Abstract
Patients with chronic lymphocytic leukemia (CLL) who relapse after allogeneic transplant may achieve durable remission following donor lymphocytes infusion (DLI), demonstrating the potency of donor-derived immunity in eradicating tumor. We sought to elucidate the antigenic basis of the effective graft-versus-leukemia (GvL) responses associated with DLI for the treatment of CLL by analyzing the specificity of plasma antibody responses developing in two DLI-treated patients who achieved long-term remission without graft-versus-host disease (GvHD). By probing high-density protein microarrays with patient plasma, we discovered 35 predominantly intracellular antigens that elicited high-titer antibody reactivity greater in post-than in pre-DLI plasma. Three antigens, C6orf130, MDS032, and ZFYVE19, were identified by both patients. Along with additional candidate antigens DAPK3, SERBP1 and OGFOD1, these proteins demonstrated higher transcript and protein expression in B cells and CLL cells compared to normal PBMC. DAPK3 and the shared antigens do not represent minor histocompatibility antigens, as their sequences are identical in both donor and tumor. While ZFYVE19, DAPK3 and OGFOD1 elicited minimal antibody reactivity in 12 normal subjects and 12 chemotherapy-treated CLL patients, 5 of 12 CLL patients with clinical GvL responses were serologically reactive to these antigens. Moreover, antibody reactivity against these antigens was temporally correlated with clinical disease regression. These B cell antigens represent promising biomarkers of effective anti-CLL immunity.
Keywords: proteomics, protein microarray, immunology, leukemia, antigen discovery
Introduction
While chronic lymphocytic leukemia (CLL) is most often controlled with combination chemotherapy (1), allogeneic hematopoietic stem cell transplantation (HSCT) remains the only potentially curative treatment for this disease (2, 3). It has been long-appreciated that these curative responses rely on the ability of donor-derived immune effectors to recognize and eliminate leukemic cells. The existence of the graft-versus-CLL response has been supported by studies demonstrating that: a) myeloablative allogeneic, but not autologous, HSCT consistently results in a plateau in survival after 1 year, and the development of undetectable minimal residual disease (4); b) disease remission is closely associated with immunologic competence (i,e, following taper of immunosuppression, or development of graft-versus-host disease (GvHD) (5, 6); and finally, c) donor lymphocyte infusion (DLI) for post-transplant relapsed CLL effectively generates donor-derived tumor immunity against CLL (7). In this procedure, infusion of donor lymphocytes without further chemotherapy or radiation therapy results in remission in 30–50% of patients with relapsed CLL (8–10).
Because of its clear clinical efficacy in the absence of additional therapy, DLI is a promising system for elucidating the mechanism of donor-derived immunity to CLL. Identifying and characterizing the antigenic targets of DLI may provide insight into the basis of the anti-tumor effect of DLI. While T cells clearly participate in GvL responses (11), several studies have implicated an important role for B cell immunity in remission of hematologic malignancies following transplant and DLI (12–14). Some B cell-defined antigens (15) have been demonstrated to elicit T cell responses, and effective tumor immunity likely comprises a coordinated humoral and cellular adaptive immune response.
To date, few CLL-specific target antigens have been identified (16–19). In the current study, we identify several novel candidate antigens through analysis of B cell responses in CLL patients with clinically evident tumor immunity but without anti-host immune reactions following DLI. By using high-density human protein microarrays probed with plasma antibody from before and after DLI, we rapidly identified CLL antigens that elicit potent antibody responses in close temporal association with clinical response to DLI. Potential uses of these antigens include immune monitoring of CLL following therapy, and even as immunogens for CLL-specific vaccines.
Materials and Methods
Preparation of cell and plasma samples
Serum or plasma samples were obtained from normal donors and patients enrolled on clinical research protocols that were approved by the Human Subjects Protection Committee at the Dana-Farber Cancer Institute (DFCI), Boston. Plasma was removed following brief centrifugation of whole blood and cryopreserved at −80°C until the time of analysis. PBMC from normal donors and patients were isolated by Ficoll/Hypaque density-gradient centrifugation, cryopreserved with 10% DMSO, and stored in vapor-phase liquid nitrogen until the time of analysis.
Processing and analysis of protein microarrays
Protein microarrays (Human ProtoArray, v3; Invitrogen, Carlsbad, CA) from one printing lot were probed with patient plasma diluted 1:150 and processed according to the manufacturer’s recommendations, as previously described (20). These microarrays contain approximately 5000 N-terminus GST-fusion human proteins generated in an insect cell line, with proteins spotted in adjacent duplicates on nitrocellulose-coated glass slides. Binding of plasma immunoglobulin to protein features on the array was detected using an AlexaFluor 647-conjuated anti-human IgG (H and L chain) (1:2000 dilution; Invitrogen Carlsbad, CA). Protein microarrays were scanned and analyzed with correction for protein concentration, as previously described (20). Significant signal change was defined using the pre- and post-treatment Z-scores, Zpre and Zpost, based on both a change in magnitude Zdelta=Zpost−max(0, Zpre) and in ratio Zmult=Zpost/max(1,Zpre) greater than a cutoff n. The cutoff n was set at 5 for significant interactions and 3 for borderline interactions. Both replicate spots were required to pass the test individually for an interaction to be considered significant. Candidate antigens were ranked based on the largest cutoff n that would identify them as significantly changed. Microarray data and calculated significance values are in the Gene Expression Omnibus repository under accession GSE11564.
Sequencing of candidate antigens from tumor and donor tissue
The genes encoding the candidate antigens were directly sequenced using the M13-forward and M13-reverse primers after gene-specific PCR amplification of genomic DNA and TA TOPO cloning (Invitrogen, Carlsbad, CA). Genomic DNA was isolated from patient tumor and donor-generated EBV cell lines using a Wizard kit (Promega, Madison, WI), following the manufacturer’s instructions. Flow cytometric analysis of patient tumor revealed all CD19+ cells to co-express CD5, so tumor cells were immunomagnetically purified to >95% purity using CD19+ microbeads (Miltenyi, Auburn, CA). Sequences were aligned using Sequencher (Gene Codes Corp., Ann Arbor, MI).
Gene expression microarray data analysis
To compare gene expression of our collection of target antigens between normal B cells and CLL cells, raw data files (.CEL) generated on Affymetrix U95v2 microarrays by Klein et al.(21) were collected and normalized using Robust Multiarray Averaging (RMA). We used Resourcerer(22) to map gene identifiers of the panel of target antigens to Affymetrix probeset IDs (PsIDs). For those targets that mapped to multiple PsIDs, the median of the expression values was used. Raw data files were also processed using Affymetrix MAS 5.0, and genes were defined as detectable if the detection call was “present” in ≥40% of samples for each cell type. The Fisher Exact test was used for comparison between groups.
Measurement of antigen-specific gene expression by quantitative PCR
To evaluate the gene expression of the candidate antigens, RNA was extracted (RNeasy kit, QIAGEN, Valencia, CA) from normal PBMC, immunomagnetically purified B cells from normal PBMC (CD19+ microbeads; Miltenyi, Auburn, CA), and from B-CLL tumor cells. CLL RNA was extracted from cryopreserved samples of peripheral blood or marrow that were known to contain >85% tumor cells. First-strand cDNA was generated from 2 µg of total RNA by using random hexamers (Pharmacia LKB Biotechnology, Piscataway, NY) and reverse transcriptase (Superscript, GIBCO BRL, Gaithersburg, MD) according to the manufacturer’s instructions. Antigen-specific gene expression was measured by quantitative real-time PCR (qRT-PCR) (Taqman Gene Expression Assays; Applied Biosystems, Foster City, CA) using a 7500 Fast Real-time PCR cycler (Applied Biosystems, Foster City, CA). Transcript expression was measured relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The significance of the difference in gene expression of target antigens between cell populations was evaluated using the exact Wilcoxon rank sum test.
Generation of cell lysates and Western blotting
Whole cell lysate was generated from patient samples by lysis with radioimmunoprecipitation assay buffer (1% NP40, 0.5% deoxycholate, 0.1% SDS, 125 mmol/L sodium chloride, 50 mmol/L HEPES [pH 7.4]) in the presence of protease and phosphatase inhibitors. Lysates (20 µg total protein per lane) were subjected to gel electrophoresis using 4%–15% gradient SDS-PAGE gels in Tris-glycine buffer, and transferred onto nitrocellulose filters in Tris-glycine buffer containing 20% methanol. Antibodies specific to DAPK3 (B2034-1, 1:500; Epitomics, Burlingame, CA), MDS032 (ARP39361, 1:1000; Aviva Systems, San Diego CA), SERBP1 (H00026135, 1:500; Abnova, Taipei, Taiwan), ZFYVE19 (H0004936, 1:400; Abnova, Taipei, Taiwan), and OGFOD1 (Ab69359, 1:100; Abcam, Cambridge MA) were used to detect protein expression of the respective antigens by Western blot, followed by incubation with horseradish peroxidase (HRP)-linked secondary antibodies. Antibody to β-actin (A1978, 1:5000, Sigma Aldrich, St. Louis, MO) was used as a control to ensure equal loading of lanes.
Plasmid acquisition and in vitro translation of candidate antigens
To facilitate protein expression for target validation and characterization, DNA sequences encoding a subset of the candidate antigens were either acquired in or cloned into Gateway (Invitrogen, Carlsbad CA) expression vectors. DNA sequences in Gateway vectors were obtained from PlasmID (Harvard Institute of Proteomics, Cambridge, MA)(23), the Ultimate ORF collection (Invitrogen, Carlsbad, CA), or the Human Orfeome Collection(24) (generous gift of Dr. Mark Vidal, Center for Cancer Systems Biology, DFCI, Boston MA). Additional DNA sequences were purchased from American Tissue Culture Collection (Manassas, VA), Open Biosystems (Huntsville, AL), or the Resource Center of the German Human Genome Project (RZPD, Berlin, Germany), or were directly PCR-amplified from CLL tumor cell cDNA, and cloned into the pDONR221 Gateway vector (Invitrogen, Carlsbad, CA). The majority of acquired cDNA clones originated from the I.M.A.G.E. Consortium (LLNL) (Supplemental Table 1)(25). The inserts of all sequences in Gateway vectors were verified by sequencing, and then shuttled into a Gateway-converted mammalian expression vector containing a T7 promoter and a C-terminus glutathione-S-transferase (GST) tag (gift of Wagner Montor, Harvard Institute of Proteomics, Cambridge, MA) using LR Clonase (Invitrogen, Carlsbad, CA). Candidate antigens were expressed in vitro with rabbit reticulocyte lysate (TNT T7 Quick Coupled Transcription/Translation, Promega, Madison, WI), using biotinylated lysine transfer RNA (Transcend tRNA, Promega, Madison, WI). 50 µl reactions using 1–2 µg of template DNA and 2.5 µl of Transcend tRNA were incubated at 30°C for 90 min on a tabletop shaker at 950 rpm. A reaction using 1.5 µg of DNA encoding luciferase was performed with each synthesis batch as a positive control.
Immunoprecipitation (IP) of candidate antigens using patient plasma
Protein was immunoprecipitated using patient plasma, as previously described (20). In brief, 5 µl of reticulocyte lysate product was immunoprecipitated with 5 µl of serum or plasma at 4°C. To mirror the short incubation time period used for the protein array probing, and to differentiate strong antibody interactions from nonspecific cross-reactivity, we limited the immunoprecipitation reaction to one hour. Immunoprecipitated products were isolated using 50% protein A sepharose CL-4B beads (GE Healthcare, United Kingdom) and visualized by immunoblot. Protein was detected using immunoperoxidase-streptavidin (1:5000 dilution; MP Biomedicals, Solon, OH). The blots were developed with Supersignal West Femto detection substrate (Pierce Biotechnology, Rockford, IL) and imaged with BioMax Light film (Kodak, Rochester, NY).
For some experiments, immunoprecipitation of a single candidate antigen against up to 48 serum or plasma samples were performed simultaneously to directly compare antigen-antibody affinity between plasma samples. Immunoprecipitation against GST was used as comparison for background serum reactivity. As a control and for comparing protein bands across immunoblots, a 1:440 dilution of one batch of luciferase reticulocyte lysate product was loaded onto 1–2 lanes on each immunoblot. Serum or plasma sample reactivity was compared against the range of reactivity evidenced by the 12 normal subjects, taking into account the difference in the strength of the control band on each western blot.
Results
DLI for CLL is associated with potent humoral immune responses
We identified 2 patients with relapsed CLL who remained in continuous remission more than 8 years after receiving CD8-depleted DLI (26). As shown in Table 1, both Patients A and B failed multiple regimens of conventional chemotherapy before receiving myeloablative T cell-depleted HSCT from HLA-matched sibling donors. Both relapsed with clinical disease within 5 years of transplantation, and received DLI without any further chemotherapy or radiotherapy. Following DLI, both patients had a clear GvL response without acute or chronic GvHD, demonstrating rapid conversion to full donor hematopoiesis and gradual disappearance of histologically evident marrow disease over 6–16 months.
Table 1.
Clinical characteristics of Patients A and B
| Pt | Age at BMT (yr) |
Patient/ Donor Sex |
HSCT type | Treatment before BMT |
Time to relapse (mo) |
Pre- DLI Tx |
Donor chimerism conversion / pathologic response (months after DLI) |
Post- DLI GvHD |
Last follow-up (yr) |
|---|---|---|---|---|---|---|---|---|---|
| A | 51 | M/F (sister) |
myeloablative MRD TCD |
Leukeran + prednisone; 6 cycles CHOP; fludarabine |
60 | None | 6/16 | None | 10 |
| B | 49 | M/F (sister) |
myeloablative MRD TCD |
6 cycles CHOP; Dexamethasone + BEAM |
50 | None | 2/6 | None | 8 |
HLA – Human leukocyte antigen; HSCT – hematopoietic stem cell transplant; MRD - matched related donor; TCD - T cell depleted.
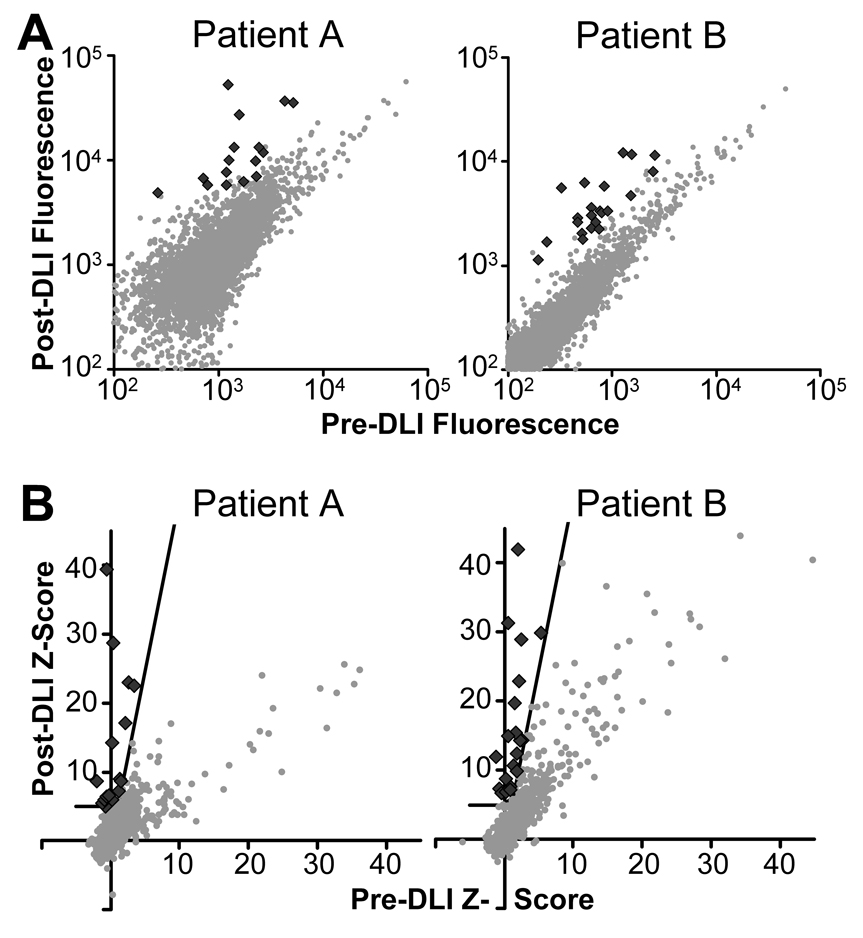
To identify the B cell targets associated with CLL tumor regression in these two patients, we used plasma collected at 0–1 months before and 9–12 months after DLI as sources of antibody for probing high-density protein microarrays. The vast majority of array proteins were bound at comparable levels by pre- and post-DLI plasma antibodies based on raw fluorescence intensity. Using a concentration-dependent analysis, significant antigen-antibody interactions were identified, shown as black diamonds in Figure 1 (20). These candidate antigens comprised less than 1% of spotted proteins. No proteins had significantly decreased antibody reactivity following immunotherapy.
Figure 1. Post-DLI plasma contains greater antigen reactivity than pre-DLI plasma.
(A) Plots of the pre- vs. post-DLI median background-subtracted fluorescence signal for all microarray proteins for Patients A and B. Each point represents the average across the two replicate protein spots on the microarray. (B) The calculated significance of the measured pre- and post-DLI fluorescence, corrected for protein concentration, for each microarray protein (20). The black line denotes the cutoff function used to determine significance. ♦ significant interaction;  other protein.
other protein.
Targets of the DLI-induced humoral immune response in patients with CLL
Analysis of the protein microarray data yielded 16 candidate DLI-associated antigens for Patient A, and 21 for Patient B. These antigens were ranked based on the largest cutoff n that would identify them as significantly changed. Although most candidate antigens were identified in only one patient, the two proteins encoded by genes ZFYVE19 and C6orf130 were recognized by both patients. A third protein, MDS032, was identified as a candidate antigen for Patient A and elicited borderline (n≥3) antibody reactivity for Patient B. The candidate antigens and the shared antigen MDS032, only borderline-significant for Patient B, are listed in Table 2. The candidate antigens as well as 26 interactions of borderline significance for Patient A and 31 for Patient B are further described in Supplemental Table 1.
Table 2.
Candidate antigens showing increased antibody reactivity post- compared to pre-DLI for Patients A and B
| Accession | Gene | Pt | Rank * |
Repro duced† |
Size (aa) |
Chrom osome |
Protein | Function |
|---|---|---|---|---|---|---|---|---|
| NM_001348 | DAPK3 | A | 1 | Y | 454 | 19p13.3 | Death-associated protein kinase 3 | Positive regulator of apoptosis |
| BC013778 | SLC7A6 OS |
A | 2 | Y | 309 | 16q22.1 | Solute carrier family 7 member 6 opposite strand |
Uncharacterized |
| NM_015640 | SERBP1 | A | 4 | Y | 387 | 1p31 | SERPINE1 mRNA binding protein 1 | Regulation of SERPINE1 mRNA stability |
| BC012596 | ARPC4 | A | 6 | - | 168 | 3p25.3 | Actin related protein 2/3 complex, subunit 4 |
Actin filament polymerization |
| NM_182692 | SRPK2 | A | 8 | - | 688 | 7q22- q31.1 |
SFRS protein kinase 2 | Spliceosome assembly and splicing factor trafficking |
| BC002755 | MKNK1 | A | 9 | Y | 465 | 1p33 | MAP kinase-interacting serine/threonine kinase 1 |
MAPK signal integration |
| NM_018070 | SSBP3 | A | 10 | - | 368 | 1p32.3 | Single stranded DNA binding protein 3 |
Transcription regulation |
| BC038105 | MPP7 | A | 11 | - | 576 | 10p11.2 3 |
Membrane protein, palmitoylated 7 | Localization of proteins to tight junctions |
| BC027729 | N/A | A | 12 | - | 130 | 3p25.1 | N/A | Uncharacterized |
| NM_004252 | SLC9A3R 1 |
A | 13 | - | 358 | 17q25.1 | Solute carrier family 9, isoform 3 regulator 1 |
Plasma scaffold protein |
| BC007347 | CHD2 | A | 14 | N | 501 | 15q26 | Chromodomain helicase DNA binding protein 2 |
Eukaryotic nucleus organization |
| NM_032567 | SPZ1 | A | 15 | Y | 430 | 5q14.1 | Spermatogenic leucine zipper protein 1 |
Cytokinesis during development; spermatogenesis |
| BC016848 | FAM131 C |
A | 16 | Y | 280 | 1p36.13 | fFamily with sequence similarity 131, member C |
Hypothetical |
| BC021092 | ZFYVE19 | A,B | 3,18 | Y,Y | 328 | 15q15.1 | Zinc finger FYVE domain- containing protein 19 |
Ubiquitin-protein ligase |
| NM_145063 | C6orf130 | A,B | 5,19 | Y,Y | 152 | 6p21.1 | Hypothetical | Uncharacterized |
| BC006005 | MDS032 | A,B | 7,35 | Y,Y | 146 | 19p13.1 1 |
Hematopoietic stem/progenitor cells protein |
Uncharacterized |
| BC036910 | LOC3888 82 |
B | 1 | Y | 240 | 22q11.2 3 |
N/A‡ | Possibly nonsense-mediated mRNA decay |
| NM_138333 | FAM122 A |
B | 2 | Y | 287 | 9q21.11 | Family with sequence similarity 122A |
Uncharacterized |
| BC031650 | SH3RF2 | B | 3 | N | 220 | 5q32 | SH3 domain containing ring finger 2 | Ubiquitin conjugation pathway |
| BC037547 | CDC20B | B | 4 | Y | 192 | 5q11.2 | Cell division cycle 20 homolog B | Regulates the anaphase promoting complex/cyclosome |
| BC001426 | UQCRH | B | 5 | Y | 91 | 1p33 | Ubiquinol-cytochrome c reductase hinge protein |
Mitochondrial |
| BC034401 | N/A | B | 6 | N | 71 | 16 | N/A | Uncharacterized |
| NM_005522 | HOXA1 | B | 7 | Y | 335 | 7p15.3 | Homeobox A1 | Development transcription regulation |
| NM_005565 | LCP2 | B | 8 | Y | 533 | 5q33.1- qter |
Lymphocyte cytosolic protein 2 | T-cell antigen receptor mediated signaling |
| BC032919 | OGFOD1 | B | 9 | Y | 542 | 16q13 | 2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
Protein metabolism |
| NM_001722 | POLR3D | B | 10 | Y | 398 | 8q21 | DNA-directed RNA polymerase III polypeptide D |
RNA polymerase |
| NM_004401 | DFFA | B | 11 | N | 331 | 1p36.3- p36.2 |
DNA fragmentation factor 45kDa alpha polypeptide |
P53-independent role in chromosome stability |
| NM_016520 | C9orf78 | B | 12 | N | 289 | 9q34.11 | Chromosome 9 open reading frame 78 |
Uncharacterized |
| NM_006251 | PRKAA1 | B | 13 | - | 550 | 5p12 | AMP-activated protein kinase catalytic subunit alpha-1 |
Cholesterol and fatty acid biosynthesis |
| NM_006002 | UCHL3 | B | 14 | N | 230 | 13q22.2 | Ubiquitin carboxyl-terminal esterase L3 |
Processing of ubiquitin and ubiquitinated proteins |
| BC026104 | PDCD4 | B | 15 | - | 469 | 10q24 | Programmed cell death 4 | Inhibits neoplastic transformation |
| NM_001219 | CALU | B | 16 | N | 315 | 7q32 | Calumenin | Endoplasmic reticulum protein folding/sorting |
| BC000108 | WWP2 | B | 17 | Y | 335 | 16q22.1 | WW domain containing E3 ubiquitin protein ligase 2 |
Ubiquitination |
| BC031228 | N/A | B | 20 | Y | 79 | 11 | N/A | Uncharacterized |
| BC047536 | SCEL | B | 21 | N | 668 | 13q22 | Sciellin | Precursor to keratinocyte cornified envelope |
Rank order by the significance of change between pre- and post-DLI reactivity.
Reactivity reproducible by IP assay. Y=successful N=unsuccessful, -=not attempted
There is support for the transcript, as a nonsense-mediated mRNA decay candidate, but not for the protein (27).
We queried publicly available databases, including Entrez, UniGene, Harvester, and SymAtlas (27–29), for information on the function of the 35 identified candidate antigens, summarized in Table 2. Of the 25 known candidate antigens, 24 are intracellular and 18 are reported as tumor-associated (27). Eight have DNA or RNA binding activity, 4 are part of the ubiquitination pathway, 2 are tumor suppressors, and 1 each are lymphocyte-associated, or function in the cell cycle, spermatogenesis or apoptosis.
Validation of serologic reactivity to candidate antigens
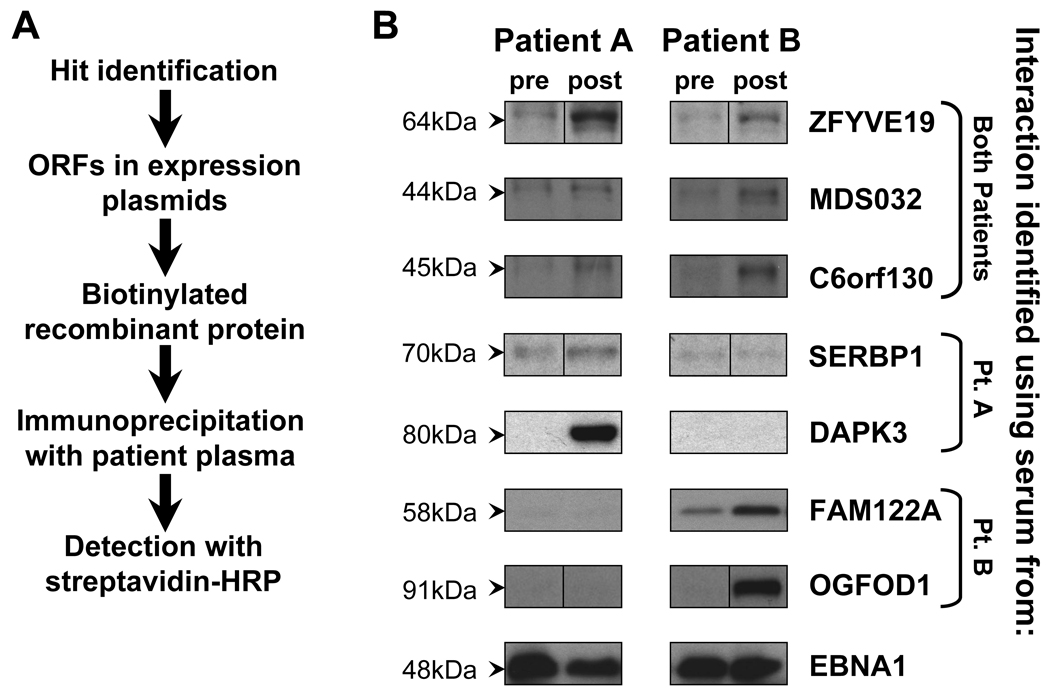
Significant microarray signals may result from true interactions, erroneous measurement, or interactions with misidentified proteins, such as due to improper synthesis or co-precipitation of additional proteins. To confirm that the serologic targets identified by microarray analysis represent true antibody-antigen interactions, we independently validated antibody reactivity against our screened candidate antigens. Figure 2A schematically summarizes our validation strategy. We expressed our candidate antigens as C-terminus GST-fusion biotinylated proteins in a cell-free mammalian expression system, a different method of protein manufacture than used on the protein microarrays. We then immunoprecipitated the recombinant proteins with patient plasma, and visualized immunoprecipitated proteins by immunoblot. In this manner, interactions between serum antibodies and properly folded and post-translationally modified antigen were detected, and nonspecific interactions were excluded on the basis of size on immunoblot.
Figure 2. Validation studies confirm greater antigen-specific antibody reactivity in post-DLI compared to pre-DLI plasma.
(A) Schematic representation of the immunoprecipation-based validation assay. (B) Validation of serologic reactivity generated by protein microarray analysis using immunoprecipitation followed by Western blot. Antigens were generated as biotinylated glutathione-S-transferase (GST) fusion proteins by in vitro transcription and translation using rabbit reticulocyte lysate, immunoprecipitated with pre- and post-DLI plasma from Patients A and B, and immunoblotted with streptavidin-HRP. Shown are ZFYVE19 (64kDa), MDS032 (44.5kDa), C6orf130 (45kDa), SERPB1 (70kDa), DAPK3 (80kDa), FAM122A (58kDa), OGFOD1 (91kDa), and EBNA1 (48kDa), with sizes including the antigen sequence on the microarray and attached GST tag. Sample full-length blots are presented in Supplemental Figure 1.
Using this assay, we successfully validated 22 of the 30 tested candidate antigen interactions, listed in Table 2. Eight interactions failed validation, and eight remained untested due to limited patient sample. Figure 2B shows representative results from these studies. Consistent with our bioinformatic analysis, post-DLI plasma from both Patients A and B immunoprecipitated greater amounts of the shared antigens C6orf130, MDS032 and ZFYVE19 compared to pre-treatment plasma. For SERBP1 and DAPK3, identified by Patient A, and FAM122A and OGFOD1, identified by Patient B, greater immunoprecipitated antigen was evidenced only by the post-DLI sample from the patient who identified the interaction. Equivalently high IP reactivity against the control EBNA1 construct was seen for all plasma samples tested. Interactions of higher significance were slightly more likely to be validated, with 88% of tested interactions with rank <10 validated, while only 57% of tested interactions with rank ≥10 were validated (Table 2).
A subset of candidate antigens are highly expressed in B lineage cells
To broadly survey the gene expression of our collection of antigens, we first compared the gene expression of our candidate antigens in primary CLL cells and normal B cell subsets using existing gene expression microarray data generated on the Affymetrix U95v2 array (21). Probesets for 18 of the 35 candidate antigens were present on this microarray. In this data, 5 of the 18 matched candidate antigens, LCP2, PDCD4, MKNK1, SRPK2, and CHD2, showed higher expression in CLL compared to normal B cells (p<0.01), but none at the level of differential expression that was found for the most significantly upregulated 33 genes reported in that study.
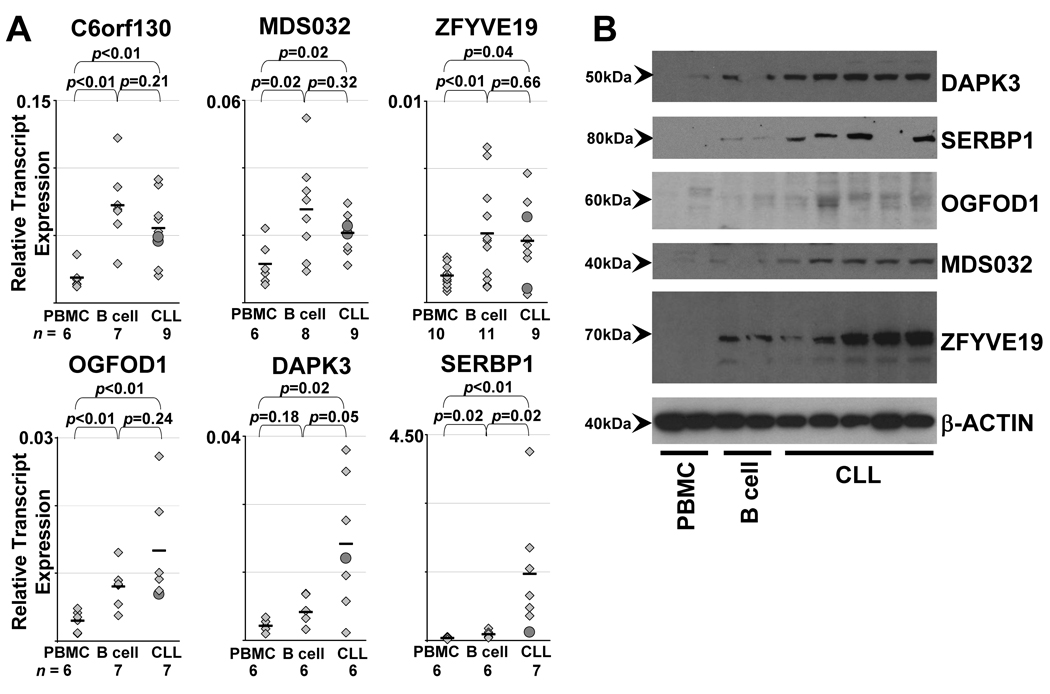
Since many of the candidate antigens were not represented on the gene expression microarray, we next directly quantified antigen-specific gene expression in normal and malignant B cells by using qRT-PCR. We performed gene expression studies for 9 candidate antigens in 3 tissue sources: normal PBMC, normal B cells and CD19+CD5+ CLL tumor cells. No significant difference in gene expression was observed among the three tissue groups for 3 of the antigens (PRKAA1, POLR3D, and SPZ1). We observed increased expression in normal and malignant B cells compared to PBMC for the three shared antigens C6orf130, MDS032, and ZFVYE19, as well as for OGFOD1, while a third pattern, in which antigen-specific gene expression is significantly increased in CLL cells compared to normal B cells, was observed for DAPK3 and SERBP1 (Figure 3A). Consistent with the notion that GvL-associated antigens may be B cell lineage genes, the three shared antigens C6orf130, MDS032, and ZFYVE19 demonstrated significantly higher expression levels in normal and malignant CD19+ B cells compared to PBMC (p<0.02). Western blotting of lysates from CLL-B cells and normal B cells compared to PBMC confirmed these results (Figure 3B). These results indicate that a subset of candidate antigens is both immunogenic and highly expressed in CLL.
Figure 3. Gene and protein expression of the shared candidate antigens in normal PBMC, normal CD19+ B cells, and CLL cells.
(A) Gene expression was measured by gene-specific quantitative real-time PCR and normalized to transcript levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Black bars denote mean values. Significance of the difference between groups by the Wilcoxon rank-sum test is shown on each graph. The number of samples in each group is given below the figures. Candidate antigens with similar relative expression in both normal and CLL B cells (C6orf130, MDS032, ZFYVE19, OGFOD1) or with increased expression in CLL cells compared to normal B cells (DAPK3, SERBP1). (B) Protein expression of candidate antigens measured by Western blot of cell lysates for PBMC (n=2), normal CD19+ B cells (n=2) and CLL cells (n=5). Samples presented are independent of the samples tested in (A). Protein lysates were loaded at 20 µg/lane. Separate Western blots were probed with commercial antibodies to DAPK3, SERBP1, OGFOD1, MDS032, ZFYVE19 and β-actin (as control).
Candidate antigens are not minor histocompatibility antigens
The immunogenicity of candidate antigens can arise from sequence differences between donor, host and tumor, or differential presentation of antigen epitopes by tumor cells. Sequence differences may exist between donor and host due to gene polymorphisms, resulting in minor histocompatibility antigens (mHA). Direct sequencing of 6–10 independent copies of both tumor and donor cell templates revealed the absence of sequence differences for ZFVYE19, MDS032, C6orf130, and DAPK3 between donor and CLL tumor cells. Thus, the selected subset of the identified antigens are not mHAs, but represents overexpressed CLL-associated or B cell-associated antigens.
Clinical response to DLI is temporally associated with antibody responses to candidate antigens
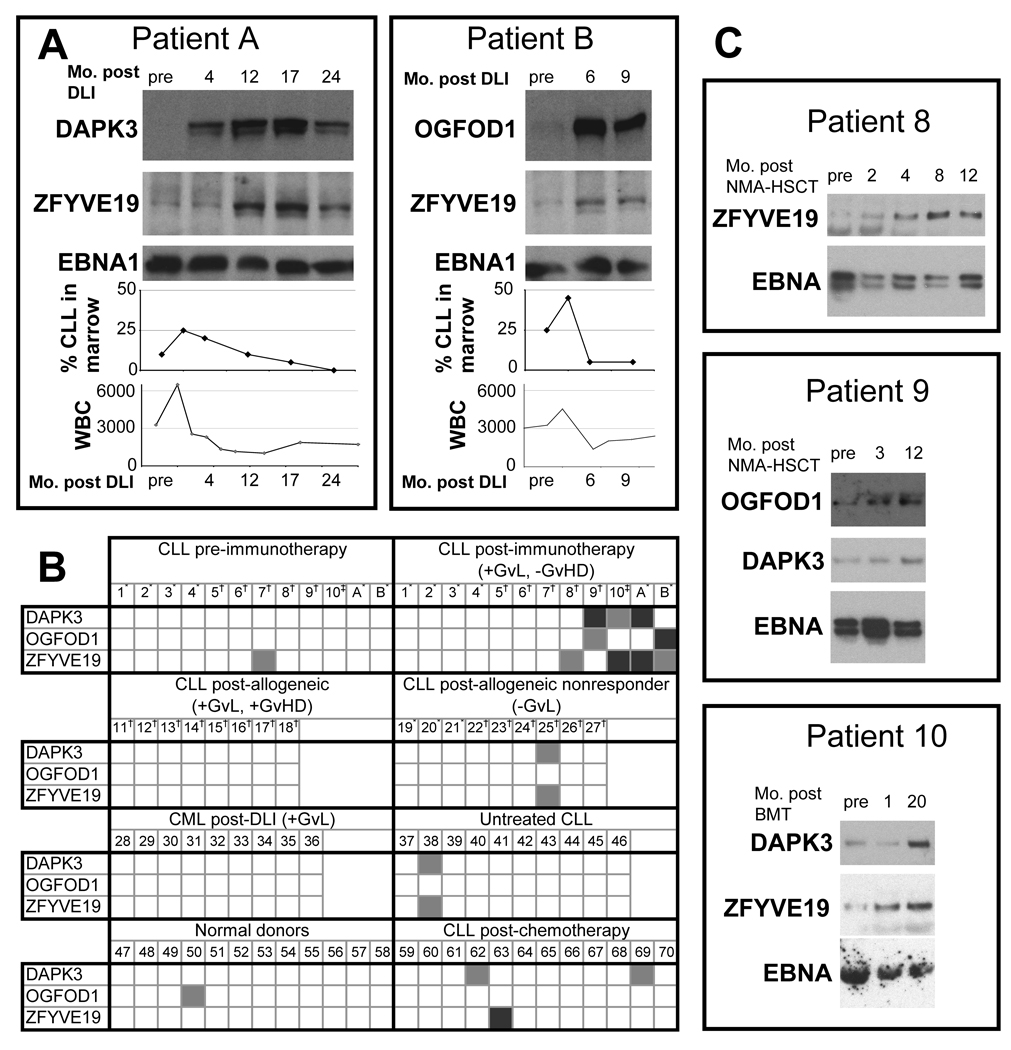
After DLI therapy, both Patients A and B demonstrated normalization of peripheral blood lymphocytosis, and disappearance of tumor cells from the marrow space (Figure 4A). Leukemia cells occupied 25% of Patient A’s marrow intertrabecular space at 2 months after DLI, but decreased to undetectable levels within 24 months. Similarly, CLL cells occupied 45% of the marrow intertrabecular space of Patient B at 3 months after DLI, but decreased to less than 5% tumor involvement by 9 months post-DLI.
Figure 4. Serologic reactivity against CLL candidate antigens.
(A) The antibody responses of Patients A and B temporally correlate with the clinical anti-tumor response. DAPK3 (80kD), OGFOD1 (91kD), ZFYVE19 (64kD) and EBNA1 (48kD) were generated as biotinylated GST-fusion proteins by in vitro transcription and translation. These were immunoprecipitated using Protein A beads with pre- and post-DLI plasma, and detected with streptavidin-HRP. The bottom graphs depict the percent of marrow intertrabecular space filled with CLL cells following DLI and the white blood cell (WBC) counts across time. (B) Plasma reactivity of multiple patient and normal subject groups against antigens DAPK3, OGFOD1 and ZFYVE19 measured using immunoprecipitation. Interactions were graded against that of normal subjects, with light gray denoting weak interactions and dark gray strong interactions. CLL-chronic lymphocytic leukemia; CML-chronic myelogenous leukemia; GvL-Graft versus Leukemia, denoted as present (+) or absent (−); GvHD-Graft versus Host Disease, denoted as present (+) or absent. Treatments included *donor lymphocyte infusion (DLI), †non-myeloablative allogeneic hematopoietic stem cell transplant (HSCT), and ‡myeloablative allogeneic HSCT. Numbers denote individual patients, with the first two groups having matched pre- and post-immunotherapy samples. (C) The antibody responses of Patients 8, 9 and 10 against antigens DAPK3, OGFOD1 and ZFYVE19 developed over time after allogeneic HSCT. Antibody responses were measured as in (A). The preparative regimen and salvage chemotherapy for HSCT resulted in disease remission, and precludes comparison of WBC and bone marrow timecourses. Full-length blots are presented in Supplemental Figure 2.
The development of humoral immunity was strongly correlated temporally with clinical response against CLL (Figure 4A). To determine whether the kinetics of the humoral immune response against the candidate antigens correlates with the clinical response to DLI, we compared the disease burden with plasma reactivity against candidate antigens at various timepoints following DLI. We examined antigen-specific antibody reactivity over time by immunoblot following IP against DAPK3 and ZFYVE19 for Patient A, and against OGFOD1 and ZFYVE19 for Patient B. Increasing antibody reactivity against the tested antigens correlated with decreasing tumor involvement in marrow. while comparable reactivity against the control EBNA1 antigen was observed for all plasma samples. For Patient A, the antibody reactivity to ZFYVE19 was constant from pre-DLI to 4 months post-DLI, then increased, peaking at 17 months and decreasing at 24 months. Antibody reactivity to DAPK3 for the same patient followed a similar pattern, increasing from its pre-DLI level from 4 months to 17 months, before decreasing at 24 months. For Patient B, the antibody reactivity to ZFYVE19 and OGFOD1 both increased at 6 and 9 months relative to the pre-DLI level, with samples for further follow-up not available.
The candidate antigens are broadly immunogenic in CLL patients with GvL
We examined patterns of serologic reactivity against GST-fusion proteins of DAPK3, OGFOD1 and ZFYVE19 by testing 84 plasma samples collected from 72 patients with hematologic malignancies or normal volunteers (Figure 4B). Five of 12 patients with long-term remission with minimal or no GvHD after HSCT developed new antibody reactivity after treatment against at least one of the three antigens. In addition to reactivity against DAPK3 and ZFYVE19 by Patient A and OGFOD1 and ZFYVE19 by Patient B, three additional patients (subjects 8–10) who underwent HSCT developed reactivity against one or more of the three antigens (Figure 4B, first 2 panels; Figure 4C). Patients 8 and 9 underwent nonmyeloablative allogeneic HSCT, while Patient 10 underwent myeloablative allogeneic HSCT. Post-treatment samples were selected at one year following transplant, when immunosuppressive medications had been tapered and GvHD was not noted.
In contrast, we observed only occasional instances of reactivity for these three antigens among normal donors or other control groups. Only 1 of 12 normal volunteers was reactive to OGFOD1, and none of 12 to the other two antigens. Only 1 of 9 post-HSCT CLL non-responder patients and 1 of 10 untreated CLL patients were reactive to both DAPK3 and ZFYVE19. Similarly, only 2 of 12 chemotherapy-treated CLL patients displayed reactivity against DAPK3; and 1 of 12 against ZFYVE19. No evidence of reactivity was observed among 9 CML patients successfully treated with DLI or among 8 allo-transplanted CLL patients who developed GvHD, suggesting that the antigens are broadly immunogenic in patients with CLL who have developed GvL responses.
Discussion
Identification of target antigens remains a priority in cancer immunology. Few CLL-associated antigens have been previously described, and of these, few have been shown to consistently generate cytotoxicity against primary CLL cells (16, 17, 19, 21, 30). To identify CLL antigens associated with the GvL effect, we have focused on naturally immunogenic antigens associated with long-lasting immunity to CLL induced by donor lymphocyte infusion. Antibody-based approaches for target antigen discovery are attractive because of their ease of use and the mounting evidence that antibody responses correlate with meaningful clinical responses to treatment. For example, Miklos et al. (12) reported a highly significant correlation between relapse-free survival and the presence of antibody responses against Y-encoded minor histocompatibility antigens. Other investigators have reported antigen-specific B cell responses as reliable markers of clinical response following vaccination (31). To this end, we dissected the immune responses of two patients who achieved long-term disease remission without clinically apparent GvHD, and identified 35 unique antigens that elicited significant antibody reactivity following, but not prior to, DLI.
While our candidate CLL-associated antigens are predominantly intracellular proteins, they are potentially reliable markers for T cell targeting. Our recent studies demonstrate that treatment-associated tumor antigens that elicit high titer B cell responses are also targets of CD8+ T cell immunity (32). Intracellular antigen may complex with antibody for enhanced delivery of antigen through FcγR-mediated pathways to antigen presenting cells, thus augmenting T cell responses (33). For example, vaccination with the intracellular tumor antigen NY-ESO-1 consistently results in antigen-specific antibody production that participates in cross-priming of this antigen to T cells (34). Our findings thus provide a rich source of candidate CLL antigens, and our future studies will explore the extent to which antigen-specific antibody responses contribute to the development of coordinated T cell responses against our most promising targets.
Our candidate antigens were identified using high-density protein microrarrays, a powerful emerging technology. This immunoproteomic screening platform enables high-throughput screening of thousands of proteins for interactions, and measures the molecular biomarker endpoints of disease more directly than DNA microarray technology (35). High-density protein microarrays offer several distinct advantages over conventional screening platforms. All proteins spotted on protein microarrays are distinct open reading frames expressed in a eukaryotic system that preserves potentially antigenic post-translational modifications. Protein microarrays also offer a degree of standardization across experiments, since the same proteins of comparable concentration are printed across multiple microarrays. On the other hand, they are unlikely to contain antigens that are specific to a tumor or to an individual, such as personal immunogenic tumor-restricted mutations. Protein microarrays are also not yet well-established; we therefore devoted extensive efforts to developing a novel bioinformatic analysis (20) as well as a reliable method for independent validation of identified antigens. Because errors in the manufacture of protein microarrays have been described which could lead to misidentified antigens on high-throughput screening (36), we developed an immunoprecipitation–based validation assay to sensitively detect the interaction of antibodies with properly folded protein antigens with size determination via immunoblot.
Overall, 22 of 30 (73%) tested antigen interactions were experimentally confirmed to elicit greater antibody reactivity in the post- compared to pre-DLI plasma. Characterization of several of the validated target antigens led us to the insight that many of the identified targets of GvL are encoded by non-mutated B cell lineage genes. Increasing numbers of mHAs have been described over the years, including some with selective B cell expression (37). Screens for GvL-associated antigens following HSCT have been biased to identify mHAs because their primary criterion for selection is differential cytolytic T cell reactivity against host tissue compared to donor cells. These studies demonstrate that allo-immunity is a fundamentally important mechanism underlying GvL. However, they neglect the potential contribution of immunity against other classes of tumor-associated antigens that have been defined in other non-transplant tumor systems, such as non-mutated differentiation, aberrantly expressed antigens (38), or even self antigens that are immunogenic when presented by tumor but are normally sequestered from immune detection (39). Our current studies therefore reveal that classes of antigen other than mHAs, namely non-mutated tumor-associated antigens, comprise a component of effective GvL responses. These results are consistent with recent studies by Nishida et al. (40) demonstrating that in addition to T cell reseonses against alloantigens, T cell reactivity against CLL-specific antigens can be detected following nonmyeloablative HSCT in association with clinical response.
The discovery that the targets of effective anti-CLL immunity include non-mutated antigens that are highly expressed in CLL suggests that these antigens may have broader anti-CLL immunological utility beyond the donor-host pairs from which they were identified. While Patients A and B developed reactivity against largely different panels of antigens, a subset of antigens elicited humoral immunity in both patients. Moreover, close to half of our cohort of 12 CLL patients with GvL responses reacted against at least 1 of the 3 candidate antigens tested. These results suggest that as a collection, these antigens would be useful for monitoring immune responses against CLL after immune-based therapies. For example, several ongoing studies are examining the effects of vaccination with whole autologous tumor in which the specific immunogens are unknown (41, 42).
We selected for further characterization three antigens identified by both patients’ sera, and three antigens found to be highly expressed in CLL. Of the shared antigens, C6orf130 is a predicted open reading frame in chromosome 6. MDS032 (USE1) is an uncharacterized hematopoietic stem/progenitor cell protein that localizes to the endoplasmic reticulum, and functions in the formation of a SNARE complex (43). ZFYVE19 is a zinc finger FYVE domain-containing protein, which has been reported as an in-frame fusion partner to the mixed lineage leukemia gene (44). The FYVE domain mediates recruitment of proteins to membranes containing phosphatidylinositol 3-phosphate, in particular endosomes. Among the 3 nonshared antigens, DAPK3, death associated protein kinase 3, plays a role in apoptosis (45) and spermatogenesis (46). SERBP1, SERPINE1 mRNA binding protein 1, is significantly overexpressed in ovarian epithelial cell tumors, with increased expression in advanced disease suggesting a role in tumor invasion and metastasis (47). OGFOD1, 2-oxoglutarate and iron-dependent oxygenase domain containing 1, has been reported in tumor cell pseudopodial protrusions (48).
In closing, our identification of non-mutated and commonly immunogenic CLL antigens reveals that immune responses following effective immunotherapy are directed, at least in part, to antigens present on CLL tumor cells. As the identified candidate antigens represent immunogenic tumor-associated antigens and not alloantigens, these may be attractive targets for immunotherapy directed at CLL outside of the context of allogeneic transplantation.
Supplementary Material
Acknowledgments
We thank Roslyn Gerwin and Beth Witten for expert technical assistance. We acknowledge the generosity of Drs. David Hill and Mark Vidal for providing plasmids and the support of the DFCI transplant team and Pasquarello Tissue Bank for providing clinical samples.
Financial Support: OM was supported by a Howard Hughes Medical Institute Medical Research Training Fellowship; CJW acknowledges support from the Department of Defense (W81XWH-07-1-0080), the Miles and Eleanor Shore Award, the NCI (5R21CA115043-2), the Early Career Physician-Scientist Award of the Howard Hughes Medical Institute, and is a Damon-Runyon Clinical Investigator supported (in part) by the Damon-Runyon Cancer Research Foundation (CI-38-07).
Footnotes
Presented in part as a simultaneous oral session at the 49th annual meeting of the American Society of Hematology, Atlanta, GA, December 9, 2007.
The authors have no competing financial interests to declare.
References
- 1.Byrd JC, Lin TS, Grever MR. Treatment of relapsed chronic lymphocytic leukemia: old and new therapies. Semin Oncol. 2006;33:210–219. doi: 10.1053/j.seminoncol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Esteve J, Villamor N, Colomer D, et al. Stem cell transplantation for chronic lymphocytic leukemia: different outcome after autologous and allogeneic transplantation and correlation with minimal residual disease status. Leukemia. 2001;15:445–451. doi: 10.1038/sj.leu.2402036. [DOI] [PubMed] [Google Scholar]
- 3.Dreger P, Corradini P, Kimby E, et al. Indications for allogeneic stem cell transplantation in chronic lymphocytic leukemia: the EBMT transplant consensus. Leukemia. 2007;21:12–17. doi: 10.1038/sj.leu.2404441. [DOI] [PubMed] [Google Scholar]
- 4.Pavletic SZ, Khouri IF, Haagenson M, et al. Unrelated donor marrow transplantation for B-cell chronic lymphocytic leukemia after using myeloablative conditioning: results from the Center for International Blood and Marrow Transplant research. J Clin Oncol. 2005;23:5788–5794. doi: 10.1200/JCO.2005.03.962. [DOI] [PubMed] [Google Scholar]
- 5.Rondon G, Giralt S, Huh Y, et al. Graft-versus-leukemia effect after allogeneic bone marrow transplantation for chronic lymphocytic leukemia. Bone Marrow Transplant. 1996;18:669–672. [PubMed] [Google Scholar]
- 6.Toze CL, Galal A, Barnett MJ, et al. Myeloablative allografting for chronic lymphocytic leukemia: evidence for a potent graft-versus-leukemia effect associated with graft-versus-host disease. Bone Marrow Transplant. 2005;36:825–830. doi: 10.1038/sj.bmt.1705130. [DOI] [PubMed] [Google Scholar]
- 7.Collins RH, Jr, Shpilberg O, Drobyski WR, et al. Donor leukocyte infusions in 140 patients with relapsed malignancy after allogeneic bone marrow transplantation. J Clin Oncol. 1997;15:433–444. doi: 10.1200/JCO.1997.15.2.433. [DOI] [PubMed] [Google Scholar]
- 8.Delgado J, Thomson K, Russell N, et al. Results of alemtuzumab-based reduced-intensity allogeneic transplantation for chronic lymphocytic leukemia: a British Society of Blood and Marrow Transplantation Study. Blood. 2006;107:1724–1730. doi: 10.1182/blood-2005-08-3372. [DOI] [PubMed] [Google Scholar]
- 9.Schetelig J, Thiede C, Bornhauser M, et al. Evidence of a graft-versus-leukemia effect in chronic lymphocytic leukemia after reduced-intensity conditioning and allogeneic stem-cell transplantation: the Cooperative German Transplant Study Group. J Clin Oncol. 2003;21:2747–2753. doi: 10.1200/JCO.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Gribben JG, Zahrieh D, Stephans K, et al. Autologous and allogeneic stem cell transplantations for poor-risk chronic lymphocytic leukemia. Blood. 2005;106:4389–4396. doi: 10.1182/blood-2005-05-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu CJ, Ritz J. The induction of tumor immunity following allogeneic stem cell transplantation. Advances in Immunology. 2006;90:129–169. doi: 10.1016/S0065-2776(06)90004-2. [DOI] [PubMed] [Google Scholar]
- 12.Miklos DB, Kim HT, Miller KH, et al. Antibody responses to H-Y minor histocompatibility antigens correlate with chronic graft-versus-host disease and disease remission. Blood. 2005;105:2973–2978. doi: 10.1182/blood-2004-09-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CJ, Yang XF, McLaughlin S, et al. Detection of a potent humoral response associated with immune-induced remission of chronic myelogenous leukemia. J Clin Invest. 2000;106:705–714. doi: 10.1172/JCI10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellucci R, Alyea EP, Chiaretti S, et al. Graft-versus-tumor response in patients with multiple myeloma is associated with antibody response to BCMA, a plasma-cell membrane receptor. Blood. 2005;105:3945–3950. doi: 10.1182/blood-2004-11-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jager E, Chen YT, Drijfhout JW, et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trojan A, Schultze JL, Witzens M, et al. Immunoglobulin framework-derived peptides function as cytotoxic T-cell epitopes commonly expressed in B-cell malignancies. Nat Med. 2000;6:667–672. doi: 10.1038/76243. [DOI] [PubMed] [Google Scholar]
- 17.Krackhardt AM, Witzens M, Harig S, et al. Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood. 2002;100:2123–2131. doi: 10.1182/blood-2002-02-0513. [DOI] [PubMed] [Google Scholar]
- 18.Mayr C, Bund D, Schlee M, et al. MDM2 is recognized as a tumor-associated antigen in chronic lymphocytic leukemia by CD8+ autologous T lymphocytes. Exp Hematol. 2006;34:44–53. doi: 10.1016/j.exphem.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Giannopoulos K, Schmitt M. Targets and strategies for T-cell based vaccines in patients with B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2006;47:2028–2036. doi: 10.1080/10428190600709721. [DOI] [PubMed] [Google Scholar]
- 20.Marina O, Biernacki MA, Brusic V, Wu CJ. A concentration-dependent analysis method for high density protein microarrays. J Proteome Res. 2008;7:2059–2068. doi: 10.1021/pr700892h. [DOI] [PubMed] [Google Scholar]
- 21.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai J, Sultana R, Lee Y, et al. RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-11-software0002. SOFTWARE0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo D, Mohr SE, Hu Y, et al. PlasmID: a centralized repository for plasmid clone information and distribution. Nucleic Acids Res. 2007;35:D680–D684. doi: 10.1093/nar/gkl898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rual JF, Hirozane-Kishikawa T, Hao T, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 26.Alyea EP, Soiffer RJ, Canning C, et al. Toxicity and efficacy of defined doses of CD4(+) donor lymphocytes for treatment of relapse after allogeneic bone marrow transplant. Blood. 1998;91:3671–3680. [PubMed] [Google Scholar]
- 27.Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebel U, Kindler B, Pepperkok R. 'Harvester': a fast meta search engine of human protein resources. Bioinformatics. 2004;20:1962–1963. doi: 10.1093/bioinformatics/bth146. [DOI] [PubMed] [Google Scholar]
- 29.Gurkan C, Lapp H, Hogenesch JB, Balch WE. Exploring trafficking GTPase function by mRNA expression profiling: use of the SymAtlas web-application and the Membrome datasets. Methods Enzymol. 2005;403:1–10. doi: 10.1016/S0076-6879(05)03001-6. [DOI] [PubMed] [Google Scholar]
- 30.Haslinger C, Schweifer N, Stilgenbauer S, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 31.Ramos TC, Vinageras EN, Ferrer MC, et al. Treatment of NSCLC patients with an EGF-based cancer vaccine: report of a Phase I trial. Cancer Biol Ther. 2006;5:145–149. doi: 10.4161/cbt.5.2.2334. [DOI] [PubMed] [Google Scholar]
- 32.Biernacki MA, Zhang GL, Zhang W, et al. Novel myeloma-associated antigens revealed in the context of successful syngeneic hematopoietic stem cell transplantation. American Society of Hematology 50th Annual meeting; 2008 Dec 6–11; San Francisco, CA. 2008. [Google Scholar]
- 33.Regnault A, Lankar D, Lacabanne V, et al. Fcgamma receptor-mediated induction of dendritic cell maturation and major histocompatibility complex class I-restricted antigen presentation after immune complex internalization. J Exp Med. 1999;189:371–380. doi: 10.1084/jem.189.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valmori D, Souleimanian NE, Tosello V, et al. Vaccination with NY-ESO-1 protein and CpG in Montanide induces integrated antibody/Th1 responses and CD8 T cells through cross-priming. Proc Natl Acad Sci U S A. 2007;104:8947–8952. doi: 10.1073/pnas.0703395104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brusic V, Marina O, Wu CJ, Reinherz EL. Proteome informatics for cancer research: from molecules to clinic. Proteomics. 2007;7:976–991. doi: 10.1002/pmic.200600965. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H, Bilgin M, Bangham R, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 37.Mullally A, Ritz J. Beyond HLA: the significance of genomic variation for allogeneic hematopoietic stem cell transplantation. Blood. 2007;109:1355–1362. doi: 10.1182/blood-2006-06-030858. [DOI] [PubMed] [Google Scholar]
- 38.Kawakami Y, Rosenberg SA. Human tumor antigens recognized by T-cells. Immunol Res. 1997;16:313–339. doi: 10.1007/BF02786397. [DOI] [PubMed] [Google Scholar]
- 39.Savage PA, Vosseller K, Kang C, et al. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science. 2008;319:215–220. doi: 10.1126/science.1148886. [DOI] [PubMed] [Google Scholar]
- 40.Nishida T, Hudecek M, Kostic A, et al. Development of tumor-reactive T cells after nonmyeloablative allogeneic hematopoietic stem cell transplant for chronic lymphocytic leukemia. Clin Cancer Res. 2009;15:4759–4768. doi: 10.1158/1078-0432.CCR-09-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wierda WG, Kipps TJ. Gene therapy and active immune therapy of hematologic malignancies. Best Pract Res Clin Haematol. 2007;20:557–568. doi: 10.1016/j.beha.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Gitelson E, Hammond C, Mena J, et al. Chronic lymphocytic leukemia-reactive T cells during disease progression and after autologous tumor cell vaccines. Clin Cancer Res. 2003;9:1656–1665. [PubMed] [Google Scholar]
- 43.Dilcher M, Veith B, Chidambaram S, Hartmann E, Schmitt HD, Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. Embo J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chinwalla V, Chien A, Odero M, Neilly MB, Zeleznik-Le NJ, Rowley JD. A t(11;15) fuses MLL to two different genes, AF15q14 and a novel gene MPFYVE on chromosome 15. Oncogene. 2003;22:1400–1410. doi: 10.1038/sj.onc.1206273. [DOI] [PubMed] [Google Scholar]
- 45.Kawai T, Matsumoto M, Takeda K, Sanjo H, Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998;18:1642–1651. doi: 10.1128/mcb.18.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H, Jiang D, Guo Z, et al. TCP10L is expressed specifically in spermatogenic cells and binds to death associated protein kinase-3. Int J Androl. 2005;28:163–170. doi: 10.1111/j.1365-2605.2005.00522.x. [DOI] [PubMed] [Google Scholar]
- 47.Koensgen D, Mustea A, Klaman I, et al. Expression analysis and RNA localization of PAI-RBP1 (SERBP1) in epithelial ovarian cancer: association with tumor progression. Gynecol Oncol. 2007;107:266–273. doi: 10.1016/j.ygyno.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 48.Jia Z, Barbier L, Stuart H, et al. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. J Biol Chem. 2005;280:30564–30573. doi: 10.1074/jbc.M501754200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.