Abstract
Pax genes are a group of critical developmental transcriptional regulators in both invertebrates and vertebrates, characterized by the presence of a paired DNA-binding domain. Pax proteins also often contain an octapeptide motif and a C-terminal homeodomain. The genome of Nasonia vitripennis (Hymenoptera) has recently become available, and analysis of this genome alongside Apis mellifera allowed us to contribute to the phylogeny of this gene family in insects. Nasonia, a parasitic wasp, has independently evolved a similar mode of development to that of the wellstudied Drosophila, making it an excellent model system for comparative studies of developmental gene networks. We report the characterization of the seven Nasonia Pax genes. We describe their genomic organization, and the embryonic expression of three of them, and uncover wider conservation of the octapeptide motif than previously described.
Keywords: Pax, Nasonia, evolution, development, octapeptide
Introduction
The jewel wasp Nasonia vitripennis is a hymenopteran that undergoes a long-germ mode of embryogenesis, similar to that of Drosophila. However, this mode of development where all segments are laid down at the same time, was likely acquired independently in the two species after diverging from their last common ancestor an estimated 280 million years ago (Savard et al., 2006). This similarity prompted the study of genes controlling the early development of Nasonia, and revealed that while many of the genes utilized in flies are also used in Nasonia, and their interactions are preserved, there are significant differences that allow development to proceed in the context of this different embryo (Pultz et al., 1999; Lynch et al., 2006a,b; Olesnicky et al., 2006; Brent et al., 2007). Since the mode of development shared by flies and wasps converged, these studies have provided the foundation for an understanding of the independent evolution of a derived developmental process from an ancestral one. Comparative studies of additional genes or gene families, particularly those that function at conserved nodes in developmental programmes, have the potential to provide similar information about the life histories of both those gene families and their host genomes.
Gene families like the Hox or Pax families contain multiple genes thought to function as critical regulators in development, and arose through expansions and subsequent diversification of function after gene duplications. These families may arise either by gene duplication within a species, creating a redundant gene without the constraints of performing essential functions, or after a speciation event, when a species is divided into two populations subject to different selective pressures, and thus parallel evolution of similar genes occurs. Sequence homology and phylogenetic analysis help to map the relationships and time-scheme of divergence between genes in such families.
The paired-domain (Pax) family of transcription factors is an ancient gene family whose members play crucial roles in developmental processes such as eye formation and neural development, organogenesis and segmentation (reviewed in (Noll, 1993). At least four ancestral Pax genes likely existed at the time of the protostome/deuterostome split (700 million years ago) (Noll, 1993; Matus et al., 2007), making the most ancestral Pax gene even older, and related genes are found in a range of organisms, from cnidarians to humans. Evolution of this family has produced different protein configurations to accompany their diverse functions.
Pax genes were originally identified in Drosophila based on their sequence homology to paralogs of the segmentation gene, paired. Members of the Pax family all contain a DNA-binding domain (the ‘paired domain’, or PD encoded by a paired box) usually located at the N-terminus, which comprises two subdomains, called PAI and RED (Jun & Desplan, 1996). The N terminal portion, the PAI domain, is the most conserved region of the PD, and contains three alpha helices used for DNA binding. This is followed by a flexible linker region, and then by the C-terminal RED domain, whose three alpha helices form a second DNA-binding domain (Jun et al., 1998). Several Pax genes share additional sequence features, including an octapeptide (OP) motif located C-terminal to the PD, whose function is unknown, and an additional whole or partial homeodomain (HD) DNA-binding motif, which can act independently from the PD. This HD is unique in that it carries an S50 at the most critical residue of the DNA binding domain. The Pax proteins are classified by virtue of which types of characteristic structures they possess: The Pax 1/9 group (in vertebrates, called Group I) possesses a PD and an OP, while the Pax B/2/5/8 subfamily (Group II) also has a partial HD. The Pax 3/7 (Group III) and 4/6 (Group IV) subfamilies all have a PD and a complete HD, but only Pax 3/7 proteins also have an OP motif. The Pax A/C group, which includes insect pox-n and cnidarian Pax A and Pax C (Matus et al., 2007), is absent from vertebrates, and its members possess a PD but not an HD. Another group exists in insects and is characterized by the loss of the PAI domain (eyegone group). These characteristic structures permit the rapid identification and classification of Pax genes given genomic sequence data.
Pax genes have been extensively studied in Drosophila, which possesses 10 Pax genes: paired (prd), gooseberry (gsb) and gooseberry-neuro (gsb-n), poxmeso (pox-m), pox-neuro (pox-n), eyeless (ey) and twin of eyeless (toy), eyegone (eyg), twin of eyegone (toe), and shaven (sv). It has been proposed that one ancestral Pax gene encoding only a PD and an OP, gave rise to the pox-m lineage (which may have given rise to pox-n, which also lacks a HD), while another possessing a HD (and presumably an OP) gave rise to the prd/gsb/gsb-n lineage, which duplicated to produce each of the three genes; similar duplications gave rise to the ey-toy and eyg-toe gene pairs found in flies. Sequence similarity among these genes is also reflected in their related functions during development. While extensive studies of these proteins in Drosophila allowed for elegant genetic dissection of functions shared by their vertebrate homologs, studies in additional models, including insects, are needed to elucidate the early events in elaboration of this important gene family. Furthermore, studies of Pax gene structure and function in additional insects will permit a greater understanding of how evolution within a single animal order can contribute to novel developmental strategies and morphological diversity.
We report the characterization of the Pax genes of Nasonia vitripennis. The genome sequence of Nasonia has recently been reported (Werren et al., 2010). We have identified and cloned the seven Pax genes from Nasonia, and used their protein sequences to construct a phylogenetic tree and make assignments to Pax subgroups. We find these genes to include all of the Pax genes found in flies, except gsb and gsb-n, which are closely related to prd, and toe. Expression of three of these genes (prd, sv and ey) is described. Lastly, our analysis reveals a wider conservation of the OP motif in insects than previously reported.
Results
The Nasonia genome assembly v.1.0 was queried for all putative Pax genes using NCBI BLAST. The 128 amino acid PD from Drosophila paired was used as a first ingression into Nasonia sequence, since all Pax genes (and only Pax genes) possess this conserved domain. Seven significant hits with E-values of less than 1e-11 were found. Primers were designed based on Gnomon gene models to attempt to clone full-length cDNAs for sequence alignment and subsequent phylogenetic tree building. Reverse transcription (RT)-PCR from embryo and adult cDNA was performed using these oligos and in three cases, nearly full-length cDNAs were obtained. In other cases, additional oligos were generated and smaller fragments of each remaining cDNA were isolated, all of which (except for sv and eyg) included a complete PD. All cDNA fragments were sequenced and then BLASTed against the Drosophila genome for identification according to sequence homology. The seven Pax genes were identified as paired (prd), shaven (sv), pox-meso (pox-m), pox-neuro (pox-n), eyeless (ey), twin of eyeless (toy), and eyegone (eyg). These assignments were confirmed through phylogenetic analysis (described below). The resulting sequences are schematically represented in Fig. 1 and were used for subsequent analysis, beginning with characterization of sequence and gene structure (schematized in Fig. S1; GenBank accession numbers are provided in the Experimental Procedures).
Figure 1.
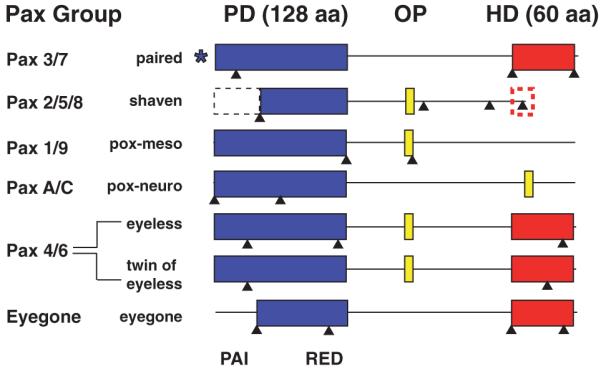
Domain structure of Nasonia Pax proteins. Pax proteins are characterized by several conserved domains, represented by boxes. The paired domain (PD), spanning 128 amino acids (blue) is characteristic of this family, and is shared by all members, while some members possess an additional octapeptide (OP) or OP-like motif (8 amino acids; yellow) and/or a partial (red dashed) or full (60 amino acids; red) homeodomain (HD). This similar protein architecture can be achieved by several different genomic locus architecture strategies (splice sites are indicated by black arrowheads). While eyegone in insects always lacks the sequence encoding the N-terminal portion of the PD, all other insect Pax genes usually possess this sequence. Nasonia shaven apparently lacks this subdomain (black dashed box). Gene models predict that the N-terminus of Nasonia paired is 475 amino acids away from the PD, which would make it exceptionally distant, and a departure from the characteristic structure. It is therefore indicated with a *.
Paired
The first Pax gene to be identified was prd. It is a Pax3/7 group gene whose role in segmentation is thought to pre-date the arthropod lineage (Davis & Patel, 1999). Like previously described orthologs from a variety of species, Nasonia prd has a complete PD and HD, but lacks an OP. The prd locus (LOC001815436) spans approximately 52 kb of genomic sequence, including a putative exon encoding a 6 amino acid sequence that resembles C-terminal remnant of an OP (VAGYIG). However, this exon was not present in the cDNA fragment that we isolated, although exons predicted upstream and downstream of that putative exon were present. The corresponding interval in the Apis and Drosophila genomes includes two introns and a small exon which are not present in Nasonia prd (or indeed, in the Nasonia genome) and none of the Apis or Drosophila coding sequence resembles VAGYIG. The PD is typically found very close to the amino-terminus of Pax genes as in the case of Drosophila prd, where its PD begins 27 amino acids from its start. Nasonia prd appears to be an exception to this trend, with its PD beginning a surprising 475 amino acids (encoded by two large exons) away from its N-terminus. We isolated only a more C-terminal fragment of this cDNA, including the third, fourth and sixth predicted exons, which we found to be contiguous. The Nasonia prd gene has a PD that is 84% identical and 97% similar to the PD of Drosophila prd and 92% identical to that in Apis.
Prd is a secondary pair rule gene in flies whose early expression depends on gap genes such as Krüppel and primary pair rule genes like even skipped (Gutjahr et al., 1993). It is also re-expressed later in embryogenesis in the head, where it plays a role in CNS development (Gutjahr et al., 1993). In the hymenopteran Apis mellifera, prd is expressed in an anterior to posterior progression, in which primary stripes form sequentially and split directly to form secondary stripes (Osborne & Dearden, 2005). We examined Nasonia prd expression and detected its earliest expression in the precellular blastoderm, where one incomplete stripe is evident before cellularization (Fig. 4A). By cellular blastoderm stage, this stripe extends completely from the ventral to the dorsal side and has broadened to several nuclei in width (Fig. 2B). As embryogenesis progresses, stripes are progressively added in an anterior to posterior fashion (Fig. 2C–E), and stripes initially alternate between stronger and weaker intensity. This is in contrast to Drosophila prd, whose initial expression is in seven primary stripes before cellularization, and whose secondary segmental expression only alternates in intensity when the secondary stripes first split from primary ones (Gutjahr et al., 1993). By the time germ band extension in Nasonia is complete, there are 16 equally intense segmental stripes of prd expression (Fig. 2F, G), corresponding to the 16 segments of Nasonia and resembling the fully extended Drosophila germ band with its 14 segmental stripes (which are fainter). This is in contrast to Apis, where anterior stripes are fading by the time the posterior-most stripes form (Osborne & Dearden, 2005). After germ band retraction during dorsal closure, prd staining is strongly evident at the anterior in the head of the embryo in both flies (Gutjahr et al., 1993) and Nasonia (Fig. 2H). Apis exhibits late stage head staining in the labrum (Osborne & Dearden, 2005).
Figure 4.
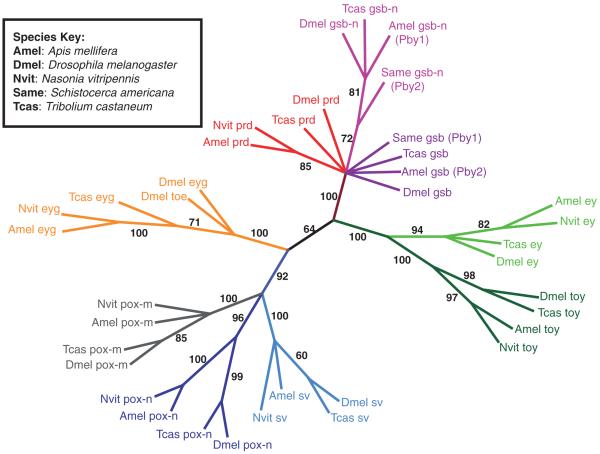
Phylogenetic tree. A phylogenetic tree was generated from insect Pax protein sequence alignments (using sequence between the start of the paired domain and the end of the homeodomain) using Maximum Parsimony bootstrap analysis with 1000 repeats and 10 add sequence repeats. Gene families are indicated by colour, and numbers adjacent to a branch indicate bootstrap support value.
Figure 2.
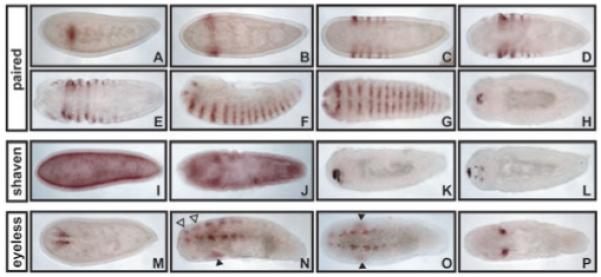
Pax gene expression in Nasonia embryos. (A–H) Nvit paired expression. (A) Lateral view of an early embryo, expressing a single partial stripe of paired (prd) expression. (B) Dorsal view of precellular blastoderm embryo with a single full prd stripe spanning the embryo. Stripes of prd are added in an anterior to posterior fashion, beginning at about cellularization (C) (dorsal view) and continuing through gastrulation (D, E, dorsal views). By full germ band extension, 16 stripes of prd expression are evident (F lateral view, G ventral view). Late prd expression is seen in the head of the embryo during dorsal closure (H, dorsal view). (I–L) Nvit shaven (sv) expression. (I) Dorsal view of basal localization of Nvit sv transcript in the embryo at cellularization. (J) Dorsal view of basal localization that persists during early gastrulation. (K–L) Dorsal views of embryos exhibiting strong expression of sv in the head during dorsal closure. (M–P) Nvit eyeless (ey) expression. (M) Dorsal view of an early embryo expressing ey in two patches of cells that prefigure the eye. (N) Ventrolateral and (O) ventral views of embryo during germ band extension, with expression in eye precursors, which persists on the dorsal side (black arrowhead), as well as in a segmentally reiterated pattern on the ventral side (open arrowheads) adjacent to the ventral midline (P) Dorsal view of late ey expression, which is restricted to the eye primordium of the head.
Shaven
Nasonia sv (or sparkling in Drosophila) is a member of the Pax B/2/5/8 subgroup, and is required in flies for both neuronal and non-neuronal cell types in the eye, and is also expressed in the nervous system (Fu & Noll, 1997; Fu et al., 1998). Drosophila sv possesses a PD, an OP and a partial HD, which are characteristic of this subgroup. However, the Nasonia sv automated gene models predicted an open reading frame consisting of a single exon encoding 96 amino acids (LOC100120909). We confirmed the presence of this exon, which contains the putative 5′end of the gene. Its sequence indicates that Nasonia sv has lost the highly conserved N-terminal portion of the PD, known as the PAI domain. In flies and beetles, the PAI domain of sv is encoded by a separate exon and splices to the RED domain exon precisely where the PD of Nasonia sv begins, providing a possible explanation for its loss. Other Drosophila Pax genes (gsb and pox-n) also possess an intron between the PAI and RED domains and it has been previously suggested that this may, indeed, indicate the disparate origins of the bipartite PAIRED domain constituents (Bopp et al., 1989). The 5′ end of the sv cDNA we isolated would encode at least six and up to 28 amino acids of non-conserved sequence before the conserved RED domain begins (depending on start codon usage), which would be a significant deviation from its typical, conserved structure.
To ascertain whether Nasonia sv has a PAI domain despite its apparent absence from genome sequence, we used genomic PCR to validate the colinearity of the sequence assembly upstream of the Nasonia sv putative start, using 1 kb amplimers with 70 bp overlap. We confirmed that no PAI sequence was encoded in at least 3.5 kb of upstream sequence. However, further upstream, our PCR data suggested some deviation from the genome sequence as assembled (data not shown), leaving open the possibility that the PAI domain sequence may appear absent from the genome because of errors in the assembly. The RED domain of Nasonia sv is highly conserved, differing by only one similar amino acid from its ortholog in the honeybee, and by only two non-similar amino acids from its ortholog in Drosophila (data not shown).
The Nasonia sv gene model is predicted to terminate just after the end of the PD, which would create a truncated protein lacking the OP and partial HD found in Tribolium and Drosophila, but which were also absent from the predicted gene in Apis. We used BLAST to interrogate the Nasonia genome using the Tribolium sv OP motif and partial HD as a query sequence, and indeed, found an OP and partial HD in the genomic region downstream of the predicted Nasonia sv locus. Using oligos corresponding to the ends of these putative domains, two additional unannotated 3′ exons were successfully cloned from cDNA, and sequencing confirmed that they contain the OP and partial HD of Nasonia sv (Fig. 1; sequence provided in Supplementary Data). This downstream sequence includes a long polyglutamine tract, which is not present in any frame in the corresponding Apis genomic sequence (data not shown). More recent gene models in Apis have identified a similar sequence that is quite conserved with the Nasonia sequence.
To determine whether expression of Nasonia sv with its unusual N terminus, resembles sv from other insect species, we used the large fragment of Nasonia sv as a probe for in situ hybridizations. Interestingly, while sv expression in early blastoderm embryos seems to be low level and ubiquitous, sv mRNA appears to be basally localized in embryos at the onset of cellularization (Fig. 2I). This localization persists even as embryos begin to gastrulate (Fig. 2J). This localization has not been reported for sv in other insects. In Drosophila, sv is expressed in the developing CNS and PNS in a segmentally repeated pattern and in the primordium of the eye (Fu & Noll, 1997). Nasonia sv is expressed in late stage embryos in the anterior (Fig. 2K, L), in what are probably precursors of head structures.
Pox-meso
Pox-meso (LOC100116076) is a member of the Pax 1/9 subgroup of the Pax gene family, whose members are distinguished by the absence of a HD. Pox-m is expressed in somatic mesoderm precursors in both flies and mouse, and has recently been shown to be important at several stages of myogenesis (Bopp et al., 1989; Duan et al., 2007). Nasonia pox-m has one predicted open reading frame of 206 amino acids, and a second predicted variant that includes a short poly-glutamine tract N-terminal to the second exon, resulting in an open-reading frame of 215 amino acids. The second of the gene’s 5 exons encodes its 128 amino acid PD. The pox-m PD is 91% identical and 97% similar to the PD of its Drosophila homolog and 99% identical and similar to that of its Apis homolog. We cloned a cDNA fragment of Nasonia pox-m that includes the short polyglutamine tract. As is characteristic of the Pax1/9 subfamily, Nasonia pox-m lacks a HD and possesses a conserved OP-like motif located about 25 amino acids from the C-terminus of its PD. The OP (HTVHDILS) is relatively well conserved among Apis, Tribolium, Nasonia and Drosophila (Fig. 3).
Figure 3.
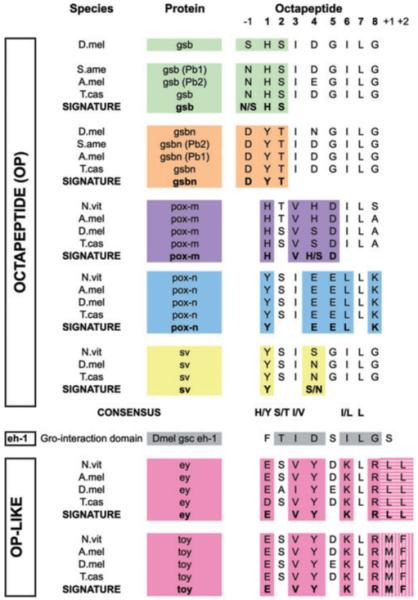
Alignment of Pax octapeptides (OPs) from insects reveals Pax subgroup signature, and uncovers a previously unreported octapeptide motif present in Pax4/6 proteins. OPs from Pax gene products are given and aligned, with adjacent conserved amino acids, in some cases. Positions within or relative to the OP are given as numbers above the alignment, where 1 is position 1 of the OP. The consensus sequence for these OPs is given below each gene family, which is coded by color: gsb (green), gsb-n (orange), pox-m (purple), pox-n (blue), shaven (yellow). Signature sequences distinctive for each Pax gene family comprising residues within the OP or in combination with adjacent amino acids, are highlighted with color and indicated in bold for each sequence group. An OP-like sequence was identified between the paired domain and homeodomain of Pax 4/6 proteins. Available insect OP-like sequences from ey and toy (pink) are aligned, with the signature sequence in bold at the bottom of each group. Two neighbouring amino acids constitute a distinguishing feature within this subgroup, indicated by shading (vertical or horizontal stripes).
Pox-neuro
Pox-neuro (LOC100122671) is a member of the PaxA/C subgroup, which is thought to have arisen as a distinct lineage from pox-m and prd/gsb/gsb-n (Noll, 1993). In flies, pox-n is expressed in a segmentally reiterated pattern, and is utilized extensively in the peripheral nervous system where it is involved in generation of polyinnervated sense organs for chemosensation (Bopp et al., 1989; Dambly-Chaudiere et al., 1992; Nottebohm et al., 1992). Similar to other PaxA/C group members, Nasonia pox-n lacks a HD, but possesses an OP motif (YSIEELLK) about 40 amino acids from its N-terminus. This motif is adjacent to an additional block of at least six amino acids that are highly conserved among homologs in Apis, Tribolium, Drosophila and Nasonia, suggesting possible functional importance of these residues in addition to the OP motif. Five exons comprise an open reading frame of 384 amino acids in the nearly full length cDNA for pox-n that we cloned. Interestingly, the second exon of this cDNA, which encodes the beginning of the conserved RED domain, begins with a sequence (ADCLQ) that is not normally found in PDs. This sequence is found in the linker region between the PAI and RED domains and directly precedes the fourth alpha helix of the PD and, therefore, is unlikely to disrupt the ordered alpha helices of the RED domain that follows it. Despite the addition of this nonconserved sequence in pox-n’s PD, the region still exhibits 82% identity and 94% similarity to the PD of its Drosophila homolog and 91% identity and 98% similarity to the PD of Apis pox-n.
Eyeless
The Pax4/6 group includes the Drosophila genes ey and toy. Drosophila ey is a master regulator of eye development, specifying tissue for eye differentiation, since ectopic expression of this gene is sufficient to induce extra eyes on leg or wing tissue (Halder et al., 1995), and mutants for this gene have reduced or absent eyes (Quiring et al., 1994). It is highly conserved both structurally and functionally; the mouse Pax6 mutant called small eye exhibits similar mutant phenotypes, despite major differences in eye type between flies and vertebrates (Hill et al., 1991). The Nasonia ey locus (LOC100116958) spans a 40 kb genomic region, over 8 predicted exons. We isolated a cDNA fragment that spans exons one to six, and sequencing revealed that the fourth and fifth of these predicted exons are not expressed. This cDNA fragment includes a polyglutamine tract of about 26 amino acids located just after the PD that is not present in any gene model (GenBank accession number GQ301537). The inclusion of this fragment was confirmed in three independent RT-PCR experiments from two different RNA preps (data not shown), suggesting that this splice form is indeed present and consistently abundant. Nasonia ey has both a complete PD of 128 amino acids and a HD of 60 amino acids. In addition, there is an OP-like sequence located between the PD and HD (Fig. 3). This sequence of eight amino acids is highly conserved among ey and toy homologs in insects. The OP-like sequence (ESVYDKLR) is 100% identical in Nasonia, Tribolium and Apis and only the serine residue (S) is different in Drosophila where it is an alanine (A) (Fig. 3). Similar to pox-n, the ey OP-like sequence is flanked by several additional highly conserved amino acids of unknown function. The PD of Nasonia ey is identical to the PD of ey in Apis and 98% identical to that of Drosophila.
The expression of Pax6 in a variety of invertebrates and vertebrates has been reported (Callaerts et al., 1997). In flies, ey expression comes on during gastrulation in a population of cells that prefigures the brain and the eye field (Quiring et al., 1994). To determine whether the expression of ey in Nasonia resembles that of other insects, we used a cDNA fragment as a template for an in situ probe and examined the expression of Nasonia ey. Nasonia ey is expressed in early embryos, in a population of cells in two spots on the dorsal side of the early embryo (Fig. 2M). This population of cells is evident on the dorsal side of gastrulating embryos (Fig. 2N black arrowhead, Fig. 2O arrowheads) during germ band extension, during which time segmental expression can also be transiently seen on the ventral side, appearing in an anterior to posterior fashion (Fig. 2N, open arrowheads and Fig. 2O). When germ band retraction is complete, strong staining of the future eye tissue on the dorsal side of the embryo is all that remains (Fig. 2P). This pattern of expression closely resembles the segmental expression in embryos and the expression in the eye primordium reported for Drosophila ey (Quiring et al., 1994).
Twin of eyeless
The second member of the Pax4/6 subgroup in wasps is the ey duplication partner toy. In flies, toy was shown to lie upstream of ey in the eye-specification pathway, binding directly to the ey enhancer via its PD, which differs from that of ey by one critical amino acid (asparagine instead of glycine at position 14). This distinction permits toy to activate ey, but does not permit ey to autoactivate (Punzo et al., 2004). Nasonia toy (LOC100118963) spans a genomic interval of 16 kb, and like its fly counterpart, possesses the critical asparagine at position 14, suggesting possible conservation of specificity. Nasonia toy possesses a complete PD and HD. While Pax4/6 proteins in other species have not been reported to possess a canonical OP, Nasonia ey and toy possess an identical OP-like motif (ESVYDKLR) (Fig. 3). The gene has a predicted open reading frame of 531 amino acids. Gene models predict the presence of 6 exons; however, the fragment we cloned lacks the third of these. The PD of Nasonia toy is highly conserved among insects. It is identical to the PD of Apis toy and 96% identical and 98% similar to the PD of its Drosophila homolog.
Eyegone
Eyegone, initially called lune, is thought to act similarly to and cooperatively with eyeless, since both genes cause eye reduction in mutant animals and ectopic eyes (in distinct locations) in overexpressors (Jang et al., 2003). Eyg is not a member of any Pax subgroup because it lacks the N-terminal PAI domain. The protein also lacks an OP motif, but has a complete S50 type HD. In flies, eyg was shown to be able to bind DNA through either its RED domain or its HD (Jun et al., 1998), demonstrating the versatility in DNA binding of Pax proteins. Nasonia eyg spans 2.8 kb (LOC100122710) in the genome, a relatively compact locus compared to most Nasonia Pax genes. Similar to the prediction for prd, the PD of eyg is predicted to begin unusually far from the protein’s N-terminus (approximately 240 amino acids away). Six predicted exons, the third and fourth of which encode a partial PD and complete HD, create an open reading frame of 649 amino acids. The Nasonia eyg partial PD is 84% identical and 91% similar to the partial PD in Drosophila eyg and 99% identical and 100% similar to the PD in Apis eyg. Phylogeny of Nasonia vitripennis (Nv) Pax genes. After initial identification of the Pax genes using BLAST, we aligned the Pax protein sequences from several insects using the program ClustalX (Fig. S2). We included our empirically derived Nasonia Pax protein sequences if the entire region between PD and HD was cloned, and used predicted Nasonia sequences in all other cases, as well as Pax protein sequences from flies, beetles, grasshoppers and honeybees in the alignment. Using the phylogenetic analysis software paup*, we generated a phylogenetic tree using Maximum Parsimony with 1000 bootstrap repeats with 10 fold sequence repetitions (Fig. 4). This tree supported the Pax gene assignments made using BLAST.
The tree seems to validate Gnomon predictions of orthology as each Nasonia sequence is grouped similarly to homologs from other insects, and in all cases, the groupings match the automated predictions/assignments for that Nasonia sequence. Nasonia Pax genes generally group more closely with their Apis counterparts than with their Drosophila ones, consistent with their shared hymenopteran lineage. The duplication events that gave rise to paired/gsb/gsb-n and to ey/toy in insects are also apparent. Our tree also places sv in the same lineage as pox-n, whose PDs are very similar, supporting a common origin that has been postulated previously (Noll, 1993). Though previous analyses of Apis (Osborne & Dearden, 2005), and Schistocerca (Davis et al., 2001) had not resolved the identities of several Pax Group III (PGIII) genes, leaving them more generally named ‘Pairberry1’ and ‘Pairberry2’, our tree grouped the gsb-n genes together (now labelled Apis gsb-n, and Schistocerca gsb-n) with relatively high confidence with their orthologs from other insects, while supporting assignments made for Tribolium gsb and gsb-n (Richards et al., 2008). Hymenopteran prd sequences grouped together. The remaining gsb sequences are not grouped based on the tree alone; however, OP signatures corresponding to each Pax family added additional confidence in assigning these to a gsb group, as well as in confirming the other PGIII assignments (discussed below).
Discussion
Here, we report the cloning and identification of the seven Pax genes of Nasonia vitripennis. We find that Nasonia shares most of the Pax gene repertoire of the well-studied fly genome, but lacks gsb and gsb-n, two of the three derivatives of an ancient lineage that also produced prd. Nasonia also lacks toe, a gene also absent from Apis and Tribolium, suggesting that it is not a recent loss from the hymenopteran lineage, but rather was likely a duplication that arose in another part of the insect order. Several Pax proteins in Nasonia possess poly-glutamine tracts, which are absent from products of orthologous genes in other insects. This seems to be a feature common to a number of proteins in Nasonia (RGK, MIR, J.A. Lynch, pers. comm.). We cloned cDNA fragments of several Pax genes that deviated significantly from initial gene models and identified regions that may be misassembled in the genome assembly, suggesting a moderate rate of error in these models, and the need for validation and caution when interpreting apparent gene loss (see below). While most Nasonia Pax genes have very similar structure to their orthologs from other insects, we find that sv appears to lack the N-terminal PAI portion of its PD, a change whose functional consequences are not known.
Sequence and expression of Nasonia shaven
Despite gene models predicting a single exon encoding a truncated protein, we have cloned a fragment of Nasonia sv that encodes a RED domain, an OP and a partial HD. While a single exon was intially also predicted for the Apis gene, we were able to interrogate the Apis genome for the same type of sequence found in Nasonia, using BLAST, and more recent gene models make clear that Apis sv possesses significant additional conserved sequence (Supplementary Data). The Apis gene possesses an intact PAI domain, which Nasonia seems to lack, as well as a RED domain. While the RED domain and OP are often encoded in the same exon, as in Nasonia sv, Apis sv is predicted to splice differently, and no OP, either on the same or an additional exon, is evident. However, there is still an N-terminal remnant of a HD that resembles the Nasonia (and Tribolium) sequence. The Apis genome may encode a protein lacking the OP, but sequence gaps in the region leave open the possibility that the missing sequence is, in fact, present in the genome; Nasonia expresses a message encoding a truncated protein, lacking an N-terminal PAI domain, but otherwise possessing all of the conserved features of sv. Among known Pax proteins, eyg is the only gene that possesses a RED domain only, and this has been shown in flies to be sufficient for DNA binding (Jun et al., 1998), suggesting that Nasonia sv could bind to DNA despite the lack of a PAI domain. Mammals possess an isoform of Pax6 (Pax6-5a) that contains an insertion within the PAI domain that causes a shift in its specificity, making it functionally equivalent to eyg (Epstein et al., 1994). Together, these data show that within the hymenopteran lineage, sv has undergone extensive changes that likely represent significant changes in function.
Nasonia sv mRNA localizes strongly to the cortex of the cellular blastoderm, in what appears to be basal cellular localization. To our knowledge, this type of mRNA localization has not been reported for Pax 2/5/8 orthologs from other species.
Pax gene octapeptides
The OP motif was first identified by virtue of its extreme conservation between human and fly Pax genes (Burri et al., 1989). The OP motif is often encoded by the same exon that encodes the C-terminal end of the PD (Noll, 1993), although there are exceptions to this rule (e.g. Nasonia pox-m). It is noteworthy that the domain structure of Pax genes is largely conserved in insects. In some Pax genes, such as pox-n, there is additional sequence adjacent to the OP (five amino acids N-terminal to the OP) that is perfectly conserved among insects surveyed (data not shown) that may be related to its function. It has been suggested that the nucleotide sequence that encodes the OP can serve as a scaffold for recruitment of methylation/demethylation machinery to the host Pax gene locus (Ziman & Kay, 1998). One report has shown that the OP of a vertebrate Pax 5 (BSAP) is able to recruit a potent co-repressor to convert it from an activator into a repressor (Eberhard et al., 2000).
Alignment of the Pax OPs from a variety of insects (Fig. 3) highlights the significant resemblance that this motif bears to a Groucho-binding motif called eh-1 (Fig. 3, grey), which is present in many developmental transcription factors, including odd-skipped, engrailed, vnd, msh, Oct, six, and goosecoid (gsc) (Goriely et al., 1996; Jimenez et al., 1997; Goldstein et al., 2005). Groucho is an important co-repressor protein that recruits histone deacetylases to bound targets to effect transcriptional silencing (reviewed in (Parkhurst, 1998). Positions two and seven of the OP [a serine/threonine (S/T) and leucine, respectively] are nearly perfectly conserved among reported OPs (Noll, 1993). Interestingly, all eh-1 domains that bind Groucho strongly in vitro possess S/T at position two and I/L at position seven (Goldstein et al., 2005). In addition, position one, usually a Y, can accommodate an F (another aromatic amino acid) with no ill effects, but mutation to E abrogates Groucho binding, showing this position to be critical for function (Jimenez et al., 1999; Eberhard et al., 2000). Conservation of insect OPs is also consistent with tolerance of H at this first position (Fig. 3). Overall, OPs from gsb/gsb-n, pox-m, pox-n, and sv share significant similarity to the gsc eh-1 in 4-6 of 8 residues, in an otherwise non-conserved region of their host proteins. It will be interesting to determine if these conserved OPs mediate protein–protein or intra-molecular domain interactions, and whether they are, like eh-1, able to contribute to transcriptional repression of target genes through recruitment of Groucho.
Pax-6 proteins, ey and toy
Our data confirm the presence of a complete PD and HD in both Nasonia ey and toy, but also identify a previously unrecognized OP-like sequence between the PD and HD in both proteins. This sequence (ESVYDKLR) not only shows conservation of the second and seventh residue, two positions that are almost invariant in bona fide OPs, but the peptide is extremely well conserved in its entirety among insects (Fig. 3). Interestingly, we find that human Pax6, but not Pax4, possesses an OP-like sequence (DGMYDKLR) that aligns well with our insect OP. This sequence is significantly divergent from known OPs, importantly at position one, where the conserved E would be predicted to abrogate its putative function in Groucho-recruitment (Eberhard et al., 2000). Its extreme conservation across species nevertheless suggests functional constraint.
Whatever its origin or function, we have observed that the OP, in addition to group specific differences in the conserved paired-box and/or HD of Pax genes, is strongly correlated with the identity of its host Pax gene (Fig. 3), most significantly for Pax Group III genes (gsb/gsb-n/prd) (discussed below).
Nasonia prd and its origins. Paired, a pair-rule gene in flies, is thought to have arisen from an ancestral gene that duplicated, giving rise to prd and a gsb/gsb-n gene, followed by another duplication to give gsb and gsb-n (Balczarek et al., 1997). There is only one gene from the prd/gsb/gsb-n lineage in Nasonia, as compared to the complete suite of three genes in Drosophila. Three PGIII genes exist in Tribolium, suggesting that two genes have been lost in the lineage leading to Nasonia. The determination of which of the three pairberry derivatives is left in Nasonia was complicated by the uncertain assignments of pairberry derivatives from other species, including Schistocerca and Apis (Davis et al., 2001; Osborne & Dearden, 2005).
We built a tree by aligning the Pax Group III (PGIII) sequences from Tribolium, Apis, Drosophila and Nasonia to determine the identity of the Nasonia gene. Apis possesses all three genes, suggesting that the loss of the other two genes has occurred within the hymenopteran lineage. Furthermore, the Nasonia gene groups with high confidence with Drosophila prd, separately from the other fly PGIII genes gsb and gsb-n. In addition, prd lacks an OP while both gsb and gsb-n possess one. Since most Pax genes possess OP or OP-like sequences, and all PGIII genes arose from a common ancestor, the ancestral pairberry gene most likely possessed an OP; the single Nasonia protein does not. Thus, we conclude that that single Nasonia PGIII gene is, indeed, prd. Nasonia therefore develops properly without the two missing genes. It is still formally possible that these genes are present in the Nasonia genome and simply absent from the genome assembly and available sequence. However, this would not change our assignment of the annotated gene as paired.
It was during this analysis that it also became clear that all available gsb and gsb-n sequences share a perfectly conserved OP which is distinct for each gene (Fig. 3). The OP for all gsb homologs is HSIDGILG, for gsb-n homologs, YTIDGILG (YTINGILG in flies) and for prd homologs the OP is absent. Furthermore, the amino acid adjacent N-terminal to all gsb OPs is N while all gsb-n homologs have a D at this position. This clear difference between gsb and gsb-n distinguishes these two genes. We tested this correlation on Apis pairberry 1 and 2, which had been previously unresolved (Osborne & Dearden, 2005), and found that one possessed a perfect gsb type OP and the other gene a perfect match to that of gsb-n. Phylogenetic analysis confirmed that the PGIII genes sorted using this method were indeed correctly assigned (Fig. 4).
The Tribolium genome also has three PGIII genes, of which one (prd) has been described (Choe et al., 2006; Choe & Brown, 2007). However the identities of the two other PGIII genes, LOC663057 and LOC663027, found in the beetle genome have not been validated, though given tentative assignments (Richards et al., 2008). The OPs of these genes are characteristic of gsb and gsb-n genes, respectively, and our phylogenetic tree groups these genes with other gsb and gsb-n homologs (Fig. 4). Furthermore, these two genes are located next to each other in the genome, a hallmark of duplicated genes, and have likely remained proximal to each other because of shared regulatory sequence between them. Taken together, these data add additional confidence to the characterization of these Apis and Tribolium PGIII genes as homologs of gsb or gsb-n.
There are only two reported pairberry genes in Schistocerca and they could not be assigned previously (Davis et al., 2001). Both of these sequences have incomplete sequences reported (30 amino acids shorter than for other PGIII genes). Phylogenetic analysis revealed that these two genes were more closely related to each other than to either gsb or gsb-n homologs from other insects, which was interpreted to mean that this duplication occurred independently in Schistocerca (Davis et al., 2001). Our analysis segregates these genes with their orthologs from other insects, with moderate bootstrap support. If we apply the OP signature as a classification criterion to the same genes in Schistocerca, we find that one paralog possesses a perfect gsb type OP and the other paralog a gsb-n one. We have therefore assigned these genes according to this homology (Fig. 4). We believe that the correlation will be borne out by phylogenetic analysis when full-length sequence is available for Schistocerca, and we propose this method of identification as a reliable tool in the classification of PGIII genes.
The sequencing of the Nasonia genome has enabled the rapid, genome-wide study of important gene families, such as the Pax family, which we report here, revealing several interesting features of these genes both within the hymenopteran lineage and others shared by all insects studied to date. Sequencing of additional insect genomes will add significantly to the power of these analyses and enable mapping of the protein domains that have transited via selection through evolution.
Experimental procedures
Pax gene identification and cloning
All Nasonia Pax genes were initially identified using NCBI TBLASTN, using the PD from Drosophila paired as the query sequence for identification of all PD containing (putative Pax) genes in Nasonia. To determine which of these genes also possessed an S50, paired-like HD, an additional search was performed using the entire region spanning the Drosophila paired PD and HD. These candidates were cloned using standard RT-PCR methodology according to manufacturer’s specifications using Superscript II (Invitrogen, Carlsbad, CA, USA) and total RNA from embryos and adults extracted using Trizol (Invitrogen). Oligos for PCR were designed based on predicted gene models (NCBI pipeline; (Kapustin et al., 2008). In cases where predictions were incorrect or incomplete, we used predicted or reported protein sequence from Tribolium and Apis genomes, which are publicly available, as references for gene structure prediction and oligo design. Sequences for comparative analysis were obtained from other insect genomes from GenBank, and subsequently, using TBLASTN, to identify or confirm the identities of predicted but un-annotated genes, on Beetlebase (http://beetlebase.org) and using the UCSC genome browser (http://genome.ucsc.edu). Partial cDNA sequences that we obtained have been deposited in GenBank, and the accession numbers assigned are as follows: prd: GQ301535; sv: GQ301540; pox-m: GQ 301536; pox-n: GQ 301539; ey: GQ301537; toy: GQ301541; eyg: GQ 301538.
Sequence alignments and phylogenetic tree construction
ClustalW 2.0 was used to generate sequence alignments of protein sequences (Thompson et al., 1994), and these were rendered using MacClade (Sinauer Associates, Sunderland, MA, USA). To improve the quality of our alignments, only sequence between the start of the PD and the end of the HD was used. Phylogenetic trees were generated using paup* (Sinauer Associates), using a Maximum Parsimony bootstrap algorithm with 1000 bootstrap replicates with 10 additional sequence replicates each. The final tree was rendered using Dendroscope (Huson et al., 2007). Branches with bootstrap values of <60 were collapsed.
Nasonia embryo collections and in situ-hybridization
Wild-type Nasonia cured of Wolbachia (AsymCx) were maintained at 22 °C, and embryos collected and fixed as previously described (Olesnicky et al., 2006). Digoxigenin labeled probes of an average length of 600 bp corresponding to cloned Nasonia Pax gene fragments were generated by in vitro transcription using either T7 or SP6 polymerase according to manufacturer’s specifications (Roche, Indianapolis, IN, USA).
Supplementary data
Shaven sequence
Sequences are given below as protein sequence. For Nasonia, this is a translation of the full-length cDNA fragment we obtained from PCR. For Apis, this is the predicted protein according to the gene model from the UCSC Genome Browser.
Nasonia shaven sequence (predicted protein sequence based on experimentally obtained cDNA sequence; assumes the first ATG is translated)
MSSARCDHCDHRTRSDTRRYLLMSVEIRRYYETGSFKAGVIGGSKPKVATPPVVDAIANYKRDNPTMFAWEIRDRLLAEGICSQDNVPSVSSINRIVRNKAAEKAKHAHQQQQQQQQQAQQQGQQQQQQQQQQQQQQQQGPVSVIAHAPATAATAPNAYSISGILGIPAHHQDPNGNSIKRKRADDDDNRDLADHSDDDLKRQRNTYNGDQLYSNLWSSKWSIKDEHKLLSEL
Apis shaven sequence (predicted)
MELPAHTTQHGALHLGVGVGVNPGDPAGSALTAIHPHPQDALALHHHNHGAATPGGMHEDSKKKRHGGVNQLGGVFVNGRPLPDVVRQRIVELAHSGVRPCDISRQLRVSHGCVSKILSRYYETGSFKAGVIGGSKPKVATPPVVEAIANYKRDNPTMFAWEIRDRLLAEGICSQDNVPSVSSINRCLLVLIADDNRDLNDHPEDDLKRQRSNYNGDQLYSNLWSSKWSIKDEHKLLSELGGGGGGGGASGGGGYYEHGGFPGNAIATSAELYDSLGTISTMTQAQTPHLYTPPIGGTIAGGTLTPLAPLTMQELKLSQTLDGANMSPYHTTGGGDYAYSAAYSQYSSAPYSGYGYGAATSGLLNSTYYYCNGDTLASSHTNSQQGGGCNNTGPENSTDVASRSPLAATRASSGASAASPTGSACTKPDHTSTPTDLYLA
Supplementary Material
Genomic organization of Pax gene loci in Nasonia. Gene models predicted by Gnomon (NCBI) are illustrated in blue (exons). Cloned cDNA fragments are illustrated in red. Locus location is given, including coordinates on genomic scaffolds.
Protein alignment of insect Pax proteins between paired domain and homeodomain. Amino acids are coloured according to type, numbers indicate relative position in sequence. Dashes indicate a gap in the alignment. For proteins that lack a homeodomain, the sequence for alignments ends after the octapeptide.
Acknowledgements
The authors would like to thank Karin Kiontke for amazing help above and beyond the call of duty, and for critical reading of the manuscript, as well as David Fitch and Harmit Singh Malik for helpful advice during preparation of this manuscript. The authors would also like to thank the Nasonia Genome Consortium for access to genome sequence and assembly information prior to publication. This work was funded by a NIH Grant number GM064864-05 to CD, NIH NRSA Postdoctoral Fellowship Grant # F32GM084563 to MIR from the National Institute of General Medical Sciences, and by an Undergraduate Fellowship from the Arnold and Mabel Beckman Foundation to RGK.
Footnotes
Conflicts of interest
The authors have declared no conflicts of interest.
References
- Balczarek KA, Lai ZC, Kumar S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Mol Biol E. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- Bopp D, Jamet E, Baumgartner S, Burri M, Noll M. Isolation of two tissue-specific Drosophila paired box genes, Pox meso and Pox neuro. EMBO J. 1989;8:3447–3457. doi: 10.1002/j.1460-2075.1989.tb08509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science. 2007;315:1841–1843. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- Burri M, Tromvoukis Y, Bopp D, Frigerio G, Noll M. Conservation of the paired domain in metazoans and its structure in three isolated human genes. EMBO J. 1989;8:1183–1190. doi: 10.1002/j.1460-2075.1989.tb03490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Evolutionary flexibility of pairrule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Miller SC, Brown SJ. A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc Natl Acad Sci USA. 2006;103:6560–6564. doi: 10.1073/pnas.0510440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Jamet E, Burri M, Bopp D, Basler K, Hafen E, et al. The paired box gene pox neuro: a determinant of poly-innervated sense organs in Drosophila. Cell. 1992;69:159–172. doi: 10.1016/0092-8674(92)90127-x. [DOI] [PubMed] [Google Scholar]
- Davis GK, Patel NH. The origin and evolution of segmentation. Trends Cell Biol. 1999;9:M68–M72. [PubMed] [Google Scholar]
- Davis GK, Jaramillo CA, Patel NH. Pax group III genes and the evolution of insect pair-rule patterning. Development. 2001;128:3445–3458. doi: 10.1242/dev.128.18.3445. [DOI] [PubMed] [Google Scholar]
- Duan H, Zhang C, Chen J, Sink H, Frei E, Noll M. A key role of Pox meso in somatic myogenesis of Drosophila. Development. 2007;134:3985–3997. doi: 10.1242/dev.008821. [DOI] [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fu W, Noll M. The Pax2 homolog sparkling is required for development of cone and pigment cells in the Drosophila eye. Genes Dev. 1997;11:2066–2078. doi: 10.1101/gad.11.16.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Duan H, Frei E, Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943–2950. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- Goldstein RE, Cook O, Dinur T, Pisante A, Karandikar UC, Bidwai A, et al. An eh1-like motif in odd-skipped mediates recruitment of Groucho and repression in vivo. Mol Cell Biol. 2005;25:10711–10720. doi: 10.1128/MCB.25.24.10711-10720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, Stella M, Coffinier C, Kessler D, Mailhos C, Dessain S, et al. A functional homologue of goosecoid in Drosophila. Development. 1996;122:1641–1650. doi: 10.1242/dev.122.5.1641. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Frei E, Noll M. Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair-rule genes. Development. 1993;117:609–623. doi: 10.1242/dev.117.2.609. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, et al. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in goosecoid mediates groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci USA. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T, Lipman D. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 2008;3:20. doi: 10.1186/1745-6150-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JA, Brent AE, Leaf DS, Pultz MA, Desplan C. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature. 2006a;439:728–732. doi: 10.1038/nature04445. [DOI] [PubMed] [Google Scholar]
- Lynch JA, Olesnicky EC, Desplan C. Regulation and function of tailless in the long germ wasp Nasonia vitripennis. Dev Genes Evol. 2006b;216:493–498. doi: 10.1007/s00427-006-0076-5. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol Dev. 2007;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Nottebohm E, Dambly-Chaudiere C, Ghysen A. Connectivity of chemosensory neurons is controlled by the gene poxn in Drosophila. Nature. 1992;359:829–832. doi: 10.1038/359829a0. [DOI] [PubMed] [Google Scholar]
- Olesnicky EC, Brent AE, Tonnes L, Walker M, Pultz MA, Leaf D, et al. A caudal mRNA gradient controls posterior development in the wasp Nasonia. Development. 2006;133:3973–3982. doi: 10.1242/dev.02576. [DOI] [PubMed] [Google Scholar]
- Osborne PW, Dearden PK. Expression of Pax group III genes in the honeybee (Apis mellifera) Dev Genes Evol. 2005;215:499–508. doi: 10.1007/s00427-005-0008-9. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM. Groucho: making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Pultz MA, Pitt JN, Alto NM. Extensive zygotic control of the anteroposterior axis in the wasp Nasonia vitripennis. Development. 1999;126:701–710. doi: 10.1242/dev.126.4.701. [DOI] [PubMed] [Google Scholar]
- Punzo C, Plaza S, Seimiya M, Schnupf P, Kurata S, Jaeger J, et al. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004;131:3943–3953. doi: 10.1242/dev.01278. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010 doi: 10.1126/science.1178028. doi:10/1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman MR, Kay PH. A conserved TN8TCCT motif in the octapeptide-encoding region of Pax genes which has the potential to direct cytosine methylation. Gene. 1998;223:303–308. doi: 10.1016/s0378-1119(98)00162-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of Pax gene loci in Nasonia. Gene models predicted by Gnomon (NCBI) are illustrated in blue (exons). Cloned cDNA fragments are illustrated in red. Locus location is given, including coordinates on genomic scaffolds.
Protein alignment of insect Pax proteins between paired domain and homeodomain. Amino acids are coloured according to type, numbers indicate relative position in sequence. Dashes indicate a gap in the alignment. For proteins that lack a homeodomain, the sequence for alignments ends after the octapeptide.