Abstract
Magnetic resonance (MR) microscopy has enormous potential for small animal cardiac imaging, because it is capable of producing volumetric images at multiple time points to accurately measure cardiac function. MR has not been used as frequently as ultrasound to measure cardiac function in the small animal because the MR methods required relatively long scan times, limiting throughput. Here, we demonstrate 4D radial acquisition in conjunction with a liposomal blood pool agent to explore functional differences in three populations of mice: 6 C57BL/6J mice, 6 DBA/2J mice, and 6 DBA/2J CSQ+ mice, all with the same gestational age and approximately the same weight. Cardiovascular function was determined by measuring both left ventricular and right ventricular end diastolic volume, end systolic volume, stroke volume, and ejection fraction. Statistical significance was observed in end diastolic volume, end systolic volume, and ejection fraction for left ventricular measurements between all three populations of mice. No statistically significant difference was observed in stroke volume in either the left or right ventricle for any of the three populations of mice. This study shows that MR imaging is capable of efficient, high-throughput four-dimensional cardiovascular phenotyping of the mouse.
Keywords: 4D, radial, mouse, heart, liposomal Gd, phenotyping
Introduction
Ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI) are all competing modalities for cardiac imaging in the clinical arena. Scaling these techniques to the mouse is challenging because compared to humans, the mouse is 3000-times smaller by weight, the mouse heart beats 10-times faster, and the mouse breathes 5-times faster. Ultrasound is the most widely used method for cardiac imaging in the mouse. While this method is technically simple, precision and accuracy have been limited because the measurements are based on one-dimensional or two-dimensional measurements that use model-based assumptions of heart size to extrapolate to three-dimensional volumes (1–3). These assumptions can break down for disease models and have low reproducibility, requiring large numbers of animals to obtain statistically significant results. Recent studies have been reported in CT, but the radiation dose associated with CT can be problematic for longitudinal studies (4,5). MRI has been scaled to the mouse and is becoming a popular alternative to ultrasound. More laboratories are able to perform MRI on small animals, but applications thus far has been limited because acquisition times are long and segmentation is not automated; and applications also depend on a skilled operator of the imaging equipment. The vast majority of work in imaging the mouse heart with MR has been performed in two dimensions (Table 1) (6–9), with only one publication noted that presents a 3D technique for imaging (10). 3D imaging techniques in the clinical arena have been shown to increase precision and accuracy of the functional cardiovascular measurements (11–14). Likewise, moving to a 3D method for imaging the mouse heart should improve precision and accuracy of cardiovascular measurements. And, the increase in signal-to-noise ratio (SNR) could be used to reduce acquisition time. A 3D, higher spatial resolution imaging technique that increases precision and accuracy should allow researchers to detect subtle variations in cardiovascular phenotypes using much smaller populations of mice than was previously feasible.
Table 1.
The spatial resolution, temporal resolution, and acquisition time reported for several publications on imaging the mouse heart.
| Reference | Slice (mm) | Resolution (mm) | Voxel (nL) | Phases acquired | Acquisition time (approx) |
|---|---|---|---|---|---|
| Williams et al. 2001 (29) | 1 | 0.234 × 0.234 | 54.7 | 10 | 80 min |
| Zhou et al. 2003 (7) | 1 | 0.234 × 0.234 | 54.7 | 8 | 20 min |
| Berr et al. 2005 (9) | 1 | 0.2 × 0.2 | 40 | 12 | 30 min |
| Ross et al. 2002 (30) | 1 | 0.2 × 0.2 | 40 | 12 | 45 min |
| Sosnovik et al., 2007 (31) | 1 | 0.15 × 0.15 | 22.5 | 10 | 80 min |
| Ruff et al., 1998 (32) | 1 | 0.117 × 0.117 | 11.7 | 12 | 13 min |
| Cassidy et al., 2004 (8) | 1 | 0.1 × 0.1 | 10 | 1 | 13 min |
| Dawson et al., 2004 (6) | 1 | 0.1 × 0.1 | 10 | 20 | 60 min |
| Feintuch et al., 2007 (10) | 0.2 | 0.20 × 0.20 | 8 | 12 | 50 min |
| Bucholz et al., 2008 (15) | 0.352 | 0.087 × 0.087 | 2.6 | 10–12 | 16 min |
We report here an application of our previous technique of 4D radial cardiac imaging (15) to a limited population of three different mice—wild type C57BL/6J mice, wild type DBA/2J mice, and a genetically modified transgenic mouse model of heart failure, the DBA/2J CSQ+ mouse (generated by the cardiac-specific over-expression of the calcium binding protein calsequestrin) (16). Even with a limited sample of mice (6 of each), we were able to detect subtle variations in end diastolic volume, end systolic volume, and ejection fraction, and were able to find statistically significant differences in the cardiovascular function of the three populations of mice, which have not previously been reported.
Methods
All MR studies were performed at the Duke Center for In Vivo Microscopy on a 7T, 120-mm bore, Magnex magnet with a GE EXCITE console (EPIC 12.4; GE Medical Systems, Milwaukee, WI). The system employs Resonance Research (Billerica, MA) shielded gradient coils that provide a maximum gradient strength of 770 mT/m and slew rate of 6160 mT/m/ms. The gradients are driven by high-power (12 KW) amplifiers (Copley Controls Model 266). The radiofrequency (RF) front-end of the GE console has been modified for operation at higher field through a second synthesizer and RF mixer that provide both up and down conversion in the RF chain via sum and difference signals. The strong gradients with high slew rates, high duty cycle, and high power amplifiers are critical for achieving the very short repetition times over small fields of view required for efficient acquisition.
A specially designed cradle anesthesia nose cone was integrated with a 3.2 cm quadrature birdcage coil designed by m2m Imaging Corp. (Cleveland, OH) to maximize SNR and improve efficiency of animal handling.
Pulse Sequence
A 3D localizer scan was performed to determine the orientation of the short axis of the left ventricle of the mouse heart. Using the localizer scan, the 4D radial sequence was oriented obliquely and the location of the short axis was defined by viewing the aortic root and acquiring 21 or more slices through the left ventricle of the heart.
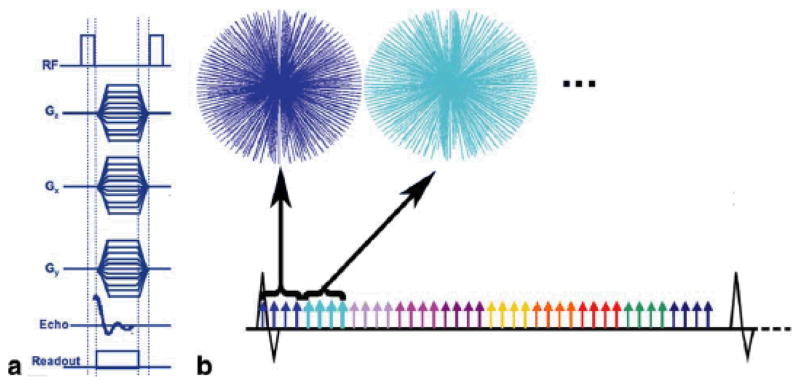
The 4D radial encoding sequence has previously been described in detail (15). Briefly, the sequence is prospectively cardiac-gated with radial encoding that samples the free induction decay from a volume excitation (Figure 1A). Gradients along all three axes define radial trajectories sampling the Fourier volume at multiple time points of the cardiac cycle. The sequence supports a range of tradeoffs in coverage, acquisition time, and spatial and temporal resolution to produce 4D datasets. For the high-throughput challenges in these studies, we chose a rapid acquisition protocol with anisotropic spatial resolution of 87 × 87 × 352 μm3 (2.7 nL) and a temporal resolution of 9.6 ms. The R-R interval was broken into 10–14 time points (Figure 1B). The sequence was repeated through ~8000 heartbeats with a total acquisition time of ~16 minutes. The end points of the 3D radial k-space were determined using a precalculated geodesic table. Geodesic spheres are regularly used to characterize the earth and result in nearly perfect isotropic sampling of the sphere. Because of the computational complexity involved in calculating geodesic tables, the end points are predetermined and imported into the pulse sequence using a wave table. Because radial acquisitions require π-times more views for complete Nyquist sampling, this would increase scan time intractably. As a result, the 3D radial space was undersampled by a factor of √2, requiring 32000 views per phase of the heart cycle. Radial acquisitions are extremely robust to undersampling, and image quality was not sacrificed. The sequence employs minimum TE (300 μs) and TR (2.4 ms) using a bandwidth of ±125 kHz and a nominal flip angle of 45°.
Figure 1.
(A) The gradient and RF waveforms for the 4D radial acquisition (4DRA) sequence. The trajectories through the Fourier volume are depicted in B). The prospective cardiac gating strategy used yields 9.6 ms temporal resolution. (B) Each color represents the cardiac phase being acquired, so for one heartbeat, four radial views were acquired per phase of the heart cycle, requiring a total of 8000 heartbeats for a complete acquisition.
Reconstruction and Data Processing
We employed a non-uniform fast Fourier transform (NUFFT) algorithm that addressed the non-uniform sampling due to radial acquisition and the slope sampling at the same time. NUFFT uses a least squares optimized kernel for interpolation that provides a balance between computation time and accuracy of interpolation to potentially allow fast and accurate results (17–19). Reconstruction time for each 3D dataset took approximately 30 seconds, and for a typical high-throughput 4D image reconstruction, the total reconstruction time was 5 minutes for the entire 4D array.
Both right and left ventricular volumes were calculated using a segmentation algorithm available with ImageJ <http://rsb.info.nih.gov/ij/>. 3D short integer stacks were imported into ImageJ, where the images were cropped to include only slices through the ventricle of interest. The top of the left ventricle was defined as the slice after the aortic root, which was visualized in all datasets. The top of the right ventricle was determined by the appearance of the right atria. Great care was taken in excising only the right ventricle as the data was acquired in the short axis plane of the left ventricle (Figure 2). Both the left and right ventricles were excised using threshold segmentation, and the total number of voxels of the left ventricle were calculated and multiplied by voxel volume to obtain volumetric measurements.
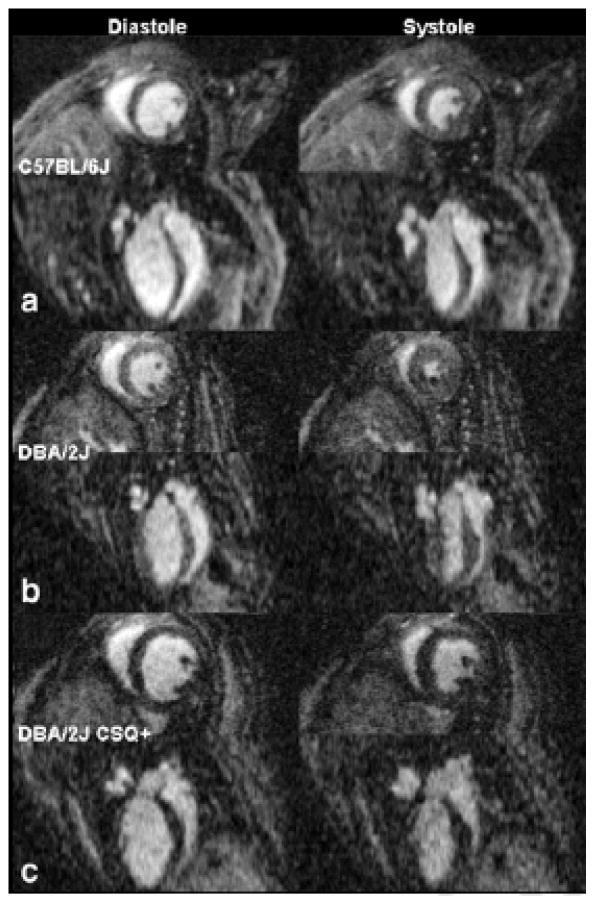
Figure 2.
The short and long axis left ventricular view for both diastole and systole are displayed for one of the six C57BL/6J mice (A), a DBA/2J mouse (B), and a DBA/2J CSQ+ mouse (C), which is a genetically modified mouse model of heart failure. In all three cases, the short axis view is on top and immediately below is the long axis image. The images on the left show diastole and the images on the right show systole.
To determine the location of end diastolic volume (EDV) and end systolic volume (ESV) for each population of mice one mouse was selected to calculate ventricular volume at all phases of the heart cycle. Using the representative mice, ED was found to occur at the first phase of the heart cycle, whereas ES occurred at the sixth phase of the heart cycle ~50 ms after the QRS wave.
Calculated values of EDV, ESV, stroke volume (SV), and ejection fraction (EF) for both left and right ventricle data were imported into SAS where ANOVA and ANOVA contrast analysis were performed. For the contrast analysis, all combinations of 2 populations of mice were compared for each measurement to verify that statistical differences obtained with ANOVA analysis were significant for each group of mice. Because the population of mice was small, statistical significance was determined to occur when p <0.01. All p-values are reported in the manuscript.
Data shown in this manuscript is available to the interested reader at CIVMSpace, <http://www.civm.duhs.duke.edu/mrm200902/index.html>, a web portal for data-sharing and collaboration at the Center for In Vivo Microscopy.
Contrast Agent
The liposomal blood pool agent used for this work has been described in detail (20). Briefly, a lipid mixture consisting of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (Genzyme Corporation, Cambridge, MA), cholesterol (Sigma, St. Louis, MO) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(poly(ethylene glycol))-2000] (Genzyme Corporation) in the ratio 55:40:5 was dissolved in ethanol at 60°C. The mixture was then hydrated with 0.5 mol/L gadodiamide solution (Omniscan® trademark of GE Healthcare, Milwaukee, WI) and stirred at 60°C. The resulting solution was sequentially extruded on Lipex thermoline extruder (Northern Lipids, Vancouver, BC, Canada) with 3 passes through 200 nm Nuclepore membrane (Whatman, Newton, MA), 4 passes through 100 nm membranes, and 10 passes through 50 nm membrane. The external phase was then cleaned using a MicroKros module (Spectrum Laboratories, Inc., Rancho Dominguez, CA) of 400-kDa cutoff.
The size distribution of the liposomes in the final formulation was determined by dynamic light scattering (DLS) using a ZetaPlus Analyzer (Brookhaven Instruments, Incorporated, Chapel House, UK). The resultant size of liposomes as determined using DLS was 72 nm with a polydispersity index of 0.12. The concentration of gadolinium (Gd) in the liposomal Gd formulation was quantified using inductively coupled plasma optical emission spectroscopy (ICPOES; Model Optima 4300D, Perkin Elmer, Norwalk, CT) operating at a wavelength of 336.223 nm. The concentration of gadolinium was 35.4 mM.
The liposomal-Gd agent used in this study has previously been optimized for use in small animal imaging. The use of small particles results in an improved relaxivity for this nanoparticle contrast agent (21). Upon in vivo administration, the agent has demonstrated uniform and stable signal enhancement for an extended period, thus enabling a longer time window for acquisition of high-spatial resolution images (22), (23). A stable and consistent CNR is obtained within minutes after equilibration of the liposomal-Gd within the blood pool. Further, the presence of polyethylene glycol (PEG) on the liposome surface and the sub-100 nm size of these particles results in a long in vivo half-life (~18 hours) of the liposomal-Gd agent (24) (25).
Animal Handling and Preparation
All animal studies were approved by the Duke University Institutional Animal Care and Use Committee. A total of 18 mice were used in this study—6 C57BL/6J mice, 6 DBA/2J mice, and 6 DBA/2J CSQ+ mice. All animals were approximately the same age and weight. The DBA/2J CSQ+ mice were genetically modified to over-express calsequestrin causing hypertrophy of all four chambers of the heart, resulting in heart failure (16). All animals were induced with isoflurane and given a tail vein injection of liposomal Gd contrast agent at a dose of 0.2 mmol/kg resulting in an average injection of 0.12 mL. Animals were free breathing and were maintained under anesthesia using an isoflurane nose cone (26). The ECG and breathing rate of the animals were monitored using an SA Instruments Inc. (Stony Brook, NY) system. Heart rate was maintained between 450 and 550 beats per minute by adjusting the mixture of isoflurane and O2. A rectal temperature probe was used to monitor core body temperature in the magnet and was adjusted to maintain stable body temperature (37 ± 0.5°C) throughout the studies. After completion of the studies, the DBA/2J and DBA/2J CSQ+ mice were survived for 4 weeks to verify survival after the MRI study.
Results
Representative images obtained at diastole and systole for three mice are displayed in Figure 2. Figure 2A shows the short axis view for diastole (top left) and the short axis view for systole (top right) for one of the six C57BL/6J mice. Below the short axis view is the long axis view for diastole and systole, respectively, for the same mouse. Despite the anisotropic acquisition (87 × 87 × 352 μm3), the long axis view displays systole and diastole with minimal artifacts. Figure 2B shows the short axis view for diastole (middle left), and systole (middle right) for one DBA/2J mouse. Below the short axis images in Figure 2B are the long axis images for both diastole and systole, respectively, for the same mouse. Qualitative differences in the size of the heart are seen between the C57BL/6J and DBA/2J mouse. Systolic volumes for the DBA/2J mouse are smaller than systolic volumes for the C57BL/6J mice. Figure 2C shows the short axis view for diastole (bottom left) and systole (bottom right) for a DBA/2J CSQ+ mouse. Located below the short axis images are the long axis images of both diastole and systole for the same mouse. Qualitative differences in heart size and end systolic volume between the wild type sibling (DBA/2J mouse in Figure 2B) and the genetically modified DBA/2J CSQ+ mice are readily observable. The heart size of the DBA/2J CSQ+ is larger than the C57BL/6J mouse, as well, and the difference between systole and diastole (ejection fraction) is much smaller for the DBA/2J CSQ+ than either of the other two mice.
The robustness of segmentation depends on high SNR and CNR images. The SNR of the blood and CNR between the blood and myocardium are plotted for each mouse strain (Figure 3). The high SNR and CNR for all 18 datasets highlight the reproducibility and robustness of the technique. Because liposomal Gd contrast is used, flow artifacts due to blood turbulence are not present in any phase of the cardiac cycle, thereby enabling semiautomated threshold segmentation of both the left and right ventricles.
Figure 3.
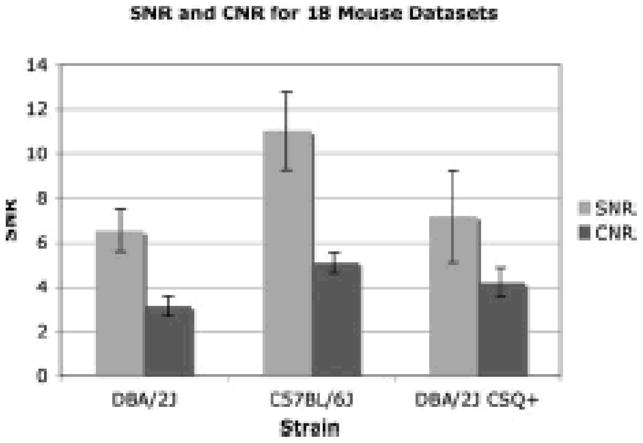
Mean SNR of the blood and mean CNR between blood and myocardium are plotted for each strain of mouse for all 18 datasets. Standard deviation for the 18 datasets is also displayed to show the robustness of the imaging technique. Images are routinely acquired with high SNR and CNR.
Because of the high CNR and SNR images, we were able for all 18 mice to segment both right and left ventricles for diastole and systole. Figure 4 shows the segmentation of a typical dataset. Figure 4A shows the top of the left ventricle, Figure 4B a mid-ventricular slice, and Figure 4C shows the apex of the left ventricle, along with the semi-automated volumes chosen through thresholding. Figure 4D shows segmentation of the top of the right ventricle, Figure 4E shows segmentation of the right mid-ventricular slice, and Figure 4F shows the segmentation of the apex of the right ventricle.
Figure 4.
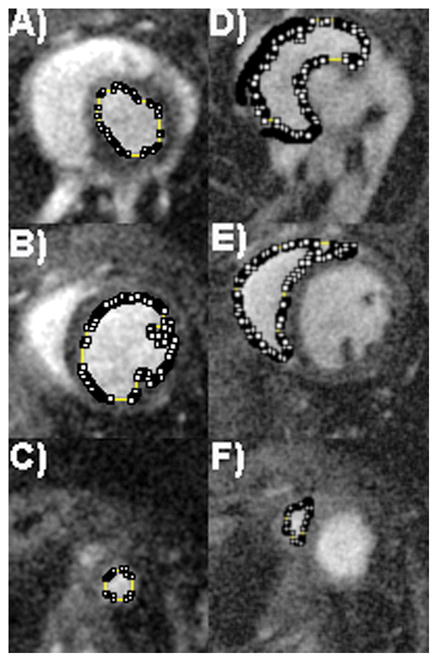
The high contrast-to-noise ratio allows easy semi-automated threshold segmentation of both the left ventricles (LV) and right ventricles (RV) for each of the 18 mice used in this study. Segmentation is shown for one C57BL/6J mouse, with left ventricular segmentation (A, B, and C), and right ventricular segmentation (D, E, and F). Segmentation of the top of the LV can be seen in (A), while (B) represents segmentation of a mid-ventricular slice through the LV, and (C) shows the apex of the LV. As images were acquired in the LV short axis plane, care was taken when segmenting out the top of the RV (D), as the delineation between right atria and ventricle must be hand-segmented. Mid-ventricular segmentation of the RV (E) is straightforward, as is the apex of the RV (F).
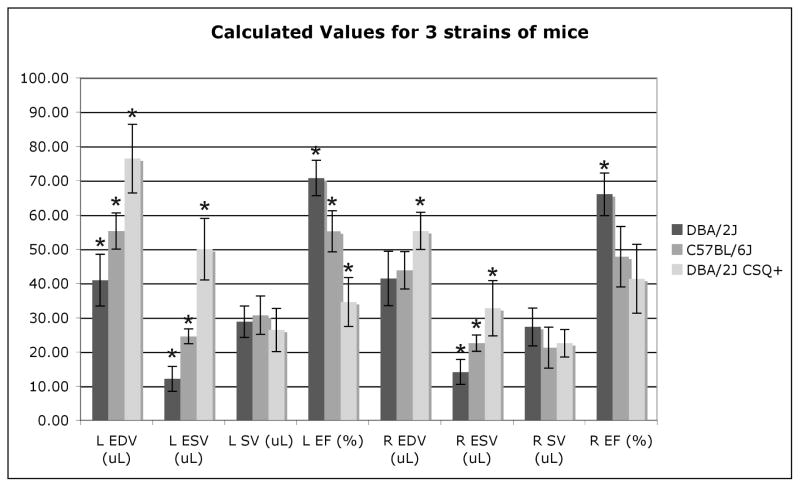
Following threshold segmentation, a voxel count was performed and multiplied by voxel volume to obtain measurements of end diastolic volume (EDV), end systolic volume (ESV), ejection fraction (EF), and stroke volume (SV) for both right and left ventricles of all 18 datasets. The results are shown in Table 2. The DBA/2J CSQ+ mouse had the largest left ventricular EDV at 76.4 ± 10.0 μL and the DBA/2J had the smallest left ventricular EDV at 41.0 ± 7.5 μL.
Table 2.
Both the left and right ventricles were segmented and multiplied by voxel volume to 1calculate end diastolic volume (EDV), end systolic volume (ESV), stroke volume (SV), and ejection fraction (EF). To validate threshold segmentation, one of the lower SNR DBA/2J mice datasets was independently segmented 6 times to determine the variability in segmentation. The results are reported in the first row of the table. The mean volume calculations for each population of 6 mice are presented in rows 2–4.
| Mouse Strain | N | Weight (g) | Left EDV (μL) | Left ESV (μL) | Left SV (μL) | Left EF (%) | Right EDV (μL) | Right ESV (μL) | Right SV (μL) | Right EF (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| DBA/2J | 1×6 | 19.5 | 29.03±0.43 | 7.01±0.31 | 22.02±0.49 | 75.86±1.08 | 30.98±0.70 | 8.69±0.94 | 22.29±1.21 | 71.94±3.10 |
| C57BL/6J | 6 | 22.9±1.2 | 55.3±5.3 | 24.6±2.1 | 30.7±5.6 | 55.2±6.0 | 43.8±5.5 | 22.6±2.4 | 21.2±6.0 | 47.8±8.8 |
| DBA/2J | 6 | 21.8±1.4 | 41.0±7.5 | 12.1±3.6 | 28.8±4.6 | 70.8±5.2 | 41.4±8.0 | 14.12±3.66 | 27.31±5.54 | 66.0±6.24 |
| DBA/2J CSQ+ | 6 | 22.7±1.1 | 76.4±10.0 | 50.0±9.0 | 26.4±6.3 | 34.6±7.1 | 55.3±5.4 | 32.8±8.1 | 22.54±3.99 | 41.3±10.1 |
The segmented volumes of both the left and right ventricle were volume-rendered using ImageJ (see Figure 5). Figure 5A shows left ventricular diastole, followed by left ventricular systole, then followed by right ventricular diastole and right ventricular systole for one of the six C57BL/6J mice. Figure 5B shows both diastole and systole for the left and right ventricle of one DBA/2J mouse, and Figure 5C shows diastole and systole for the left and right ventricle of one DBA/2J CSQ+ mouse. 3D visualization makes it easier to appreciate both volume and shape differences between systole and diastole and between strains.
Figure 5.
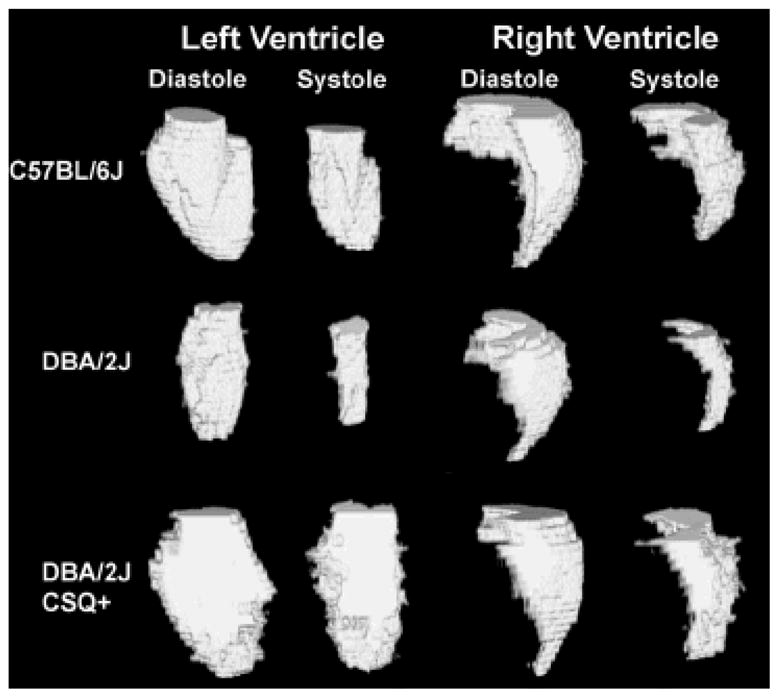
An example dataset for each of the three strains of mice evaluated in this paper that were threshold-segmented. Volume rendering was performed using ImageJ to show the excised left and right ventricles for both systole and diastole for one (A) C57BL/6J mouse, (B) a DBA/2J mouse, and (C) a DBA/2J CSQ+ mouse.
To determine the reproducibility of segmentation of the datasets, one of the lowest SNR DBA/2J datasets was segmented six times for both left and right ventricular volumes to determine the variability associated with segmentation (Table 2, row 1). For the left ventricular segmentation, coefficient of variability of the EDV, ESV, SV, and EF was 0.015, 0.044, 0.022, and 0.014. For right ventricular segmentation, the coefficient of variability in segmentation of the EDV, ESV, SV, and EF was 0.022, 0.109, 0.054, and 0.044. Variability in segmentation was approximately two-times higher for the right ventricle because the delineation of atria and ventricle were more subjective and the orientation was not in the short axis plane of the right ventricle. The variability seems independent of the volume size (systole or diastole), because the smaller ESV of the DBA/2J had approximately the same standard deviation as the EDV for the same mouse for both the right and left ventricular measurements. Table 2 shows the calculated values of ventricular function for the independently segmented datasets. The variability associated with left ventricular segmentation of one dataset (six times) was 10% of the inter-specimen variability. The variability associated with right ventricular segmentation was 20% of the inter-specimen variability. Calculation of EF is dependent on both error of systolic and diastolic measurements, and therefore, it is expected for this error to increase. Left ventricular (LV) EF segmentation variability was 20% of the inter-specimen variability while right ventricular (RV) EF segmentation variability corresponded to 50% of the inter-specimen variability.
ANOVA and ANOVA contrast analysis were performed on the three populations of mice to determine statistically significant differences. The results of the ANOVA analysis and ANOVA contrast analysis for each group are presented in Figure 6. An asterisk (*) indicates statistical significance between groups (p<0.01). ANOVA analysis indicated differences between mice were statistically significant for LV EDV (p<0.0001), LV ESV (p<0.0001), LV EF (p<0.0001), RV EDV (p<0.004), RV ESV (p<0.0001), and RV EF (p<0.0004). For all three populations of mice, the SV for both right ventricle and left ventricle did not show any significant difference, but the EF for the right ventricle was lower than the EF for the left ventricle for both the C57BL/6J and the DBA/2J mouse. Importantly, left ventricular EF in the DBA/2J CSQ+ mouse was markedly lower (p<0.0001) than the other two normal mice, indicating severe dilated cardiomyopathy. Left ventricular EDV, ESV, and EF were statistically significantly different for all three populations of mice (p<0.007), while only right ventricular ESV was statistically significantly different for all three populations (p<0.005). Only the CSQ+ mouse had a statistically significant difference in right ventricular EDV (p<0.007), and only the DBA/2J wild type mice had a statistically significantly higher EF (p<0.002) than either C57BL/6J or DBA/2J CSQ+ mice. The increased variability associated with the measurement of right ventricular function could contribute to the reduced ability to make statistically significant conclusions between populations of mice.
Figure 6.
The calculated values in Table 2 show mean, as well as standard deviation. An asterisk* indicates all significant results (p<0.01) as determined by ANOVA contrasts.
Discussion
A need exists for an imaging tool that allows rapid phenotyping of the mouse heart with the ability to make statistically significant conclusions on the smallest population of mice possible. The main competitors in the microscopy market are ultrasound, MRI, and microCT. MicroCT has high contrast-to-noise, but the ability to perform routine cardiovascular measurements is limited, because only two research centers are capable of performing cardiovascular measurements (27,28), and the ability to perform longitudinal studies is even more limited due to animal-handling requirements and radiation dose. Ultrasound is the most commonly used imaging modality for imaging the mouse heart because it is very fast and capable of performing multiple longitudinal studies, but the 1 and 2D measurements have very low reproducibility with high error in measurements and as a result, a large population of animals is required to generate statistical significance. Furthermore, small changes in heart function that might be observed between two wild type populations may never generate statistically significant differences using ultrasound simply because the error in the measurements for ultrasound is so great. Magnetic resonance microscopy has excellent soft tissue contrast, but thus far, has suffered from limited temporal resolution and long acquisition times, significantly decreasing throughput. Decrease in throughput reduces the ability to perform longitudinal studies on a population of mice required to generate statistically significant results.
The 4D technique presented here supports rapid (16-minute) 4D images of the whole heart at high spatial resolution of 87 × 87 × 352 μm3 and high temporal resolution (9.6 ms). This represents a reduction in acquisition time by a factor of 4 with a simultaneous increase in spatial resolution by a factor of 3 over previously published techniques (6–10). The blood pool agent reduces flow related artifacts in bright blood imaging that compromise segmentation (9) and increases the SNR of the blood pool relative to myocardium. This SNR increase allows for short TE imaging that results in higher spatial resolution with faster acquisition time, significantly improving ability to perform rapid cardiovascular phenotyping of the mouse. The high contrast-to-noise and signal-to-noise of the images allows routine semi-automated threshold segmentation with very low coefficient of variability (e.g. 1.5% LV EDV, and <5% for RV EF). While segmentation of the right ventricles is more susceptible to error due to data acquisition along the short axis of the left ventricle, measurements of right ventricular function obtained from left ventricle short axis images produce small enough variability to generate statistically significant results. Because of the high spatial resolution, small differences in cardiovascular function can be determined to be statistically significant for fewer animals than would be possible with an ultrasound study. Left ventricular mass was not measured as the DBA/2J and DBA/2J CSQ+ mice were survived to verify the potential for longitudinal studies in hypertrophic animals. Future work would benefit from including this parameter as a means for measuring accuracy of this 4D imaging technique.
The results of this study indicate that the C57BL/6J mouse has a slightly hypertrophic heart compared to the DBA/2J wild type mouse. The C57BL/6J mice as compared to the DBA/2J have larger hearts with higher EDV (p<0.0066), ESV (p<0.0019), and lower EF (p<0.0005), while SV remains statistically insignificantly different. The transgenic DBA/2J CSQ+ mice were shown to be significantly hypertrophic compared to either the C57BL/6J (p<0.0003) or the DBA/2J (p<0.0001) for both EDV and ESV; with statistically significant lower EF than either the DBA/2J (p<0.0001) or C57BL/6J (p<0.0001) mice. As expected, stroke volume was preserved in the DBA/2J CSQ+ mice—consistent with the known mechanisms of adaptation in dilated cardiomyopathy, which results in marked increases in chamber volumes allowing for maintenance of stroke volume in the normal range.
Conclusion
A 4D radial acquisition was applied to a population of 18 mice representing 3 different strains: 6 C57BL/6J wild type mice, 6 DBA/2J wild type mice, and 6 DBA/2J CSQ+ heart failure mice. The 4D acquisition with high spatial resolution (87 × 87 × 352 μm3), high temporal resolution (9.6 ms), and fast acquisition time (16 minutes) allowed for rapid measurement of heart function in all 18 mice for both left and right ventricular volumes. The high spatial and temporal resolution increased the reproducibility of the technique allowing rapid phenotyping of the mouse models. Using the liposomal Gd contrast agent allowed for high CNR and SNR images that could be reliably segmented with a semi-automated threshold segmentation algorithm in ImageJ. The relatively small population size used in the study was enough to determine statistical significance between all three groups for EDV, ESV, and EF for the left ventricle and ESV for the right ventricle. SV was not found to be statistically significant for any of the three populations of mice between either left or right ventricle. Right ventricular heart function indicated that the DBA/2J mouse was statistically lower for EDV and statistically higher for EF than the other two mouse populations. This technique represents a significant step forward in characterization and quantification of heart function for application to cardiovascular phenotyping using MRI.
Acknowledgments
This work was performed at the Duke Center for In Vivo Microscopy, an NCRR Biomedical Technology Research Center (P41 RR005959), and NCI Small Animal Imaging Resource (U24 CA092656). This study was also supported by NHLBI R01 HL056687 to HAR). We thank Gary Cofer for his help with coil design, Sally Gewalt and Dr. Jiayu Song for help with reconstruction, Dr. Laurence Hedlund for advice with animal support and handling, Jeff Brandenburg for his help setting up the website materials, and Sally Zimney for editorial assistance.
References
- 1.Manning W, Wei J, Katz S, Litwin S, Douglas P. In vivo assessment of LV mass in mice using high-frequency cardiac ultrasound: necropsy validation. Am J Physiol. 1994;266(4 pt 2):H1672–1675. doi: 10.1152/ajpheart.1994.266.4.H1672. [DOI] [PubMed] [Google Scholar]
- 2.Youn H, Rokosh G, Lester S, Simpson P, Schiller N, Foster E. Two-dimensional echocardiography with a 15-MHz transducer is a promising alternative for in vivo measurement of left ventricular mass in mice. J Am Soc Echocardiogr. 1999;12(1):70–75. doi: 10.1016/s0894-7317(99)70175-6. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz E, Pollick C, Dow J, Patterson M, Birnbaum Y, Kloner R. A small animal model of non-ischemic cardiomyopathy and its evaluation by transthoracic echocardiography. Cardiovasc Res. 1998;39(1):216–223. doi: 10.1016/s0008-6363(98)00009-1. [DOI] [PubMed] [Google Scholar]
- 4.Badea CT, Hedlund LW, Boslego MJF, Mao L, Rockman HA, Johnson GA. Cardiac micro-CT for morphological and functional phenotyping of muscle LIM protein null mice. Molecular Imaging. 2007;6(4):261–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Nahrendorf M, Badea C, Hedlund LW, Figueiredo JL, Sosnovik DE, Johnson GA, Weissleder R. High-resolution imaging of murine myocardial infarction with delayed-enhancement cine micro-CT. Am J Physiol Heart Circ Physiol. 2007;292(6):H3172–3178. doi: 10.1152/ajpheart.01307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson D, Lygate CA, Saunders J, Schneider JE, Ye X, Hulbert K, Noble JA, Neubauer S. Quantitative 3-dimensional echocardiography for accurate and rapid cardiac phenotype characterization in mice. Circulation. 2004;110(12):1632–1637. doi: 10.1161/01.CIR.0000142049.14227.AD. [DOI] [PubMed] [Google Scholar]
- 7.Zhou R, Pickup S, Glickson JD, Scott CH, Ferrari VA. Assessment of global and regional myocardial function in the mouse using cine and tagged MRI. Magn Reson Med. 2003;49(4):760–764. doi: 10.1002/mrm.10423. [DOI] [PubMed] [Google Scholar]
- 8.Cassidy PJ, Schneider JE, Grieve SM, Lygate C, Neubauer S, Clarke K. Assessment of motion gating strategies for mouse magnetic resonance at high magnetic fields. J Magn Reson Imaging. 2004;19(2):229–237. doi: 10.1002/jmri.10454. [DOI] [PubMed] [Google Scholar]
- 9.Berr S, Roy RJ, French BA, Yang Z, Gilson W, Kramer CM, Epstein F. Black Blood Gradient Echo Cine Magnetic Resonance Imaging of the Mouse Heart. Magn Reson Med. 2005;53:1074–1079. doi: 10.1002/mrm.20487. [DOI] [PubMed] [Google Scholar]
- 10.Feintuch A, Zhu Y, Bishop J, Davidson L, Dazai J, Bruneau B, Henkelman R. 4D cardiac MRI in the mouse. NMR Biomed. 2007;20(3):360–365. doi: 10.1002/nbm.1164. [DOI] [PubMed] [Google Scholar]
- 11.Rettmann D, Saranathan M, Wu K, Azevedo C, Bluemke D, Foo T. High temporal resolution breathheld 3D FIESTA CINE imaging: validation of ventricular function in patients with chronic myocardial infarction. J Magn Reson Imaging. 2007;25(6):1141–1146. doi: 10.1002/jmri.20923. [DOI] [PubMed] [Google Scholar]
- 12.Peters D, Ennis D, Rohatgi P, Syed M, McVeigh E, Arai A. 3D breath-held cardiac function with projection reconstruction in steady state free precession validated using 2D cine MRI. J Magn Reson Imaging. 2004;20(3):411–416. doi: 10.1002/jmri.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amano Y, Herfkens R, Shifrin R, Alley M, Pelc N. Three-dimensional cardiac cine magnetic resonance imaging with an ultrasmall superparamagnetic iron oxide blood pool agent (NC100150) J Magn Reson Imaging. 2000;11(2):81–86. doi: 10.1002/(sici)1522-2586(200002)11:2<81::aid-jmri2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 14.Barger A, Grist T, Block W, Mistretta C. Single breath-hold 3D contrast-enhanced method for assessment of cardiac function. Magn Reson Med. 2000;44(6):821–824. doi: 10.1002/1522-2594(200012)44:6<821::aid-mrm1>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 15.Bucholz E, Ghaghada K, Qi Y, Mukundan S, Johnson GA. Four-dimensional MR microscopy of the mouse heart using radial acquisition and liposomal gadolinium contrast agent. Magn Reson Med. 2008;60(1):111–118. doi: 10.1002/mrm.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho M-C, Rapacciuolo A, Koch WJ, Kobayashi Y, Jones LR, Rockman HA. Defective beta -adrenergic receptor signaling precedes the development of dilated cardiomyopathy in transgenic mice with calsequestrin overexpression. J Biol Chem. 1999;274(32):22251–22256. doi: 10.1074/jbc.274.32.22251. [DOI] [PubMed] [Google Scholar]
- 17.Song J, Liu Q. Improving non-cartesian MRI reconstruction through discontinuity subtraction. Int J Bio Imag. 2006:1–9. doi: 10.1155/IJBI/2006/87092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sha L, Guo H, AWS An improved gridding method for spiral MRI using nonuniform fast Fourier transform. J Magn Reson. 2003;162(2):250–258. doi: 10.1016/s1090-7807(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 19.Liu QH, Nguyen N. An accurate algorithm for nonuniform fast Fourier transform (NUFFT’s) IEEE Microwave and Guided Wave Letters. 1998;8(1):18–20. [Google Scholar]
- 20.Ghaghada K, Bockhorst K, Mukundan JS, Annapragada A, Narayana P. High resolution vascular imaging of the rat spine using liposomal blood pool MR agent. AJNR Am J Neuroradiol. 2007;28(1):48–53. [PMC free article] [PubMed] [Google Scholar]
- 21.Ghaghada K, Hawley C, Kawaji K, Annapragada A, Mukundan SJ. T1 relaxivity of core-encapsulated gadolinium liposomal contrast agents--effect of liposome size and internal gadolinium concentration. Acad Radiol. 2008;15(10):1259–1263. doi: 10.1016/j.acra.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bucholz E, Ghaghada K, Qi Y, Srinivasan M, Johnson GA. Four-dimensional MR microscopy of the mouse heart using radial acquisition and liposomal gadolinium contrast agent. Magn Reson Med. 2008;60:111–118. doi: 10.1002/mrm.21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayyagari AL, Zhang X, Ghaghada KB, Annapragada A, Hu X, Bellamkonda RV. Long-circulating liposomal contrast agents for magnetic resonance imaging. Magn Reson Med. 2006;55(5):1023–1029. doi: 10.1002/mrm.20846. [DOI] [PubMed] [Google Scholar]
- 24.Allen T, Hansen C, Martin F, Redemann C, Yau-Young A. Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo. Biochim Biophys Acta. 1991;1066(1):29–36. doi: 10.1016/0005-2736(91)90246-5. [DOI] [PubMed] [Google Scholar]
- 25.Gabizon A, Huang L, Martin F, Barenholz Y. Doxorubicin encapsulated in polyethylene-glycol coated liposomes: initial clinical-pharmacokinetic studies in solid tumors. In: Lasic DD, Martin FJ, editors. Stealth liposomes. Boca Raton, FL: CRC Press; 1995. pp. 245–257. [Google Scholar]
- 26.Howles GP, Nouls JC, Qi Y, Johnson GA. Rapid production of specialized animal handling devices using computer-aided design and solid freeform fabrication. J Magn Reson Imaging. 2009;30(2):466–471. doi: 10.1002/jmri.21821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badea C, Fubara B, Hedlund L, Johnson G. 4D micro-CT of the mouse heart. Molecular Imaging. 2005;4(2):110–116. doi: 10.1162/15353500200504187. [DOI] [PubMed] [Google Scholar]
- 28.Drangova M, Ford NL, Detombe SA, Wheatley AR, Holdsworth DW. Fast Retrospectively gated quantitative four-dimensional (4D) cardiac micro computed tomography imaging of free-breathing mice. Invest Radiol. 2007;42(2):85–94. doi: 10.1097/01.rli.0000251572.56139.a3. [DOI] [PubMed] [Google Scholar]
- 29.Williams S, Gerber H, Giordano F, Peale F, Bernstein L, Bunting S, Chien K, Ferrara N, van Bruggern N. Dobutamine stress cine-MRI of cardiac function in the hearts of adult cardiomyocyte-specific VEGF knockout mice. J Magn Reson Imaging. 2001;14:374–382. doi: 10.1002/jmri.1197. [DOI] [PubMed] [Google Scholar]
- 30.Ross AJ. Serial MRI evaluation of cardiac structure and function in mice after reperfused myocardial infarction. Magn Reson Med. 2002;47(6):1158–1168. doi: 10.1002/mrm.10166. [DOI] [PubMed] [Google Scholar]
- 31.Sosnovik D, Dai G, Nahrendorf M, Rosen B, RS Cardiac MRI in mice at 9.4 tesla with a transmit-receive surface coil and a cardiac-tailored intensity-correction algorithm. J Magn Reson Imaging. 2007;26:279–287. doi: 10.1002/jmri.20966. [DOI] [PubMed] [Google Scholar]
- 32.Ruff J, Wiesmann F, Hiller K-H, Voll S, von Kienlin M, Bauer WR, Rommel E, Neubauer S, Haase A. Magnetic resonance microimaging for noninvasive quantification of myocardial function and mass in the mouse. Magn Res Med. 1998;40:43–48. doi: 10.1002/mrm.1910400106. [DOI] [PubMed] [Google Scholar]