP was set to comprise a symmetric region around 1.0 × 103 fluorescence units, which contained approximately 4 -6% of all the SL cells. The upper and lower ends of the gate were set to coincide roughly with the boundaries of the intermediate peak between the positive and negative peaks on CD150 histogram. Typically, this region would fall directly below the fraction of SL-GMP with the highest CD150 expression. MCP were sorted as previously described (Chen et al., 2005). Re-analysis of the sorted cells showed that each population was cleanly isolated (Figure S4A). All antibodies were conjugated in-house or purchased from eBioscience, Becton Dickinson, or Biolegend.
Clonal in vitro culture of progenitors
Single cells were sorted using a FACS Aria (Becton Dickinson) into 96-well round bottom plates containing growth media supplemented with 10% FCS, and 10 ng/ml each of IL-1α, IL-3, IL-5, IL-6, IL-7, IL-9, IL-10, IL-11, GM-CSF, TPO, EPO, SCF, and Flt3L. After seven days in culture at 37°C, half of each well was removed from culture and the remaining half was supplemented with fresh media and growth factors. The half that was removed was split into two parts, half was analyzed by flow cytometry on a FACS Aria (Becton Dickinson) and the other half was used for cytospin followed by May-Grunwald-Giemsa staining.
Low cell number in vivo reconstitution
20-30 progenitor cells from the mice described above, together with 105 congenic host whole bone marrow cells, were injected i.v. into lethally-irradiated (800 rads) congenic recipients. Peripheral blood was drawn at indicated time points and red blood cells were removed using Lympholyte (Cedarlane Labs) to facilitate leukocyte analysis by flow cytometry.
Single cell multi-gene quantitative real-time PCR
Single cells were sorted using a FACS Aria (Becton Dickinson) into tubes containing a 10 ul mix of CellsDirect One-Step qRT-PCR Kit (Invitrogen) and a 0.2× pool of TaqMan probes (Applied Biosystems) for the genes of interest. Gene probes used were Gata-1 (Mm01352636_m1), Gata-2 (Mm03053564_s1), PU.1 (Mm00488140_m1), Gfi-1 (Mm00515855_m1), CEBPα (Mm00514283_s1), Mpo (Mm01298424_m1), EpoR (Mm00438760_m1), T1/ST2 (Mm00516117_m1), Cpa3 (MC-CPA) (Mm00483940_m1), and GAPDH (Mm99999915_g1). Cells underwent reverse transcription followed by 22 cycles of pre-amplification. The resulting amplified cDNA was analyzed for quantitative gene expression levels with the aforementioned TaqMan probes and TaqMan Gene Expression Master Mix (Applied Biosystems) on an ABI-7900HT (Applied Biosystems). Results were analyzed using the dCt method normalized to GAPDH. Relative expression = log(2-(Ct_gene-Ct_GAPDH))+11, where 11 is an arbitrary value to make all relative expression values greater than zero.
Supplementary Material
Acknowledgments
We thank members of the I.L.W. and S.J.G. laboratories for helpful discussions, particularly Rong Lu, Thomas Serwold, and Jun Seita. We also thank Libuse Jerabek for excellent lab management, Christina Muscat for antibody conjugation and Adriane Mosely for animal husbandry.
This work was supported by NIH grants AI23990, AI070813 & CA72074 (to S.J.G.), and CA086065 & HL058770 (to I.L.W.), a National Science Foundation Graduate Research Fellowship (to C.B.F.), an NIH Pathway to Independence award 5K99HL087936 (to C.-C.C.), and a Human Frontiers Science Program postdoctoral fellowship (to M.D.).
Footnotes
Supplemental Information: Supplemental information includes four supplemental figures and four supplemental tables, and can be found with this article online at …
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Baroni E, Biffi M, Benigni F, Monno A, Carlucci D, Carmeliet G, Bouillon R, D'Ambrosio D. VDR-dependent regulation of mast cell maturation mediated by 1,25-dihydroxyvitamin D3. J Leukoc Biol. 2007;81:250–262. doi: 10.1189/jlb.0506322. [DOI] [PubMed] [Google Scholar]
- Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak AM, Seder RA, Paul WE, Morgan ES, Galli SJ. Effects of interleukin-3 with or without the c-kit ligand, stem cell factor, on the survival and cytoplasmic granule formation of mouse basophils and mast cells in vitro. Am J Pathol. 1994;144:160–170. [PMC free article] [PubMed] [Google Scholar]
- Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, Gale NW, Radtke F, Fehling HJ, Rodewald HR. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- Kalesnikoff J, Galli SJ. New developments in mast cell biology. Nat Immunol. 2008;9:1215–1223. doi: 10.1038/ni.f.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura Y. Heterogeneity of mast cells and phenotypic change between subpopulations. Annu Rev Immunol. 1989;7:59–76. doi: 10.1146/annurev.iy.07.040189.000423. [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Shimada M, Hatanaka K, Miyano Y. Development of mast cells from grafted bone marrow cells in irradiated mice. Nature. 1977;268:442–443. doi: 10.1038/268442a0. [DOI] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Zon LI, Mutter G, Orkin SH. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990;344:444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- Ogawa M. Effects of hemopoietic growth factors on stem cells in vitro. Hematol Oncol Clin North Am. 1989;3:453–464. [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Rao KN, Brown MA. Mast cells: multifaceted immune cells with diverse roles in health and disease. Ann N Y Acad Sci. 2008;1143:83–104. doi: 10.1196/annals.1443.023. [DOI] [PubMed] [Google Scholar]
- Reynolds DS, Stevens RL, Gurley DS, Lane WS, Austen KF, Serafin WE. Isolation and molecular cloning of mast cell carboxypeptidase A. A novel member of the carboxypeptidase gene family. J Biol Chem. 1989;264:20094–20099. [PubMed] [Google Scholar]
- Rodewald HR, Dessing M, Dvorak AM, Galli SJ. Identification of a committed precursor for the mast cell lineage. Science. 1996;271:818–822. doi: 10.1126/science.271.5250.818. [DOI] [PubMed] [Google Scholar]
- Rodrigues NP, Janzen V, Forkert R, Dombkowski DM, Boyd AS, Orkin SH, Enver T, Vyas P, Scadden DT. Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood. 2005;106:477–484. doi: 10.1182/blood-2004-08-2989. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zuniga-Pflucker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Suda T, Suda J, Ogawa M. Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci U S A. 1983;80:6689–6693. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping AJ, Pina C, Castor A, Hong D, Rodrigues NP, Lazzari L, May GE, Jacobsen SE, Enver T. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113:2661–2672. doi: 10.1182/blood-2008-06-161117. [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Allsopp RC, Weissman IL. Changes in integrin expression are associated with altered homing properties of Lin-/loThy1.1-/loSca-1+c-kit+ hematopoietic stem cells following mobilization by cyclophosphamide/granulocyte colony-stimulating factor. Exp Hematol. 2002;30:176–185. doi: 10.1016/s0301-472x(01)00777-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.