The crystal structure of perdeuterated diisopropyl fluorophosphatase is reported and compared with the hydrogenated structure. Diffraction guidelines for neutron crystallography experiments are summarized.
Keywords: diisopropyl fluorophosphatase, perdeuteration
Abstract
The signal-to-noise ratio is one of the limiting factors in neutron macromolecular crystallography. Protein perdeuteration, which replaces all H atoms with deuterium, is a method of improving the signal-to-noise ratio of neutron crystallography experiments by reducing the incoherent scattering of the hydrogen isotope. Detailed analyses of perdeuterated and hydrogenated structures are necessary in order to evaluate the utility of perdeuterated crystals for neutron diffraction studies. The room-temperature X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase) is reported at 2.1 Å resolution. Comparison with an independently refined hydrogenated room-temperature structure of DFPase revealed no major systematic differences, although the crystals of perdeuterated DFPase did not diffract neutrons. The lack of diffraction is examined with respect to data-collection and crystallographic parameters. The diffraction characteristics of successful neutron structure determinations are presented as a guideline for future neutron diffraction studies of macromolecules. X-ray diffraction to beyond 2.0 Å resolution appears to be a strong predictor of successful neutron structures.
1. Introduction
Neutron crystallography is one of the only techniques that can be used to experimentally determine the position of H atoms in a three-dimensional structure and thus provides complementary information to the location of heavier atoms obtained from X-ray crystallographic analyses (Niimura & Bau, 2008 ▶). The unique information available from neutron crystallography has provided important insights into the mechanisms of enzymes (Adachi et al., 2009 ▶; Bennett et al., 2006 ▶; Blakeley et al., 2008 ▶; Blum et al., 2009 ▶; Coates et al., 2001 ▶, 2008 ▶; Katz et al., 2006 ▶; Kossiakoff & Spencer, 1981 ▶; Kovalevsky et al., 2008 ▶; Tamada et al., 2009 ▶; Yagi et al., 2009 ▶; Tomanicek et al., 2010 ▶). Until recently, elucidating neutron structures has required large amounts of sample in order to grow large crystals (typically >1 mm3) and long data-collection periods. Recent developments in instrumentation and data-collection techniques at both reactor and spallation neutron sources (Blakeley et al., 2008 ▶; Langan et al., 2008 ▶) have significantly reduced the sample-size requirements and data-acquisition times for neutron data sets (Blum et al., 2007 ▶). Concurrently, recent improvements in structure-refinement software and strategies, such as the use of X-ray and neutron crystallographic data in maximum-likelihood refinement in nCNS and PHENIX, have helped to address the limitations of the low data-to-parameter ratios and low data resolutions that are often associated with neutron crystallographic data (Adams et al., 2009 ▶). Despite these developments, neutron crystallography remains a demanding technique.
A remaining limiting factor in neutron crystallographic analyses of proteins is incoherent scattering from hydrogen, which adds to the scattering background and attenuates the diffracted intensities. These effects can greatly reduce the signal-to-noise level and therefore the accuracy and resolution of neutron crystallographic data because of the large numbers of H atoms present in proteins (around one-half of all atoms) and solvent. For this reason, hydrogenated protein crystals are typically soaked in deuterated mother liquor prior to data collection to replace accessible and labile H atoms with D atoms, a process called deuteration. Because deuterium has a smaller incoherent scattering cross section than hydrogen (2.05 b for D compared with 80.27 b for H; 1 b = 100 fm2) and a larger coherent scattering length (6.6 fm compared with −3.7 fm), deuteration can increase the neutron scattering power of a crystal and also the signal-to-noise ratio of the diffracted intensities. However, only about 25% of H atoms in proteins are labile and can be replaced by deuterium. Furthermore, some of these labile H atoms are exchanged on timescales of the order of weeks or months. Despite being labile, certain backbone amides in β-sheets or within the hydrophobic core of proteins remain unexchanged even after long periods of time. To replace the remaining H atoms and obtain a fully deuterated (perdeuterated) protein requires expression of the gene of interest in an organism that is adapted for growth in deuterated media.
Complete perdeuteration of proteins, which was first demonstrated for staphylococcal nuclease (Gamble et al., 1994 ▶) and myoglobin (Shu et al., 2000 ▶) and has subsequently been applied to aldose reductase (Hazemann et al., 2005 ▶; Blakeley et al., 2008 ▶) and type III antifreeze protein (Petit-Haertlein et al., 2009 ▶), has allowed the collection of neutron diffraction data from crystals as small as ∼0.1 mm3. Encouraged by these results, several investigators are exploring perdeuteration as a means of making neutron crystallographic analyses feasible for proteins that can only be grown to small crystal volumes or to improve the resolution of neutron data collected from deuterated crystals of hydrogenous protein (Artero et al., 2005 ▶; Budayova-Spano et al., 2006 ▶; Liu et al., 2007 ▶; Meilleur et al., 2004 ▶, 2005 ▶; Tuominen et al., 2004 ▶). Although the development of perdeuteration protocols and an economical user-friendly infrastructure for the production of perdeuterated proteins has been a major focus of research and development efforts, there remain few examples of neutron structures solved using perdeuterated proteins. It is well known that major functional changes often occur upon deuteration. Excessive amounts of D2O are toxic to higher organisms and growth of bacteria in D2O greatly alters their metabolic processes and growth rates. A careful comparative analysis of hydrogenated and perdeuterated protein structures is crucial in order to understand the sometimes subtle structural changes that can occur upon deuteration and to verify the assumptions underlying the use of perdeuteration in neutron crystallography.
The enzyme diisopropyl fluorophosphatase (DFPase; EC 3.1.8.2) from the squid Loligo vulgaris (Fig. 1 ▶) is a potent and well characterized Ca2+-dependent phosphotriesterase that is capable of hydrolyzing a variety of organophosphorus compounds that act as irreversible inhibitors of acetylcholinesterase (AChE). Substrates include diisopropyl fluorophosphate (DFP) and a range of highly toxic G-type organophosphorus (OP) nerve agents such as sarin (GB), soman (GD) and cyclosarin (GF) that pose a threat to military personnel and civilian populations (Blum et al., 2006 ▶, 2008 ▶; Scharff et al., 2001 ▶). To elucidate the mechanism of DFPase, the 2.2 Å neutron structure of DFPase has been determined (PDB code 3byc) using a deuterated crystal of 0.43 mm3 in volume (Blum et al., 2009 ▶). In the present study, crystals of perdeuterated DFPase have been grown in an attempt to improve the quality of the neutron crystallographic data and to obtain more detailed information on the active-site environment. The 2.1 Å resolution room-temperature (RT) X-ray structure of perdeuterated DFPase (d-DFPase) was solved as a prerequisite for a planned joint refinement of neutron and X-ray diffraction data.
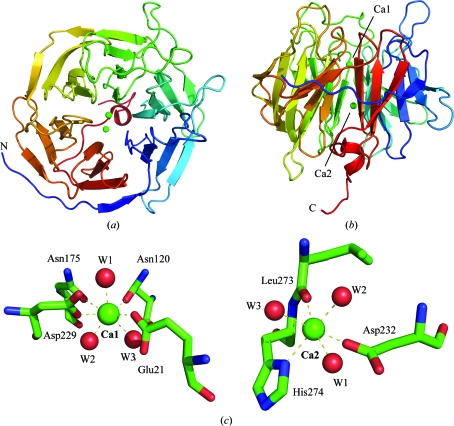
Figure 1.
Overall structure of DFPase. (a) Top view along the propeller axis. (b) Side view showing the central structural calcium ion and the catalytic calcium ion at the bottom of the binding pocket. Colouring is according to primary sequence from blue (N-terminus) to red (C-terminus). (c) The coordination environment around the catalytic calcium (Ca1) and the structural calcium (Ca2).
This study addresses the perdeuteration process by thoroughly characterizing the structural changes that occur upon substitution of D in place of H in proteins under conditions close to those used for typical neutron diffraction experiments. We compare the solvent environment around DFPase, as this is the first example of a protein for which high-resolution X-ray data for hydrogenated and perdeuterated versions crystallized under very similar conditions have been collected at RT. Thus, we are able to analyze differences in the solvent environment that result from perdeuteration, which is crucial as more proteins are perdeuterated as part of preparations for neutron diffraction. The diffraction limit of successful neutron structure determinations is analyzed by comparing the nominal resolution of the crystals using X-rays and neutrons and a set of empirical guidelines is presented.
2. Materials and methods
2.1. Expression and purification of d-DFPase
The expression and purification of hydrogenous DFPase (h-DFPase) as well as the RT X-ray crystal structure (PDB code 2gvw) have been reported previously (Blum et al., 2006 ▶). d-DFPase was expressed according to the same protocol, exchanging TB culture medium for a special perdeuterated culture medium developed at Los Alamos National Laboratory called Altone. Altone is based on a medium developed by Stone et al. (1987 ▶) but without glucose and glycerol supplements and with the addition of a number of trace elements [2%(w/v) hydrolysate of the perdeuterated algae Scenedesmus obliquus, 1.1 g ND4Cl, 100 mg MgSO4, 3.0 g KD2PO4, 3.2 g Na2DPO4, 2 mg CaCl2, 2 mg FeCl3, 2.8 mg D3BO3, 900 µg MnCl2·4D2O, 80 µg CaSO4·5D2O, 150 µg Na2MoO4·2D2O and 220 µg ZnSO4·7D2O per litre of D2O]. After cell harvest and lysis, the protein-purification process was conducted in hydrogenous buffer solutions.
2.2. MALDI–TOF MS
MALDI–TOF MS measurements were performed with an Autoflex III instrument equipped with a modified pulsed all-solid-state laser (355 nm, smartbeam technology, Bruker Daltonics, Bremen, Germany). Samples were spotted on an MTP384 polished steel target plate. External mass calibration of the instrument was carried out using the protein standards trypsinogen and protein A. The accompanying flexControl software (v.3.0, build 173) was used for controlling and flexAnalysis (v.3.0, build 96) was applied for data post-processing. Hydrogenous DFPase and its perdeuterated analogue were analyzed in the linear positive-ion mode (m/z 10 000–70 000; laser intensity 41% at 200 Hz) following the common protocol of the dried-droplet technique mixing equal volumes (1 µl) of sample and matrix solution [sinapinic acid, saturated solution in acetonitrile/0.1%(v/v) TFA, 33:66(v:v)]. Sinapinic acid and protein calibration standard II containing protein A and trypsinogen were from Bruker (Bremen, Germany).
2.3. Crystallization, data collection and refinement
Crystals of d-DFPase were prepared by vapour diffusion against hydrogenated mother liquor composed of 11% PEG 6K, MES pH 6.5. One large crystal (∼1.5 × 1.5 × 1.0 mm) was mounted in a quartz capillary and exchanged against deuterated mother liquor by vapour diffusion. The capillary was exchanged once prior to the start of screening for neutron diffraction at the Protein Crystallography Station (PCS) at LANSCE; X-ray data collection from an identical crystal was started approximately two months after initial mounting. In comparison, for the neutron structure of h-DFPase the capillary-mounted crystal was exchanged for deuterated mother liquor for less than one month before data collection and the subsequent nuclear density maps showed full exchange of solvent atoms.
X-ray diffraction data were collected to 2.1 Å resolution at RT on an in-house Rigaku rotating-anode source equipped with a Saturn 92 CCD detector using 0.5° oscillations. The data were processed using MOSFLM (Leslie, 1992 ▶) and SCALA (Evans, 2006 ▶), which are both part of the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶), and molecular replacement was run in CNS v.1.2 (Brünger et al., 1998 ▶) using the 2gvw structure with waters and ions removed. The structure was refined using a combination of positional, individual temperature-factor and simulated-annealing refinement protocols in CNS v.1.2, with model building in Coot (Emsley & Cowtan, 2004 ▶), and refined to an R free of 21.9%. Statistics are reported in Table 1 ▶. Alignment of the perdeuterated structure with 2gvw was performed using LSQMAN (Kleywegt, 1996 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the last shell.
| Source | Rotating anode, λ = 1.5418 Å |
| Resolution (Å) | 2.1 |
| Temperature | RT |
| Unit-cell parameters (Å) | a = 43.1, b = 83.2, c = 87.4 |
| Space group | P212121 |
| Rmerge | 0.088 (0.577) |
| 〈I/σ(I)〉 | 17.3 (2.9) |
| Reflections | 18983 |
| Completeness (%) | 99.8 (100.0) |
| Rfree/Rcryst | 0.218/0.176 |
| Atoms | |
| Protein | 2460 |
| Ca | 2 |
| Waters | 189 |
| 〈B〉 (Å2) | |
| Overall | 28.5 |
| Protein | 27.5 |
| Solvent | 42.2 |
| Ions | 20.0 |
| R.m.s.d. bond lengths (Å) | 0.0056 |
| R.m.s.d. bond angles (°) | 1.431 |
| Luzzati error (Å) | 0.22 |
3. Results
As d-DFPase and h-DFPase crystallized under very similar conditions with regards to pH and PEG concentration, diffracted to similar resolution at RT and were refined to similar R free values, a detailed comparative analysis of the two X-ray structures is possible. RT structures are likely to be the most relevant for neutron diffraction data collection, as nearly all neutron data sets have been collected at RT. The structures of h-DFPase and d-DFPase are compared on the basis of criteria including overall protein structure (including r.m.s.d. values and B factors), solvent structure (through an analysis of hydrogen-bond distances and properties) and metal coordination.
3.1. Structure of perdeuterated DFPase
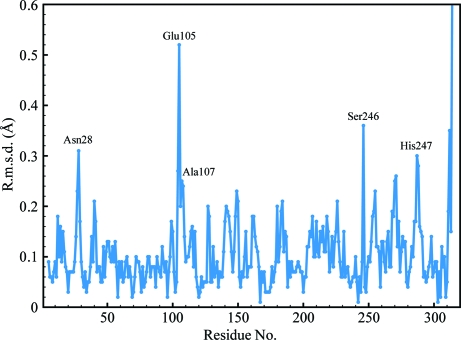
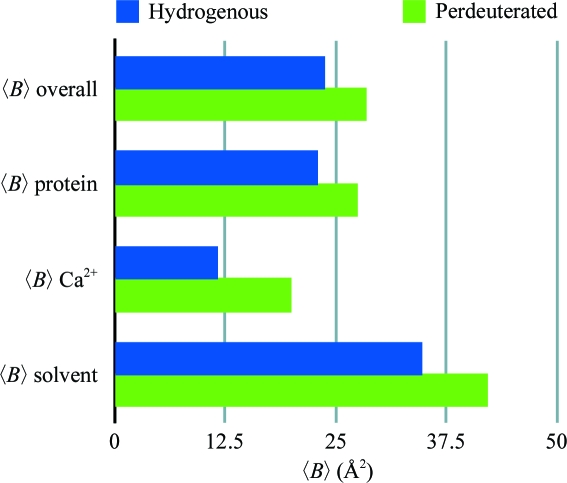
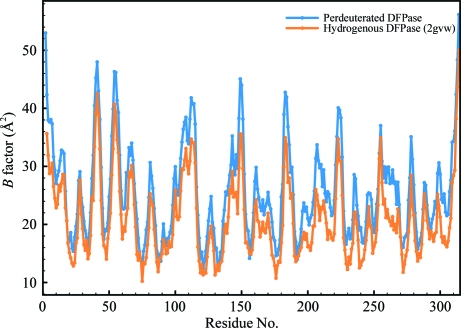
The RT structures of d-DFPase and h-DFPase (PDB code 2gvw) differed by an r.m.s.d. of 0.125 Å over the Cα atoms, which is well within the Luzzati error of 0.22 Å. The structures are therefore essentially identical. There are several outliers within the structure, including Asn28, Glu105, Ala107, Ser246 and His287, with a maximum Cα r.m.s.d. of 0.52 Å for Glu105 (Fig. 2 ▶). All of these residues are surface-exposed and visual inspection of the aligned structures of perdeuterated and hydrogenous DFPase did not reveal any significant differences. Next, we examined the temperature-factor distribution of the structure. The average temperature factor was slightly higher in d-DFPase and was around 4.7 Å2 higher than in h-DFPase (Fig. 3 ▶). The main-chain B factors of the structure presented here and of 2gvw were found to mirror each other closely, although the average B factors of d-DFPase were consistently higher than those of h-DFPase. One area, between residues 260 and 270, is worth noting: it corresponds to a β-strand involved in hydrogen bonding to the N-terminal β-strand and has somewhat higher B factors than its counterpart in the hydrogenated structure (Fig. 4 ▶).
Figure 2.
Plot of Cα r.m.s.d. values between d-DFPase and h-DFPase.
Figure 3.
Comparison of average B factors for d-DFPase and h-DFPase.
Figure 4.
Plot of B factors for Cα atoms of individual residues in perdeuterated (blue) and hydrogenous (orange) DFPase.
3.2. Calcium coordination
Two calcium ions are coordinated to DFPase: an internal structural calcium (Ca2) located in the central water tunnel of the protein and a catalytic calcium (Ca1) located at the base of the active site (Fig. 1 ▶ c). The coordination distances for h-DFPase (2gvw) and d-DFPase and also for the joint refined neutron structure (3byc) are summarized in Table 2 ▶. Ca—D2O coordination distances are generally longer than the corresponding Ca—H2O distances by an average of 0.12 Å for waters around the active-site calcium and an average of 0.04 Å for waters around the structural calcium. Calcium coordination distances for the amino acids are very slightly longer for the perdeuterated structure. Overall, the calcium coordination is biased towards longer distances for the perdeuterated structure. The replacement of H2O by D2O is not expected to affect the coordination to calcium, which is an interaction between the oxygen and the calcium, but the differences in coordination distances may reflect the slightly different electronic properties of the D2O molecule. In this structure, the interactions between water and the internal Ca are shorter, while the interactions between water and the active-site Ca are slightly longer. In a study of crystalline small-molecule hydrates, longer Ca–water coordination distances are indicative of three hydrogen-bond interactions (two donor, one acceptor) involving the waters, while shorter distances are found for waters with only two hydrogen-bond interactions (two donor) (Einspahr & Bugg, 1980 ▶).
Table 2. Calcium coordination environment.
Distances are given in Å.
| Perdeuterated | Hydrogenated (2gvw) | Neutron (3byc) | |
|---|---|---|---|
| Ca1–21 | 2.35 | 2.37 | 2.42 |
| Ca1–120 | 2.49 | 2.42 | 2.45 |
| Ca1–175 | 2.30 | 2.33 | 2.39 |
| Ca1–229 | 2.42 | 2.33 | 2.37 |
| Ca1–W1 (active site) | 2.61 | 2.48 | 2.52 |
| Ca1–W2 (tunnel 1) | 2.44 | 2.33 (W34) | 2.41 |
| Ca1–W3 (tunnel 2) | 2.63 | 2.50 (W32) | 2.45 |
| Ca2–274 | 2.51 | 2.45 | 2.43 |
| Ca2–273 | 2.35 | 2.32 | 2.23 |
| Ca2–232 | 2.23 | 2.16 | 2.16 |
| Ca2–W1 | 2.35 | 2.32 (W55) | 2.38 |
| Ca2–W2 | 2.33 | 2.30 (W15) | 2.43 |
| Ca2–W3 | 2.42 | 2.36 (W52) | 2.42 |
3.3. Solvent structure and hydrogen bonding
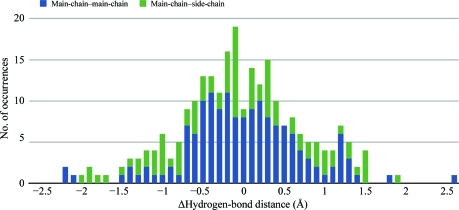
189 solvent molecules were modelled in the perdeuterated DFPase structure. From a structural alignment of h-DFPase and d-DFPase a subset of 120 waters was analyzed corresponding to pairs that were within 1.0 Å of each other and common to both structures. The difference distances between the corresponding waters were calculated and binned into a histogram. Most difference distances fall within the Luzzati error and r.m.s.d. of the two structures, indicating that perdeuteration has no systematic effect on the solvent structure. We next examined the potential hydrogen-bonding partners of these 120 water molecules and the corresponding donor–acceptor distances between the heavy atoms. A statistical distribution of these hydrogen-bond distances is shown in Fig. 5 ▶ as a histogram of the difference in distances between hydrogen-bonding pairs (perdeuterated distance − hydrogenated distance). The distribution is Gaussian around 0, indicating that there is also no systematic difference in hydrogen-bond distances between the two structures. Nevertheless, the chemical properties of D2O are different from those of H2O. The O—D bond is shorter and more stable owing to the lower zero-point energy of the O—D bond compared with the O—H bond. This is coupled with a slightly longer distance between the heavy atoms of the donor–acceptor pair. The resolution of the structure is not sufficient to ascertain these differences and can be further analyzed using higher resolution data (Efimova et al., 2007 ▶).
Figure 5.
Statistical distribution of hydrogen-bond distances shown as a histogram of the difference in distances between hydrogen-bonding pairs (perdeuterated distance − hydrogenated distance) for main-chain–main-chain and main-chain–side-chain interactions.
3.4. Neutron diffraction tests of perdeuterated DFPase
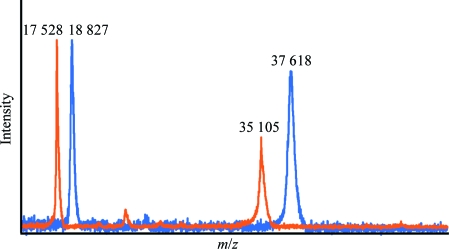
Crystals of d-DFPase were grown in hydrogenated mother liquor and back-exchanged against deuterated mother liquor prior to screening at the PCS at LANSCE. All perdeuterated crystals that were tested did not diffract neutrons beyond low resolution, despite the large sample sizes. A MALDI mass-spectrometric analysis of a crystal used in this study following neutron and X-ray data collection showed that the protein was virtually completely deuterated, with a molecular mass of 37 617.9 Da for the perdeuterated protein compared with 35 104.9 Da for the hydrogenated protein (Fig. 6 ▶). As the protein was fully perdeuterated, the lack of neutron diffraction cannot be explained by a low observed signal-to-noise ratio or by incoherent scattering by unexchanged H atoms. It should be noted that a smaller crystal (0.43 mm3) of hydrogenated DFPase was used for the neutron structure determination, so the size of the perdeuterated crystal (2.25 mm3) was more than sufficient for strong neutron diffraction.
Figure 6.
MALDI–TOF MS spectrum of hydrogenous and perdeuterated DFPase indicating full deuteration. The signals at 17 528 and 18 827 are protein molecules carrying a double charge.
4. Discussion
This study illustrates that perdeuterated protein can be routinely prepared in suitable quantities for crystallization. By solving and refining the X-ray structures of h-DFPase and d-DFPase, which were prepared and solved under strictly controlled conditions with respect to crystallization conditions, space group and unit-cell parameters, temperature of data collection, the refinement program used and the final R free and R cryst, we have been able to make a detailed comparison of the two structures. Both structures are at high resolution, although the perdeuterated protein diffracted to somewhat lower resolution than the hydrogenated protein. The major difference lies in the solvent–Ca coordination distances, which are consistently longer for the perdeuterated structure. We conclude from this comparative study that perdeuteration does not affect the protein or the solvent structure in the case of DFPase. A review of the literature shows that perdeuteration has no significant effect on overall structure in small molecules and proteins (Fisher & Helliwell, 2008 ▶), although in the case of haloalkane dehalogenase differences in the crystallization pH led to a number of conformational changes in the active site (Liu et al., 2007 ▶).
Perdeuteration for use in neutron diffraction was first described by Gamble et al. (1994 ▶) for staphylococcal nuclease; it was subsequently applied as a proof-of-principle in 2001 to re-solve myoglobin and has most recently been used for determination of the aldose reductase structure. A number of studies have described the X-ray characterization of crystals of perdeuterated protein for neutron diffraction. Unexpectedly, crystals of d-DFPase did not diffract neutrons beyond 6 Å resolution despite their large size. For screening purposes, these crystals were exposed to the neutron beam for only a few hours until it became clear that they would not diffract neutrons to sufficiently high resolution to justify full data collection. With the longer exposure times of 12–24 h typically used for full neutron data collection at the PCS this resolution could probably have been improved towards 4 Å. The aldose reductase neutron structure utilized a 0.15 mm3 crystal, compared with the 2.25 mm3 crystal used for this study. Analysis of the X-ray structure of d-DFPase showed that nearly all of the values of typical structural parameters such as B factor and solvent structure are virtually identical to those of hydrogenated protein and do not easily explain the lack of neutron diffraction. Nevertheless, several factors may have contributed to the lack of neutron diffraction. The cross-section of the neutron beam at PCS is approximately 7 mm2, compared with a 200 × 200 µm beam size for the X-ray beam. As such, the crystal may not be sufficiently macroscopically ordered to diffract neutrons well. In contrast, the scattering contributions over a smaller area, such as that of the X-ray beam, would reflect a greater amount of local order in the crystal.
As such, each perdeuterated crystallization scheme will need to be evaluated on a case-by-case basis. The neutron structure determination of DFPase was achieved by using a comparatively small hydrogenated crystal of 0.43 mm3 that was exchanged for deuterated mother liquor. As deuteration facilities are available at neutron sources equipped for macromolecular crystallography, it is advisable to prepare crystals of both the hydrogenated and perdeuterated protein when possible.
To investigate empirical parameters that may predict a successful neutron protein structure, we analyzed currently available neutron structures in the literature and the PDB and compiled a table detailing the diffraction limits and B factors of the crystals used in these structure determinations (Table 3 ▶). Compared with X-ray diffraction, neutron diffraction from macromolecules suffers from a loss of resolution, which is a consequence of the low flux and low signal-to-noise ratio. In every case, the X-ray diffraction limit of these crystals was better than 2.0 Å resolution and in about half of the cases the crystals diffracted to better than 1.0 Å resolution. The resolution of the respective neutron structure was in all cases worse than the X-ray resolution. The ratio of the resolutions of the neutron and X-ray structures at RT was calculated as an indicator of the relative loss of resolution, with an average ratio of 1.3 (neutron/X-ray). The largest observed value for this ratio is 1.8 for BPTI. The value of this ratio for h-DFPase is 1.2, which is close to the average.
Table 3. Resolution limits and temperature factors of successful neutron structures of proteins.
Neutron resolutions are the maximal resolution of refinement; X-ray resolutions are given as the refined resolution reported in the PDB or in the literature for that particular space group. When available, resolutions obtained from frozen crystals (at synchrotrons) or at RT are reported. 〈B〉 values reported are for the refined X-ray structures.
| Protein | Space group | Neutron resolution (Å) | X-ray resolution (Å) | 〈B〉 (Å2) |
|---|---|---|---|---|
| Aldose reductase | P21 | 2.2 (2r24; perdeuterated) | 0.80 (2qxw; 15 K; perdeuterated) | 9.8 |
| 0.66† (1us0) | 8.6 | |||
| 1.75 (2r24) | 17.0 | |||
| Amicyanin | P21 | 2.2 (not in PDB) | 1.31 (1aac) | 13.0 |
| 0.75† (2ov0) | 10.3 | |||
| BPTI | P212121 | 1.8 (5pti) | 1.0 (5pti) | 19.1 |
| 1.09‡ (1bpi) | 10.3 | |||
| Carbonic anhydrase | P21 | 2.4 (not in PDB) | 1.5 | |
| 1.05† (2ili) | 15.2 | |||
| γ-Chymotrypsin | P42212 | 2.0 (not in PDB) | 1.98 | |
| 1.60 (1gct) | 17.1 | |||
| Concanavalin A | I222 | 2.2 (2yz4) | 0.94† (1nls) | 15.1 |
| 2.5 (1c57) | 1.20‡ (1jbc) | 15.2 | ||
| 2.5 (1xqn; 15 K) | ||||
| Crambin | P21 | 1.25 (not in PDB) | 0.83‡ (1cbn) | 4.5 |
| 0.88 (1crn; refined to 0.945 Å) | 6.9 | |||
| 0.54† (1ejg) | 3.9 | |||
| DFPase | P212121 | 2.2 (3byc) | 1.86 (2gvw) | 23.8 |
| 0.85† (1pjx) | 12.6 | |||
| DHFR (MTX + NADPH) | P61 | 2.2 (2inq) | 1.70 (4dfr) | 33.4 |
| DsrD | P212121 | 2.4 (1wq2) | 1.20† (1ucr) | 21.0 |
| Deoxyhemoglobin | P21 | 2.1 (2dxm) | 1.74 (4hhb) | 24.8 |
| 1.25§ (2dn2) | 23.4 | |||
| Elastase (FR130180) | P212121 | 1.65 (3hgn) | 1.20§ (3hgn) | 15.5 |
| 0.94† (3hgp) | 10.3 | |||
| Endothiapepsin (gem-diol) | P21 | 2.0 (2vs2) | 1.57 (2jji) | 17.3 |
| 1.0† (2jjj) | 9.8 | |||
| Endothiapepsin (h261) | P21 | 2.1 (1gkt) | 1.10† (1oex) | 15.1 |
| HIV-1 PR (KNI-272) | P21212 | 1.9 (2yze) | 1.4 (2zye) | 22.4 |
| 0.93† (3fx5) | 17.2 | |||
| Insulin (porcine) | H3 | 2.2 (3ins) | 1.5 (3ins, 4ins) | 22.5, 30.6 |
| I213 | 2.5 (2zpp, 2efa) | 1.7 (9ins) | 28.5 | |
| Lysozyme (Gallus gallus) | P43212 | 2.0 (1io5) | 0.94† (1iee) | 14.4 |
| 1.33§ (193l) | 19.3 | |||
| P1 | 1.7 (1lzn) | 0.65† (2vb1) | 7.7 | |
| 0.95§ (4lzt) | 16.4 | |||
| Myoglobin | P21 | 1.5 (1l2k) | 1.40 (1mbd; oxy-Mb) | 21.9 |
| 2.0 (1cq2; perdeuterated) | 1.60 (1mbo; oxy-Mb) | 21.1 | ||
| 1.8 (2mb5; CO-Mb) | 1.21† (3e55, 3e5i, 3e5o; CO-Mb) | 15.4, 15.2, 15.8 | ||
| Proteinase K | P43212 | 2.3 (not in PDB) | 1.9 | |
| 0.98† (1ic6) | 10.6 | |||
| 1.5§ (2prk) | 11.2 | |||
| Photoactive yellow protein | P63 | 2.5 (2qws) | 1.25 (2zoh, 2zoi) | 17.9, 14.7 |
| 1.1 (1otb) | 21.6 | |||
| 0.82† (1nwz) | 12.8 | |||
| RNAse A | P21 | 1.7 (3a1r) | 1.05† (1kf3) | 23.8 |
| RNAse A (vanadate) | P21 | 2.0 (6rsa) | 1.3 (1ruv) | 11.9 |
| Rubredoxin (Pyrococcus furiosus) (WT) | P212121 | 1.5 (1vcx) | 0.95‡ (1brf) | 12.2 |
| Rubredoxin (P. furiosus) (mut) | P212121 | 1.6 (1iu6) | 1.5 (1iu5) | 15.9 |
| Subtilisin | P212121 | 2.0 (not in PDB) | 1.8 (2st1) | 11.1 |
| Thaumatin | P41212 | 2.0 (not in PDB) | 1.1‡ (1rqw) | 15.8 |
| Toho-1 E166A/R274N/R276N β-lactamase | P3221 | 2.1 (2wyx) | 0.94† (2zq7) | 10.5 |
| 1.8§ (1bza; E166A) | 22.0 | |||
| Trypsin (MIP) | P212121 | 1.8 (1ntp) | 1.23† (1ppz) | 17.8 |
| 1.34 (5ptp) | 16.0 | |||
| D-Xylose isomerase | I222 | 1.8 (2gve) | 1.60 (1xib) | 17.9 |
| 0.94† (2glk) | 13.5 | |||
| 0.86† (1muw) | 12.5 |
Synchrotron radiation, frozen crystal.
Rotating anode, frozen crystal.
Synchrotron radiation, RT.
In the context of the structure presented here, the X-ray diffraction limit of d-DFPase is 2.1 Å and would appear to predict suboptimal neutron diffraction to 2.7–3.8 Å (by multiplying the X-ray diffraction limit by 1.3–1.8), which is in qualitative agreement with the observed neutron diffraction. In contrast, the highest resolution neutron diffraction data were obtained from crystals of crambin, which diffracted X-rays to beyond 0.88 Å at in-house sources and diffracted neutrons to 1.2 Å resolution. An analysis of the average B factor of the X-ray structures can also be used as a measure of crystal quality irrespective of the greatly varying sizes of macromolecular crystals (Arai et al., 2004 ▶). In the case of successful neutron structures, the average B factors of the corresponding X-ray structures are generally below 30 Å2, with the exceptions being insulin and DHFR (Table 3 ▶). The average B factor of d-DFPase is 28.5 Å2 (Fig. 3 ▶), which, although high, is within the range observed for neutron crystal structures.
The following empirical guidelines for successful neutron structure determinations are suggested. In previous work we noted that crystal sizes should exceed 0.3 mm3, although potentially smaller sample sizes of ∼0.1 mm3 have been shown to be feasible using perdeuterated sample. From the analysis presented here, derived from 40 years of neutron macromolecular crystallography, the X-ray diffraction limit for neutron samples should be better than 2.0 Å and preferably better than 1.5 Å resolution. These factors appear to be strong predictors for the success of a neutron experiment and may be useful to investigators interested in utilizing the technique. Furthermore, given the significantly increased demand for neutron sources, these guidelines may be used to allocate the optimal use of beam time. These guidelines will be revised with the development of new-generation neutron sources and improved detector technology.
Supplementary Material
PDB reference: perdeuterated diisopropyl fluorophosphatase, 3kgg
Acknowledgments
We thank Mary Jo Waltman for technical assistance. This project was funded by the German Ministry of Defence under grant E/UR3G/6G115/6A801 and the Hessisches Ministerium für Wissenschaft und Kultur. ST and BLH were supported by NSF 446218. The PCS is funded by the Office of Science and the Office of Biological and Environmental Research of the US Department of Energy. PL and BPS were partly supported by an NIH–NIGMS-funded consortium (1R01GM071939-01) between Los Alamos National Laboratory and Lawrence Berkeley National Laboratory to develop computational tools for neutron protein crystallography.
References
- Adachi, M. et al. (2009). Proc. Natl Acad. Sci. USA, 106, 4641–4646.
- Adams, P. D., Mustyakimov, M., Afonine, P. V. & Langan, P. (2009). Acta Cryst. D65, 567–573. [DOI] [PMC free article] [PubMed]
- Arai, S., Chatake, T., Suzuki, N., Mizuno, H. & Niimura, N. (2004). Acta Cryst. D60, 1032–1039. [DOI] [PubMed]
- Artero, J.-B., Härtlein, M., McSweeney, S. & Timmins, P. (2005). Acta Cryst. D61, 1541–1549. [DOI] [PubMed]
- Bennett, B., Langan, P., Coates, L., Mustykimov, M., Schoenborn, B., Howell, E. E. & Dealwis, C. (2006). Proc. Natl Acad. Sci. USA, 103, 18493–18498. [DOI] [PMC free article] [PubMed]
- Blakeley, M. P., Ruiz, F., Cachau, R., Hazemann, I., Meilleur, F., Mitschler, A., Ginell, S., Afonine, P., Ventura, O. N., Cousido-Siah, A., Haertlein, M., Joachimiak, A., Myles, D. & Podjarny, A. (2008). Proc. Natl Acad. Sci. USA, 105, 1844–1848. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Koglin, A., Rüterjans, H., Schoenborn, B., Langan, P. & Chen, J. C.-H. (2007). Acta Cryst. F63, 42–45. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Loehr, F., Richardt, A., Rüterjans, H. & Chen, J. C.-H. (2006). J. Am. Chem. Soc.128, 12750–12757. [DOI] [PubMed]
- Blum, M.-M., Mustyakimov, M., Rüterjans, H., Kehe, K., Schoenborn, B. P., Langan, P. & Chen, J. C.-H. (2009). Proc. Natl Acad. Sci. USA, 106, 713–718. [DOI] [PMC free article] [PubMed]
- Blum, M.-M., Timperley, C. M., Williams, G. R., Thiermann, H. & Worek, F. (2008). Biochemistry, 47, 5216–5224. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Budayova-Spano, M., Fisher, S. Z., Dauvergne, M.-T., Agbandje-McKenna, M., Silverman, D. N., Myles, D. A. A. & McKenna, R. (2006). Acta Cryst. F62, 6–9. [DOI] [PMC free article] [PubMed]
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Coates, L., Tuan, H., Tomanicek, S., Kovalevsky, A., Mustyakimov, M., Erskine, P. & Cooper, J. (2008). J. Am. Chem. Soc.130, 7235–7237. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Efimova, Y. M., Haemers, S., Wierczinski, B., Norde, W. & van Well, A. A. (2007). Biopolymers, 85, 264–273. [DOI] [PubMed]
- Einspahr, H. & Bugg, C. E. (1980). Acta Cryst. B36, 264–271.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fisher, S. J. & Helliwell, J. R. (2008). Acta Cryst. A64, 359–367. [DOI] [PubMed]
- Gamble, T. R., Clauser, K. R. & Kossiakoff, A. A. (1994). Biophys. Chem.53, 15–25. [DOI] [PubMed]
- Hazemann, I., Dauvergne, M. T., Blakeley, M. P., Meilleur, F., Haertlein, M., Van Dorsselaer, A., Mitschler, A., Myles, D. A. A. & Podjarny, A. (2005). Acta Cryst. D61, 1413–1417. [DOI] [PubMed]
- Katz, A. K., Xinmin, L., Carrell, H. L., Hanson, B. L., Langan, P., Coates, L., Schoenborn, B. P., Glusker, J. P. & Bunick, G. J. (2006). Proc. Natl Acad. Sci. USA, 103, 8342–8347. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J. (1996). Acta Cryst. D52, 842–857. [DOI] [PubMed]
- Kossiakoff, A. A. & Spencer, S. A. (1981). Biochemistry, 20, 6462–6474. [DOI] [PubMed]
- Kovalevsky, A. Y., Katz, A. K., Carrell, H. L., Hanson, L., Mustyakimov, M., Fisher, S. Z., Coates, L., Schoenborn, B. P., Bunick, G. J., Glusker, J. P. & Langan, P. (2008). Biochemistry, 47, 7595–7597. [DOI] [PMC free article] [PubMed]
- Langan, P., Fisher, Z., Kovalevsky, A., Mustyakimov, M., Sutcliffe Valone, A., Unkefer, C., Waltman, M. J., Coates, L., Adams, P. D., Afonine, P. V., Bennett, B., Dealwis, C. & Schoenborn, B. P. (2008). J. Synchrotron Rad.15, 215–218. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Liu, X., Hanson, B. L., Langan, P. & Viola, R. E. (2007). Acta Cryst. D63, 1000–1008. [DOI] [PubMed]
- Meilleur, F., Contzen, J., Myles, D. A. A. & Jung, C. (2004). Biochemistry, 43, 8744–8753. [DOI] [PubMed]
- Meilleur, F., Dauvergne, M.-T., Schlichting, I. & Myles, D. A. A. (2005). Acta Cryst. D61, 539–544. [DOI] [PubMed]
- Niimura, N. & Bau, R. (2008). Acta Cryst. A64, 12–22. [DOI] [PubMed]
- Petit-Haertlein, I., Blakeley, M. P., Howard, E., Hazemann, I., Mitschler, A., Haertlein, M. & Podjarny, A. (2009). Acta Cryst. F65, 406–409. [DOI] [PMC free article] [PubMed]
- Scharff, E. I., Koepke, J., Fritzsch, G., Lücke, C. & Rüterjans, H. (2001). Structure, 9, 493–502. [DOI] [PubMed]
- Shu, F., Ramakrishnan, V. & Schoenborn, B. P. (2000). Proc. Natl Acad. Sci. USA, 97, 3872–3877. [DOI] [PMC free article] [PubMed]
- Stone, D. B., Curmi, P. M. & Mendelson, R. A. (1987). Methods Cell Biol.28, 215–229. [DOI] [PubMed]
- Tamada, T., Kinoshita, T., Kurihara, K., Adachi, M., Ohhara, T., Imai, K., Kuroki, R. & Tada, T. (2009). J. Am. Chem. Soc.131, 11033–11040. [DOI] [PubMed]
- Tomanicek, S. J., Blakeley, M. P., Cooper, J., Chen, Y., Afonine, P.V. & Coates, L. (2010). J. Mol. Biol.396, 1070–1080. [DOI] [PubMed]
- Tuominen, V. U., Myles, D. A. A., Dauvergne, M.-T., Lahti, R., Heikinheimo, P. & Goldman, A. (2004). Acta Cryst. D60, 606–609. [DOI] [PubMed]
- Yagi, D., Yamada, T., Kurihara, K., Ohnishi, Y., Yamashita, M., Tamada, T., Tanaka, I., Kuroki, R. & Niimura, N. (2009). Acta Cryst. D65, 892–899. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: perdeuterated diisopropyl fluorophosphatase, 3kgg
PDB reference: perdeuterated diisopropyl fluorophosphatase, 3kgg