SUMMARY
To identify Haemophilus ducreyi transcripts that are expressed during human infection, we used selective capture of transcribed sequences (SCOTS) with RNA isolated from pustules obtained from three volunteers infected with H. ducreyi and with RNA isolated from broth grown bacteria used to infect volunteers. With SCOTS, competitive hybridization of tissue-derived and broth-derived sequences identifies genes that may be preferentially expressed in vivo. Among the three tissue specimens, we identified 531 genes expressed in vivo. Southern blot analysis of 60 genes from each tissue showed that 87% of the identified genes hybridized better with cDNA derived from tissue specimens than with cDNA derived from broth grown bacteria. RT-PCR on 9 additional pustules confirmed in vivo expression of 10 of 11 selected genes in other volunteers. Of the 531 genes, 139 were identified in at least two volunteers. These 139 genes fell into several functional categories, including biosynthesis and metabolism, regulation, and cellular processes such as transcription, translation, cell division, DNA replication and repair, and transport. Genes involved in anaerobic or aerobic respiration indicated that H. ducreyi likely encounters both microenvironments within the pustule. Other genes suggest an increase in DNA damage and stress in vivo. Genes involved in virulence in other bacterial pathogens and 32 genes encoding hypothetical proteins were identified, which may represent novel virulence factors. We identified three genes, lspA1, lspA2 and tadA, known to be required for virulence in humans. This is the first study to broadly define transcripts expressed by H. ducreyi in humans.
INTRODUCTION
Haemophilus ducreyi is the causative agent of chancroid, a genital ulcer disease. H. ducreyi facilitates both the acquisition and transmission of human immunodeficiency virus (HIV)-1 and contributes to the HIV-1 pandemic in certain regions of Africa and Asia (Steen, 2001).
To study the pathogenesis of H. ducreyi infection, we developed an experimental model of infection in which healthy adult volunteers are infected on the upper arm with 101 to 102 CFU of H. ducreyi. The clinical course of disease in the model accurately mimics natural disease through the pustular stage (Al-Tawfiq et al., 1998; Palmer et al., 1998; Spinola et al., 2002). The histology of experimental pustules and naturally acquired ulcers are nearly identical, and H. ducreyi maintains the same general relationship with host cells during experimental and natural infection (Bauer et al., 2006; Bong et al., 2002a). Pustules (n=10) that have been quantitatively cultured contain 1.8 × 105 ± 3.6 × 105 total CFU (mean ± std. dev.) of H. ducreyi, indicating that the bacteria replicate in the model.
Studies of virulence factors in H. ducreyi have relied on identification of gene function in vitro, followed by mutant/parent comparison trials in human or other models of disease. Of 20 putative virulence factors (Janowicz et al., 2006b; Spinola et al., 2002), 9 genes or gene clusters have been shown to be required for full virulence in humans (Al-Tawfiq et al., 2000; Bong et al., 2001; Fortney et al., 2000; Fulcher et al., 2006; Janowicz et al., 2004; Janowicz et al., 2006a; Spinola et al., 2003b and unpublished observations). Although these trials yield important information about the roles of specific putative virulence determinants in disease, this approach is impractical for identifying broad categories of genes important to pathogenesis or the survival of the organism. In other bacterial systems, identification of in vivo expressed genes has led to the discovery of virulence factors that may not have been identified otherwise because the genes were not expressed in vitro, their annotation did not suggest a role in virulence, or the complexity of their in vivo niche could not be adequately reproduced in vitro (Rediers et al., 2005). Thus, we sought a broader approach to examining H. ducreyi gene expression in vivo.
Several molecular techniques have been developed to identify in vivo expressed bacterial genes (Chiang et al., 1999; Mahan et al., 2000; Rediers et al., 2005; Shelburne & Musser, 2004). In selecting a method for our model, we were limited by both practical and regulatory constraints. Biosafety and Food and Drug Administration regulations preclude us from using promoter-trap or mutagenesis strategies that utilize plasmids, mobilizable cassettes, or reporter genes in humans. The amount of bacteria in a pustule yields too little RNA for microarray analysis, employing methodologies currently in use. However, H. ducreyi transcripts have been amplified from pustules by RT-PCR (Throm & Spinola, 2001). Selective capture of transcribed sequences (SCOTS) was designed to identify scarce bacterial mRNA in the presence of large amounts of eukaryotic RNA (recently reviewed in Daigle et al., 2002). With SCOTS, in vivo-derived cDNAs are captured onto bacterial chromosomal DNA by hybridization. Importantly, prehybridization of chromosomal DNA with in vitro-derived cDNAs reduces capture of in vivo-derived sequences that are also transcribed in vitro. SCOTS has been employed successfully to examine gene expression in multiple organisms and conditions: in Salmonella typhi, Mycobacterium tuberculosis, and M. avium after infection of macrophages (Daigle et al., 2001; Graham & Clark-Curtiss, 1999; Hou et al., 2002); in animal models of Actinobacillus pleuropneumoniae and Escherichia coli (Baltes & Gerlach, 2004; Dozois et al., 2003); and in Helicobacter pylori in human gastric biopsies (Graham et al., 2002). The technique has also been used to examine differential gene expression in Listeria monocytogenes in response to temperature changes (Liu et al., 2002) and to compare gene expression between related bacterial species or strains (Dozois et al., 2003; Morrow et al., 1999).
In the present study, we employed the SCOTS procedure to capture H. ducreyi transcripts that are expressed during experimental human infection. This study has led to identification of several genes that are required for infection in humans.
METHODS
Tissues
Pustules for SCOTS were obtained by biopsy from three women who had participated in human challenge trials (Table S1). Nine additional pustules were obtained from seven men and two women for RT-PCR (Table S1). All pustules were obtained 6 to 9 days after inoculation when the subjects reported pain. Informed consent for participation and for HIV serology was obtained from the volunteers in accordance with the human experimentation guidelines of the U. S. Department of Health and Human Services and the Institutional Review Board of Indiana University-Purdue University at Indianapolis.
RNA preparation
Whole biopsies were immediately placed in RNAlater (Qiagen). After 30 min, tissues were homogenized in Buffer RLT provided in the RNeasy Fibrous Tissue kit (Qiagen) using either a tissue homogenizer or a bead beater (Biospec Products) with 2.4 mm Zirconia beads (Biospec). RNA was extracted with the RNeasy Fibrous Tissue kit following the manufacturer’s directions, except that lysozyme (400 µg ml−1) was added to the proteinase K digestion step to lyse the bacteria. In vitro-derived RNA was obtained from a culture of H. ducreyi 35000HP (HP, human passaged) (Al-Tawfiq et al., 1998) grown in broth and used to inoculate human volunteers, as described (Al-Tawfiq et al., 1998; Spinola et al., 1994; Spinola et al., 1996). At mid-logarithmic phase (OD660 = 0.2), 4 ml of bacteria were collected by centrifugation and suspended in 1 ml Ultraspec RNA (Biotecx Laboratories, Inc.). Total RNA was extracted following the manufacturer’s directions. After isolation, all RNA samples were treated with DNase (Ambion) following the manufacturer’s directions. Integrity of the RNA samples was determined with an Agilent Bioanalyzer (Agilent Technologies).
cDNA synthesis
RNA was converted to double-stranded cDNA by random priming with the Advantage RT-for-PCR kit (Clontech Laboratories), following the manufacturer’s instructions, and second-strand synthesis as described previously (Froussard, 1992; Hou et al., 2002). The primers had defined 5’ sequences followed by nine random 3’ bases. Primers with different 5’ sequences were used to differentially tag the 5’ ends of cDNA derived from tissue (5’-GACACTCTCGAGACATGAGCGGTACC-3’) and from broth-grown bacteria (5’-CCTCTGAAGGTTCCAGAATCGATAG-3’). After synthesis, each cDNA pool was amplified by PCR using primers consisting of the appropriate 5’ tag without the random 3’ sequences. The amplimers were precipitated and suspended in 10 mM N-(2-hydroxyethyl)piperazine-N’-(3-propanesulfonic acid)-1 mM ethylenediaminetetraacetic acid to a final concentration of 375 ng µl−1.
rDNA plasmid construction
A 5.8-kbp locus containing 5S, 16S, and 23S rRNA genes was amplified by PCR from 35000HP using the following primers: (forward) 5’-TTGCAAAGAGGGAAAGATATCCTATAATGG-3’ and (reverse) 5’-TAGTACCCAATTACCTTATTTGTTGTTTGG-3’. The amplicon was cloned into the TA cloning vector pCR-XL-TOPO (Invitrogen) to make pSCOTS1 and propagated in E. coli TOP10F’ (Invitrogen).
SCOTS
The SCOTS procedure (diagrammed in Fig. S1 and refs. (Daigle et al., 2002; Faucher et al., 2006; Graham & Clark-Curtiss, 1999; Graham et al., 2002; Liu et al., 2002; Morrow et al., 1999)) was followed as described by Hou et al. (Hou et al., 2002). Graham and Clark-Curtiss showed that 3 rounds of SCOTS was sufficient to effectively eliminate eukaryotic sequences and normalize the relative levels of transcripts in each cDNA pool (Graham & Clark-Curtiss, 1999). Because the M. tuberculosis chromosome contains one set of rRNA genes, while the H. ducreyi chromosome contains 6 rRNA gene clusters, we increased the molar ratio of blocking rDNA 10-fold from the original protocol, to ensure an excess of blocking rDNA (Graham & Clark-Curtiss, 1999). With this modification, a pilot study confirmed that 3 rounds of SCOTS eliminated detectable rRNA-derived sequences from our cDNA pools (data not shown).
Each round of SCOTS utilized 1.2 µg of biotinylated, chromosomal DNA, 66 µg of rDNA, and 3 µg of the appropriate cDNA pool. In order to obtain sufficient material and to minimize bias introduced by individual PCR reactions, the first round of SCOTS for each cDNA pool consisted of 10 independent reactions that were pooled after PCR amplification. Two reactions were carried out and pooled in each of the subsequent rounds.
The tissue-derived and broth-derived cDNAs were each subjected to three rounds of SCOTS separately to generate 4 cDNA pools. The three tissue-derived cDNA pools were individually subjected to three rounds of competitive SCOTS in which the chromosomal DNA was prehybridized with the broth-derived cDNA pool to block capture of genes expressed both in vitro and in vivo. The final pool of selectively captured cDNAs was cloned, and the clones were screened for inserts on Luria-Bertani (LB) agar plates supplemented with ampicillin (50 µg ml−1), 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside, and isopropyl β-D-1-thiogalactopyranoside. Individual clones containing inserts were picked into LB broth in 96-well plates. Following overnight culture at 37°C, the plates were subjected to plasmid isolation and automated sequencing in a 96-well format, using universal primers (M13 forward and reverse), exactly as described previously (Munson Jr. et al., 2004). Three to four 96-well plates of clones were sequenced from each of the three cDNA libraries. Sequences of cloned inserts were compared with the H. ducreyi genome using the BLASTN algorithm.
Southern Analysis
Inserts from cDNA clones were amplified by PCR using M13 primers, separated electrophoretically, and transferred to nitrocellulose by capillary action. Normalized, tissue-derived and broth-derived cDNA pools were labeled using the PCR DIG Probe Synthesis kit (Roche Molecular Biochemicals, Indianapolis, Ind.) to use as probes. Blots were hybridized with the labeled probes and developed using the DIG Easy Hyb system (Roche Molecular Biologicals).
RT-PCR
RNA was extracted from pustules as described above or as described previously (Throm & Spinola, 2001). RT-PCR was performed to amplify H. ducreyi genes shown previously to be expressed in vivo as described (Throm & Spinola, 2001) or on the genes listed in Table 1. Each PCR reaction was performed alongside controls receiving the same amount of RNA template but no reverse transcriptase; all such controls were negative. Each reaction was performed once on each tissue indicated in Table 1. Primers are found in Table S3.
Table 1.
Verification of In Vivo Gene Transcription
| Gene ID | Annotation | Tissues in which cDNA or mRNA were detected (not detected) by |
Overall Results* | |
|---|---|---|---|---|
| SCOTS | RT-PCR | |||
| HD0192 | hypothetical lipoprotein |
238† (240, 250) | 231, 276, 281 | 4/6 |
| HD0286 | hypothetical protein |
240 (238, 250) | 160, 164, 276 (142, 231) | 4/8 |
| HD0646 | conserved hypothetical protein |
238, 240 (250) | 142, 231, 276 (164, 232) | 5/8 |
| HD0805 | conserved hypothetical protein |
238 (240, 250) | 160, 164, 231 (142, 232, 276) |
4/9 |
| HD1170 | outer membrane protein P4 |
250 (238, 240) | 231, 276, 281 | 4/6 |
| HD1280 | possible serine protease homolog |
238 (240, 250) | 231 (164, 281) | 2/6 |
| HD1589 | lipoprotein NlpI homolog |
238 (240, 250) | 276, 281 | 3/5 |
| HD1629 | outer membrane lipoprotein LolB |
250 (238, 240) | 249, 252 (231, 276, 281) | 3/8 |
| HD1655 | conserved hypothetical protein (poss. OMP) |
238, 250 (240) | (142, 160, 164, 231, 276, 281) |
2/9 |
| HD1808 | HflC protein | 238 (240, 250) | 281 (231) | 2/5 |
| HD1829 | probable outer membrane protein |
238 (240, 250) | 231 (276, 281) | 2/6 |
No. of tissues in which the transcript was detected / total no. of tissues examined
Subject number
RESULTS AND DISCUSSION
RNA isolation
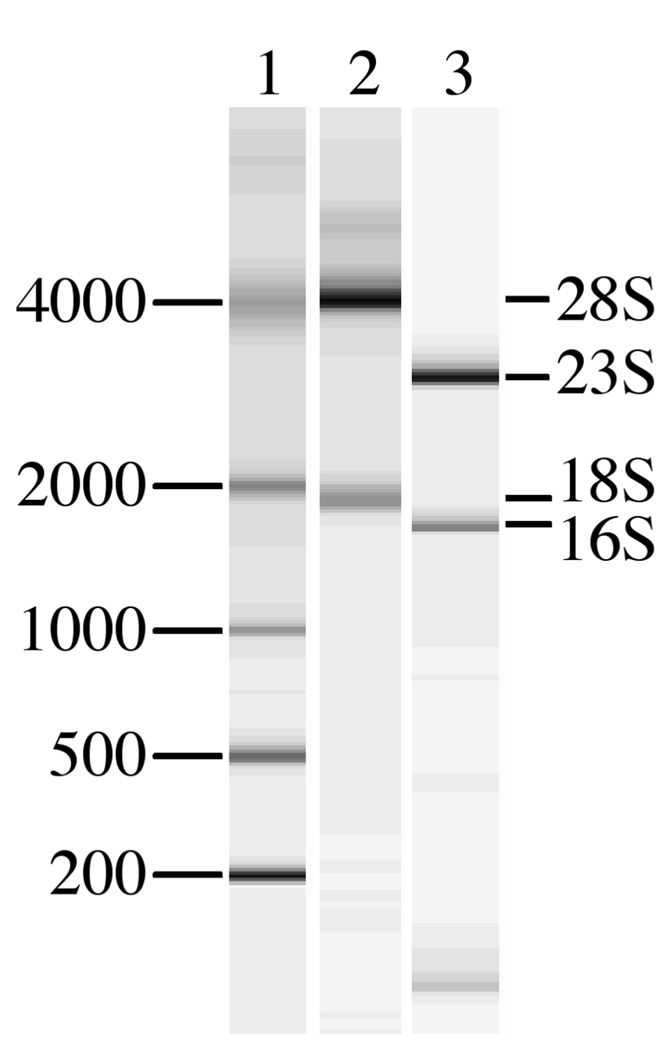
RNA was obtained from pustules of 3 volunteers (subjects 238, 240, and 250), who were experimentally infected with H. ducreyi (Table S1). Pustules contain necrotic cells that may release RNases; RNAlater was required to stabilize the specimens. The integrity of RNA in the samples was determined by analysis with an Agilent Bioanalyzer, which provides a spectrophotometric tracing and a virtual gel image of the RNA preparation (Fig. 1). The eukaryotic RNA was intact (Fig. 1), but the prokaryotic RNA levels were too low to visualize in these samples. Amplification of mRNA from several H. ducreyi genes, including cdtB, losB, dsrA, and lspA1, by RT-PCR was successful (data not shown), suggesting that the prokaryotic RNA was also intact. Intact RNA was also derived from a broth culture of 35000HP that was used to infect volunteers (Fig. 1). The four samples were independently subjected to SCOTS to produce four cDNA pools. The in vitro derived cDNA pool was then used to block capture of each in vivo derived cDNA pool in competitive SCOTS. Competitive SCOTS theoretically identifies transcripts that may be exclusively expressed or more abundant in one condition than in another. Increased transcript abundance may be due to higher rates of transcription, variations in mRNA degradation, and quality of the RNA samples. Thus, our results likely were not biased by differences in the quality of RNA among the samples.
Fig. 1.
Composite Agilent Bioanalyzer virtual gel image of RNA samples used in the study. Lane 1, RNA ladder with band sizes in bp. Lane 2, representative tissue-derived RNA sample. Lane 3, broth-derived RNA sample. Major eukaryotic and prokaryotic rRNA species are indicated.
Sequence analysis of clones obtained after competitive SCOTS
We sequenced inserts of 924 clones obtained after competitive SCOTS. All inserts contained H. ducreyi DNA. Only one insert was derived from H. ducreyi rRNA, indicating a blocking efficiency of > 99%. DNA in 8 inserts was derived from tRNAs. A few clones contained sequences that mapped to intergenic regions or pseudogenes; the remaining clones had inserts containing DNA homologous to known or putative open reading frames in the H. ducreyi genome.
The 924 clones contained sequences from 531 genes, representing nearly one third of the genome. We were somewhat surprised by the large number of captured transcripts, as H. ducreyi only infects humans and has no other known environmental niche and presumably might not require the complex regulation observed in bacteria that infect humans and infect other species or occupy environmental niches. However, within the human host, H. ducreyi infects a variety of tissues including stratified squamous or mucosal epithelium and lymph nodes, external sites such as the foreskin and labia, and internal sites such as the vagina and cervix. The cutaneous immune response to the organism is complex and consists of serum components, PMNs, macrophages, T cells, and dendritic cells. In addition, H. ducreyi infects men and women, who differ in their susceptibility to infection (Bong et al., 2002b; Spinola et al., 2003a). The H. ducreyi genome contains at least two intact systems that could sense extracytoplasmic stress, specifically the Cpx two-component system and sigma E (RpoE) system. Thus, H. ducreyi likely regulates its gene expression in response to different host pressures and microenvironments, as has been shown for other strict human pathogens such as Bordetella pertussis, which utilizes the bvg two-component system to regulate virulence genes.
Confirmation that the genes identified by SCOTS are expressed in vivo
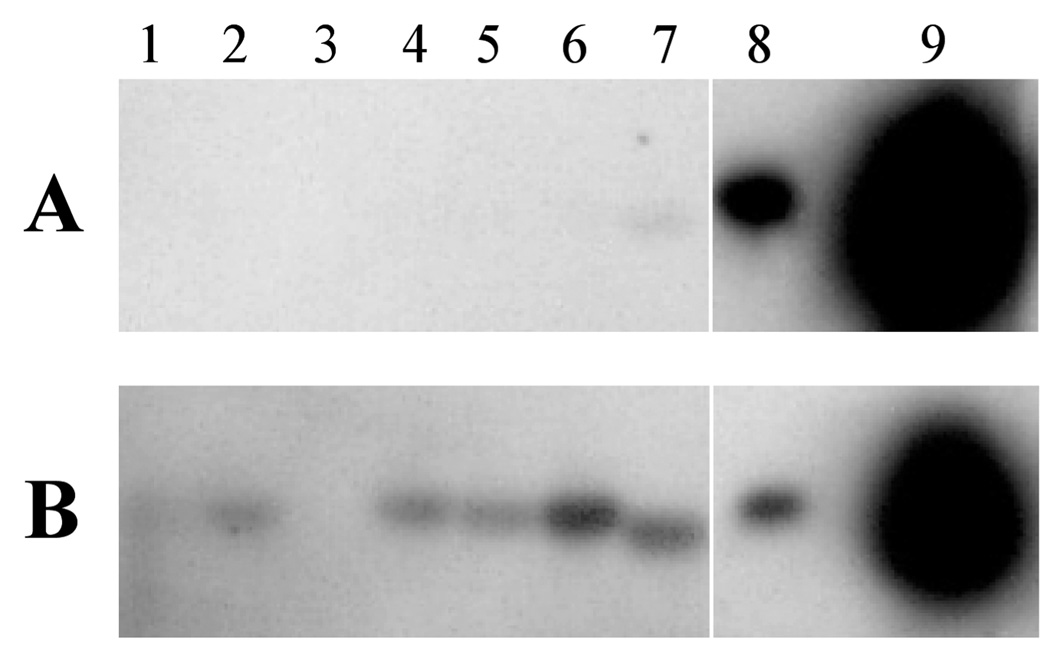
To confirm that the identified genes were expressed in vivo, 60 clones from each tissue were analyzed by Southern blot, which has been used as a confirmatory test in most other SCOTS-based studies (Baltes & Gerlach, 2004; Daigle et al., 2001; Graham & Clark-Curtiss, 1999; Haydel et al., 2002; Hou et al., 2002; Morrow et al., 1999). Duplicate blots of cloned inserts were probed with normalized cDNA pools from tissues or broth cultures after three rounds of SCOTS, and the signals from the two probes were compared. Representative results are shown in Fig. 2. Of 180 probed clones, 87% hybridized with the normalized tissue-derived H. ducreyi cDNA but either did not hybridize with the normalized broth-derived H. ducreyi cDNA (Fig. 2, lanes 1,2, 4–6) or showed a much stronger signal with the tissue-derived cDNA than with the broth-derived cDNA (Fig. 2, lane 7). These data show that the competitive SCOTS procedure preferentially captured sequences that were present in the in vivo derived cDNA pool. Of the clones that did not show a stronger signal with the tissue probe, only one (Fig. 2, lane 8) showed a similar signal with both probes, while the remainder showed no signal with either probe and were therefore inconclusive (Fig. 2, lane 3).
Fig. 2.
Composite southern blot representative of analysis of sequences identified by SCOTS. Cloned inserts were amplified by PCR, electrophoretically transferred, and probed with broth-derived cDNA (panel A) or tissue-derived cDNA (panel B) as described in Materials and Methods. Lanes 1–7, amplified inserts of SCOTS derived clones; lane 8, cloned insert expressed in broth and in vivo; lane 9, unlabeled probe as positive control.
To further confirm the SCOTS data, we also performed RT-PCR on RNA isolated from H. ducreyi-infected pustules obtained from 9 additional volunteers (Table S1). Ten of 11 (91%) of the genes captured by SCOTS were amplified from one or more additional tissues (Table 1). The one gene not detected by RT-PCR was identified in two of the subjects who provided tissue for SCOTS. The data suggest that genes identified by SCOTS are transcribed in multiple volunteers.
We considered but could not use quantitative real-time RT-PCR (qRT-PCR) to confirm our findings. qRT-PCR requires the ability to normalize results from each sample to an objective reference reflecting total bacterial genomes or RNA levels or to a gene constitutively expressed under the conditions being compared (Shelburne & Musser, 2004). Our in vivo RNA is almost entirely eukaryotic, while our in vitro RNA is entirely prokaryotic RNA. RNAlater was required to stabilize the tissue samples, which were processed entirely for RNA. Thus, we have no information about the number of bacteria or bacterial genomes in the samples. Unfortunately, multiple attempts to process biopsies simultaneously for DNA and RNA so that we could quantitate bacterial genomes yielded degraded RNA. The identity of genes constitutively expressed by H. ducreyi in vitro and in vivo is unknown. In the absence this information, 16S rRNA is commonly used to normalize bacterial RNA samples. However, 16S rRNA levels are highly dependent on growth conditions and nutrient availability (Gourse et al., 1996). The doubling time of 35000HP in broth is approximately 90 min, while the estimated doubling time of 35000HP in lesions is 16.5 ± 3.8 h (Throm & Spinola, 2001). Thus, we cannot assume that the 16S rRNA content of the bacteria grown in vitro is similar to that of the bacteria in a pustule. Although the chief limitation of SCOTS is that it is qualitative and we did not prove that the captured transcripts were upregulated in vivo, the captured genes must be expressed in vivo.
Redundancy of gene expression among volunteers
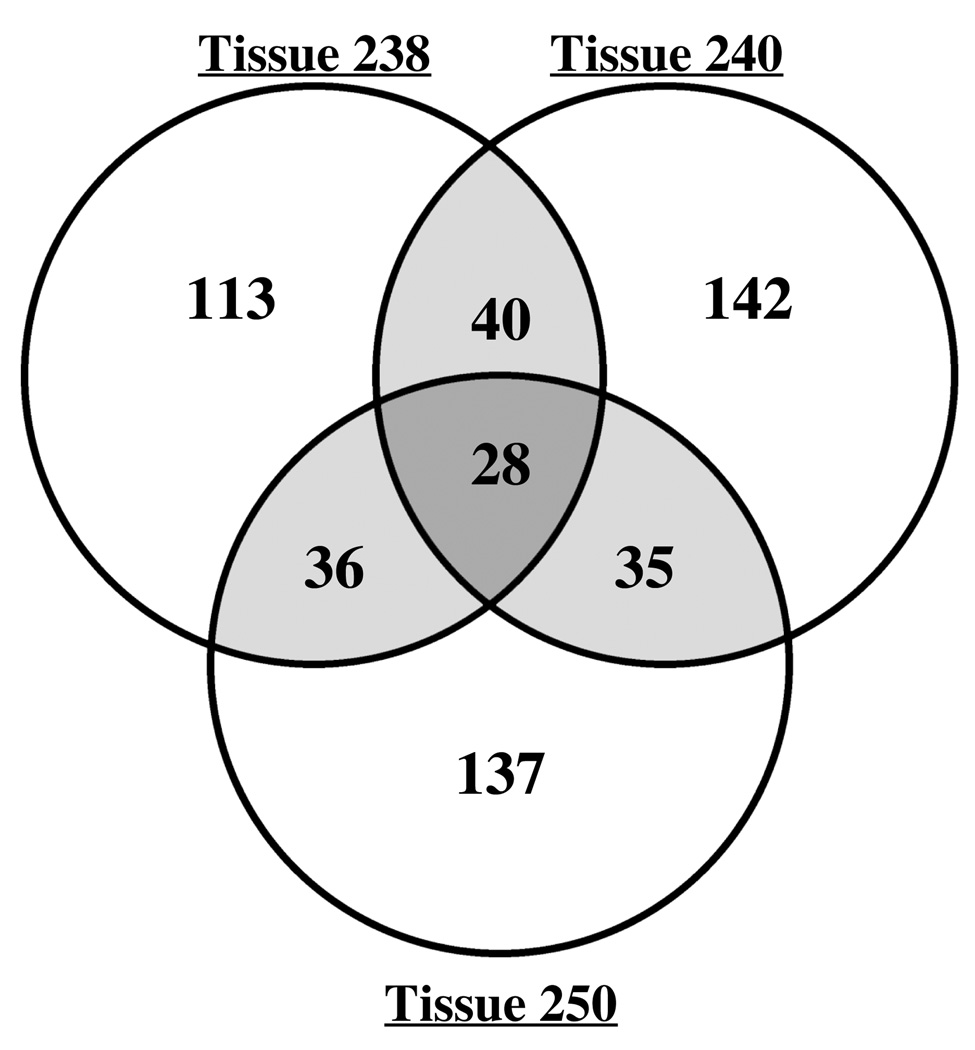
Fig. 3 shows the distribution of the 531 identified genes among the three volunteers. Although the majority of transcripts were identified in cDNA from only one volunteer, 139 transcripts were identified in at least two volunteers, and 28 transcripts were identified as expressed in all three volunteers (Fig. 3). Analysis of the functional categories of the identified genes demonstrated similar patterns of gene expression in all three volunteers (Table 2).
Fig. 3.
Venn diagram of the distribution of genes identified by SCOTS in tissues from 3 volunteers. The number of genes identified in each tissue is indicated; shaded areas show the overlap of genes identified in more than one tissue.
Table 2.
Functional Categories of in Vivo Expressed Genes
| No. of Genes identified in: | |||||
|---|---|---|---|---|---|
| Category | Tissue 238 | Tissue 240 | Tissue 250 | Any Tissue | at least Two Volunteers |
| Adherence-related Genes | 2 | 2 | 2 | 2 | 2 |
| Bacteriophage genes | 8 | 14 | 7 | 20 | 7 |
| Biosynthesis/Metabolism | 76 | 76 | 75 | 175 | 40 |
| Cell Division | 5 | 3 | 5 | 10 | 3 |
| DNA Replication/Repair | 10 | 22 | 28 | 47 | 13 |
| Electron Transport | 7 | 11 | 7 | 15 | 7 |
| Hypothetical Proteins | 57 | 64 | 51 | 133 | 32 |
| Outer Membrane Proteins | 3 | 1 | 5 | 6 | 3 |
| Regulation | 5 | 3 | 6 | 12 | 2 |
| Ribosomal Proteins | 8 | 4 | 6 | 15 | 3 |
| Secreted Proteins | 1 | 0 | 2 | 2 | 1 |
| Stress Response | 5 | 1 | 5 | 9 | 2 |
| Toxins | 0 | 2 | 1 | 2 | 1 |
| Transcription | 4 | 5 | 6 | 10 | 4 |
| Translation | 11 | 11 | 10 | 22 | 9 |
| Transport/Uptake | 15 | 26 | 20 | 51 | 10 |
| Total no. of Genes | 217 | 245 | 236 | 531 | 139 |
With the exception of microarray analysis, none of the published in vivo expression procedures, including SCOTS as well as promoter-trap and mutagenic strategies, is comprehensive for the entire genome (Rediers et al., 2005; Shelburne & Musser, 2004). Data from SCOTS are limited to the number of clones sequenced. Thus, we do not know whether 139 genes represents the true level of overlap of gene expression from host to host, or whether analysis of additional clones would have yielded greater overlap. RT-PCR of additional tissues showed that many transcripts captured in only one tissue in SCOTS were present in additional volunteers, suggesting that we would have found more overlap had we sequenced more clones.
Analysis of genes expressed in at least two volunteers
Of the 139 genes identified in at least two tissues (Table S2), 68 of 74 (92%) tested hybridized to a greater extent with tissue-derived than with broth-derived cDNA. Although the RT-PCR data suggested that there may be greater overlap among the three volunteers, observed host effects on H. ducreyi infection indicate that some of the detected genes could be expressed only in a subset of volunteers (Bong et al., 2002b; Spinola et al., 2003a). We therefore conservatively confined our examination of the potential functional significance of in vivo expressed genes to those from more than one volunteer.
The 139 genes identified in at least 2 volunteers fell into a variety of functional categories, based on homology with previously characterized bacterial genes (Table 2, final column). The most populous group was the broad category of biosynthetic and metabolic genes. There were also genes involved in a number of cellular processes, including cell division, DNA replication and repair, transcription, and translation. Some identified genes, including those encoding heat-shock proteins or transporters, can play important roles in responses to stress or nutrient deprivation. Additionally, 2 regulatory genes were expressed in vivo, including the sensor kinase of a 2-component regulator. We also identified 32 genes encoding hypothetical proteins. Homologs of at least 21 of these genes (shown in bold in Table S2) have been identified by promoter-trap strategies as in vivo expressed during infections caused by other pathogens (Rediers et al., 2005). Overall, these data are consistent with in vivo bacterial gene induction analyses, in which many categories of genes are affected in addition to traditional virulence factors (Rediers et al., 2005; Shelburne & Musser, 2004) and suggest that H. ducreyi responds to the host in a global fashion for in vivo survival.
Six identified electron transport genes are involved in anaerobic respiration in other bacterial systems, including nitrate reductases napA and nrfA, L-lactate dehydrogenase lldD, anaerobic glycerol-3-phosphate dehydrogenase subunits glpB and glpC, and torY, a c-type cytochrome involved in trimethylamine N-oxide reduction. Similarly, two metabolic genes, pflB encoding pyruvate formate-lyase and ansB encoding L-asparaginase II, are homologous to genes that are induced under anaerobic conditions in E. coli (Jennings & Beacham, 1990; Sawers & Bock, 1988). In contrast, we also identified atpA, an aerobic electron transfer chain subunit, suggesting that some aerobic respiration may also occur. Typically, the environment within pustules is anaerobic (Hays & Mandell, 1974). However, H. ducreyi are found both deep within the pustule and at the pustule surface, where the environment may be more aerobic (Bauer & Spinola, 2000; Bauer et al., 2001). Expression of both aerobic and anaerobic respiration genes could reflect differential responses within the bacterial population to microenvironments with different oxygenation levels in vivo.
Six genes identified in this study are involved in amino acid biosynthesis, including argA, argC, and argE from the arginine biosynthetic pathway, and carB, which is involved in synthesis of arginine and pyrimidines. Arginine synthetic genes have been identified during in vivo expression studies of several other bacterial pathogens (Rediers et al., 2005). Notably, homologs of argA and carAB, were identified during in vivo expression studies in Vibrio cholerae and Salmonella enterica serovar Typhimurium, respectively (Camilli & Mekalanos, 1995; Mahan et al., 1993). Mutational analysis in these pathogens demonstrated that argA and carAB were required for virulence of V. cholerae and S. enterica, respectively, in mouse models of infection (Camilli & Mekalanos, 1995; Mahan et al., 1993). These data suggest that H. ducreyi, among other pathogens, may require increased arginine in vivo, which is a semi-essential amino acid for mammals and thus likely to be in limited supply in tissues. In addition to its utility as an amino acid, arginine is a substrate in other synthesis pathways, such as polyamine production (Cunin et al., 1986). Possibly, one or more arginine-requiring biosynthetic pathway(s) is induced in vivo, leading to an increased arginine requirement.
Eight genes identified in vivo are involved in DNA repair, including mutL, mutS, radA, recB, recD, recG, ung, and uvrA, suggesting that H. ducreyi encounters DNA-damaging stresses during human infection. Six of the identified DNA repair genes were also identified during in vivo expression studies with other pathogens (Rediers et al., 2005). Additionally, the recBCD-encoded exonuclease V is required for repair of double-stranded DNA breaks and has been shown to be necessary in S. enterica for both survival in macrophages and virulence in mice (Cano et al., 2002). Thus, DNA repair could be an important mechanism for H. ducreyi survival in vivo.
In addition to genes involved in various cellular processes or regulation, we identified several genes encoding bacterial components that have been characterized by our laboratory and others as potential virulence determinants, including the large supernatant proteins LspA1 or LspA2, the cytolethal distending toxin subunit CdtB, tight adherence protein TadA, and outer membrane proteins, including MOMP, predicted outer membrane protein HD1655, predicted outer membrane lipoprotein HD1094, and a homolog of H. influenzae outer membrane protein OMP P1 (Table 2 and Table S2). The LspA proteins, LspA1 and LspA2, share 86% sequence identity and confer antiphagocytic activity to the organism. (Vakevainen et al., 2003). We identified clones with inserts corresponding to identical regions shared by LspA1 and LspA2. Although expression of MOMP and CdtC are not required for pustule formation in humans (Throm et al., 2000; Young et al.), expression of LspA1 and LspA2 and the TadA-containing operon are required for full virulence in the human model of H. ducreyi infection (Janowicz et al., 2004; Spinola et al., 2003b). Prompted by the findings from this study, we constructed mutants in HD1844, which encodes WecA, the first enzyme in the enterobacterial common antigen pathway, and in HD0192 (Table 1), which encodes a hypothetical lipoprotein. Both mutants were partially attenuated for pustule formation in humans (K.E. Banks, K.R. Fortney, B. Baker, S.D. Billings, B.P. Katz, R.S. Munson, Jr., and S. M. Spinola, unpublished observations; D.M. Janowicz, K.R. Fortney, B.W. Zwickl, B.P. Katz, S.M. Spinola, and M.E. Bauer, unpublished observations). Thus, several captured transcripts were relevant to pathogenesis. Identification of momp, lspA1 or lspA2, cdtB, and tadA also showed that SCOTS detected some genes H. ducreyi expresses in vitro (Nika et al., 2002; Stevens et al., 1999; Throm & Spinola, 2001; Ward et al., 1998). Southern blot analysis confirmed that momp, lspA2, and tadA were more abundant in the in vivo derived cDNA pool (data not shown), suggesting that expression of these genes may be upregulated in vivo.
HD1895, a gene encoding a HmwC-like protein, was identified in this screen. In H. influenzae, HmwC is involved in glycosylation of the high molecular weight (HMW) adhesins, and a copy of this gene is present in both HMW gene clusters in H. influenzae (Grass et al., 2003; St. Geme & Grass, 1998). The N-terminal portions of LspA1 and LspA2 share homology with HMW1A and HMW2A (Ward et al., 1998), but there is no evidence of glycosylation of the LspA proteins, and the genes up- and down-stream of HD1895 are not likely surface-associated proteins. Thus, the substrate for the putative glycosyl transferase HmwC is unknown.
Several genes encoding enzymes involved in lipooligosaccharide (LOS) biosynthesis were identified, including galE, kdkA, rfaE, and lpxK. Sialic acid is a component of the LOS of H. ducreyi. HD1669 and HD1670 were identified in the screen and have recently been shown to encode members of an ABC transporter that transports sialic acid (Post et al., 2005). HD1842 and HD1844 encode proteins that also are involved in complex carbohydrate biosynthesis. These genes are part of a gene cluster with strong homology to genes involved in enterobacterial common antigen synthesis.
H. ducreyi has 3 Mu-like bacteriophage gene clusters. Genes in these regions have not been characterized, and no bacteriophage production or activity has been observed in H. ducreyi. However, 7 genes from two of these clusters were identified in our study (Table S2).
Our results were similar to those reported in a recent SCOTS-based study of in vivo gene expression by A. pleuropneumoniae in a porcine model of infection (Baltes & Gerlach, 2004). Baltes and Gerlach identified 46 A. pleuropneumoniae genes expressed in lung specimens pooled from 4 pigs. The H. ducreyi genome contains homologs of 28 of the 46 A. pleuropneumoniae genes identified in the porcine study, and 12 of the 28 homologs were also expressed in our 3 human volunteers in vivo. These homologs included a gene encoding an outer membrane protein, 6 biosynthetic genes, two regulatory genes, a stress response gene, and genes encoding two transporters. While H. ducreyi and A. pleuropneumoniae have different host ranges and cause different diseases, they are members of the Pasteurellaceae, and both are extracellular pathogens that rely on similar strategies such as resistance to phagocytosis for in vivo survival.
In summary, our data suggest that H. ducreyi gene expression undergoes multiple changes within the human host compared with in vitro broth culture. These data also indicate that one or more regulatory mechanisms of H. ducreyi functions in vivo and define a number of candidates for genes important to in vivo survival or virulence of the organism, including several hypothetical genes as well as genes required for virulence in other bacteria. Future studies will include defining regulatory mechanisms, identifying host factors to which the organism responds, and defining the roles in virulence of genes expressed in vivo.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI27863 and AI31494 from the National Institute of Allergy and Infectious Diseases (NIAID). The human challenge trials were supported by Public Health Service grant MO1RR00750 to the General Clinical Research Center at Indiana University. D.M.J. was supported by NIH grants T32 AI007637 and K08 AI074657 from NIAID. M.E.B. was partially supported by the Indiana Genomics Initiative (INGEN) of Indiana University, which is supported in part by Lilly Endowment, Inc.
We thank Josephine Clark-Curtiss for training on the SCOTS procedure, many helpful discussions, and critical review of the manuscript. We thank Joan Hou for training and trouble-shooting on the SCOTS procedure, Carisa Townsend for technical assistance, Genshi Zhao, James Graham, and France Daigle for helpful discussions, and Tricia Humphreys, Barbara Van Der Pol, and Byron Batteiger for critical review of the manuscript.
Abbreviations
- SCOTS
selective capture of transcribed sequences
Footnotes
Publisher's Disclaimer: This is an author manuscript that has been accepted for publication in Microbiology, copyright Society for General Microbiology, but has not been copyedited, formatted or proofed. Cite this article as appearing in Microbiology. This version of the manuscript may not be duplicated or reproduced, other than for personal use or within the rule of ‘Fair Use of Copyrighted Materials’ (section 17, Title 17, US Code), without permission from the copyright owner, Society for General Microbiology. The Society for General Microbiology disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by any other parties. The final copy-edited, published article, which is the version of record, can be found at http://mic.sgmjournals.org/, and is freely available without a subscription.
REFERENCES
- Al-Tawfiq JA, Thornton AC, Katz BP, Fortney KR, Todd KD, Hood AF, Spinola SM. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J Infect Dis. 1998;178:1684–1687. doi: 10.1086/314483. [DOI] [PubMed] [Google Scholar]
- Al-Tawfiq JA, Fortney KR, Katz BP, Elkins C, Spinola SM. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J Infect Dis. 2000;181:1049–1054. doi: 10.1086/315309. [DOI] [PubMed] [Google Scholar]
- Baltes N, Gerlach GF. Identification of genes transcribed by Actinobacillus pleuropneumoniae in necrotic porcine lung tissue by using selective capture of transcribed sequences. Infect Immun. 2004;72:6711–6716. doi: 10.1128/IAI.72.11.6711-6716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer ME, Spinola SM. Localization of Haemophilus ducreyi at the pustular stage of disease in the human model of infection. Infect Immun. 2000;68:2309–2314. doi: 10.1128/iai.68.4.2309-2314.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer ME, Goheen MP, Townsend CA, Spinola SM. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect Immun. 2001;69:2549–2557. doi: 10.1128/IAI.69.4.2549-2557.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer ME, Townsend CA, Ronald AR, Spinola SM. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microb Infect. 2006;8:2465–2468. doi: 10.1016/j.micinf.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Bong CTH, Throm RE, Fortney KR, Katz BP, Hood AF, Elkins C, Spinola SM. A DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect Immun. 2001;69:1488–1491. doi: 10.1128/IAI.69.3.1488-1491.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bong CTH, Bauer ME, Spinola SM. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microb Infect. 2002a;4:1141–1148. doi: 10.1016/s1286-4579(02)01639-8. [DOI] [PubMed] [Google Scholar]
- Bong CTH, Harezlak J, Katz BP, Spinola SM. Men are more susceptible to pustule formation than women in the experimental model of Haemophilus ducreyi infection. Sexually Trans Dis. 2002b;29:114–118. doi: 10.1097/00007435-200202000-00009. [DOI] [PubMed] [Google Scholar]
- Camilli A, Mekalanos JJ. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano DA, Pucciarelli MG, García-del Portillo F, Casadesús J. Role of the RecBCD recombination pathway in Salmonella virulence. J Bacteriol. 2002;184:592–595. doi: 10.1128/JB.184.2.592-595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Mekalanos JJ, Holden DW. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol. 1999;53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- Cunin R, Glansdorff N, Piérard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiological Reviews. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle F, Graham JE, Curtiss R., III Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS) Mol Microbiol. 2001;41(5):1211–1222. doi: 10.1046/j.1365-2958.2001.02593.x. [DOI] [PubMed] [Google Scholar]
- Daigle F, Hou JY, Clark-Curtiss JE. Microbial gene expression elucidated by selective capture of transcribed sequences (SCOTS) Methods Enzymol. 2002;358:108–122. doi: 10.1016/s0076-6879(02)58083-6. [DOI] [PubMed] [Google Scholar]
- Dozois CM, Daigle F, Curtiss R., III Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc Natl Acad Sci USA. 2006;103:1903–1911. doi: 10.1073/pnas.0509183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortney KR, Young RS, Bauer ME, Katz BP, Hood AF, Munson RS, Jr, Spinola SM. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect Immun. 2000;68:6441–6448. doi: 10.1128/iai.68.11.6441-6448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froussard P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher RA, Cole LE, Janowicz DM, Toffer KL, Fortney KR, Katz BP, Orndorff PE, Spinola SM, Kawula TH. Expression of Haemophilus ducreyi collagen binding outer membrane protein NcaA is required for virulence in swine and human challenge models of chancroid. Infect Immun. 2006;74:2651–2658. doi: 10.1128/IAI.74.5.2651-2658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Gaal T, Bartlett MS, Appleman JA, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Ann Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JE, Peek RM, Jr, Krishna U, Cover TL. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology. 2002;123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass S, Buscher AZ, Swords WE, Apicella MA, Barenkamp SJ, Ozchlewski N, St. Geme Jw., III The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involves in lipooligosaccharide biosynthesis. Mol Microbiol. 2003;48:737–751. doi: 10.1046/j.1365-2958.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- Haydel SE, Benjamin WH, Jr, Dunlap NE, Clark-Curtiss JE. Expression, autoregulation, and DNA binding properties of the Mycobacterium tuberculosis TrcR response regulator. J Bacteriol. 2002;184:2192–2203. doi: 10.1128/JB.184.8.2192-2203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays RC, Mandell GL. pO2, pH, and redox potential of experimental abscesses. Proc Soc Exp Biol Med. 1974;147:29–30. doi: 10.3181/00379727-147-38275. [DOI] [PubMed] [Google Scholar]
- Hou JY, Graham JE, Clark-Curtiss JE. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS) Infect Immun. 2002;70:3714–3726. doi: 10.1128/IAI.70.7.3714-3726.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowicz DM, Fortney KR, Katz BP, Latimer JL, Deng K, Hansen EJ, Spinola SM. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect Immun. 2004;72:4528–4533. doi: 10.1128/IAI.72.8.4528-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowicz DM, Leduc I, Fortney KR, Katz BP, Elkins C, Spinola SM. A DltA mutant of Haemophilus ducreyi is partially attenuated in its ability to cause pustules in human volunteers. Infect Immun. 2006a;74:1394–1397. doi: 10.1128/IAI.74.2.1394-1397.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowicz DM, Luke NR, Fortney KR, Katz BP, Campagnari AA, Spinola SM. Expression of OmpP2A and OmpP2B is not required for pustule formation by Haemophilus ducreyi in human volunteers. Microb Pathog. 2006b;40:110–115. doi: 10.1016/j.micpath.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Jennings MP, Beacham IR. Analysis of the Escherichia coli gene encoding L-asparaginase II, ansB, and its regulation by cyclic AMP receptor. J Bacteriol. 1990;172:1491–1498. doi: 10.1128/jb.172.3.1491-1498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Graham JE, Bigelow L, Morse PD, II, Wilkinson BJ. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl Environ Microbiol. 2002;68:1697–1705. doi: 10.1128/AEM.68.4.1697-1705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan MJ, Slauch JM, Mekalanos JJ. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- Mahan MJ, Heithoff DM, Sinsheimer RL, Low DA. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Ann Rev Genet. 2000;34:139–164. doi: 10.1146/annurev.genet.34.1.139. [DOI] [PubMed] [Google Scholar]
- Morrow BJ, Graham JE, Curtiss R., III Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect Immun. 1999;67:5106–5116. doi: 10.1128/iai.67.10.5106-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson RS, Jr, Harrison A, Gillaspy A, et al. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect Immun. 2004;72:3002–3010. doi: 10.1128/IAI.72.5.3002-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nika JR, Latimer JL, Ward CK, Blick RJ, Wagner NJ, Cope LD, Mahairas GG, Munson JRS, Hansen EJ. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect Immun. 2002;70:2965–2975. doi: 10.1128/IAI.70.6.2965-2975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Schnizlein-Bick CT, Orazi A, John K, Chen C-Y, Hood AF, Spinola SM. The immune response to Haemophilus ducreyi resembles a delayed-type hypersensitivity reaction throughout experimental infection of human subjects. J Infect Dis. 1998;178:1688–1697. doi: 10.1086/314489. [DOI] [PubMed] [Google Scholar]
- Post DM, Mungur R, Gibson BW, Munson RS., Jr Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect Immun. 2005;73:6727–6735. doi: 10.1128/IAI.73.10.6727-6735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rediers H, Rainey PB, Vanderleyden J, De Mot R. Unraveling the secret lives of bacteria: use of in vivo expression technology and differential fluorescence induction promoter traps as tools for exploring niche-specific gene expression. Microbiology and Molecular Biology Reviews. 2005;69:217–261. doi: 10.1128/MMBR.69.2.217-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers G, Bock A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, Musser JM. Virulence gene expression in vivo. Curr Opin Microbiol. 2004;7:283–289. doi: 10.1016/j.mib.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Spinola SM, Wild LM, Apicella MA, Gaspari AA, Campagnari AA. Experimental human infection with Haemophilus ducreyi. J Infect Dis. 1994;169:1146–1150. doi: 10.1093/infdis/169.5.1146. [DOI] [PubMed] [Google Scholar]
- Spinola SM, Orazi A, Arno JN, Fortney K, Kotylo P, Chen C-Y, Campagnari AA, Hood AF. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J Infect Dis. 1996;173:394–402. doi: 10.1093/infdis/173.2.394. [DOI] [PubMed] [Google Scholar]
- Spinola SM, Bauer ME, Munson RS., Jr Immunopathogenesis of Haemophilus ducreyi infection (chancroid) Infect Immun. 2002;70:1667–1676. doi: 10.1128/IAI.70.4.1667-1676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinola SM, Bong CTH, Faber AL, et al. Differences in host susceptibility to disease progression in the human challenge model of Haemophilus ducreyi infection. Infect Immun. 2003a;71:6658–6663. doi: 10.1128/IAI.71.11.6658-6663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinola SM, Fortney KR, Katz BP, Latimer JL, Mock JR, Vakevainen M, Hansen EJ. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect Immun. 2003b;71:7178–7182. doi: 10.1128/IAI.71.12.7178-7182.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Geme JW, III, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- Steen R. On eradicating chancroid. Bull WHO. 2001;79:818–826. [PMC free article] [PubMed] [Google Scholar]
- Stevens MK, Latimer JL, Lumbley SR, Ward CK, Cope LD, Lagergård T, Hansen EJ. Characterization of a Haemophilus ducreyi mutant deficient in expression of cytolethal distending toxin. Infect Immun. 1999;67:3900–3908. doi: 10.1128/iai.67.8.3900-3908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throm RE, Al-Tawfiq JA, Fortney KR, Katz BP, Hood AF, Hansen EJ, Spinola SM. Evaluation of an isogenic MOMP-deficient mutant in the human model of Haemophilus ducreyi infection. Infect Immun. 2000;68:2602–2607. doi: 10.1128/iai.68.5.2602-2607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throm RE, Spinola SM. Transcription of candidate virulence genes of Haemophilus ducreyi during infection of human volunteers. Infect Immun. 2001;69:1483–1487. doi: 10.1128/IAI.69.3.1483-1487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakevainen M, Greenberg S, Hansen EJ. Inhibition of phagocytosis by Haemophilus ducreyi requires expression of the LspA1 and LspA2 proteins. Infect Immun. 2003;71:5994–6003. doi: 10.1128/IAI.71.10.5994-6003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CK, Lumbley SR, Latimer JL, Cope LD, Hansen EJ. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol. 1998;180:6013–6022. doi: 10.1128/jb.180.22.6013-6022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RS, Fortney KR, Gelfanova V, et al. Expression of cytolethal distending toxin and hemolysin are not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect Immun. 2001;69:1938–1942. doi: 10.1128/IAI.69.3.1938-1942.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.