E. coli d-alanyl-d-alanine ligase has been crystallized in complex with ADP and d-alanyl-d-alanine and in complex with ATP and d-alanyl-d-alanine.
Keywords: peptidoglycans, d-alanyl-d-alanine ligase, DdlB
Abstract
A recombinant form of Escherichia coli DdlB (EcDdlB) has been prepared and cocrystallized with ADP and d-alanyl-d-alanine to represent the ternary complex of EcDdlB. Furthermore, EcDdlB has been cocrystallized under the same conditions with the ligands ATP and d-alanyl-d-alanine, representing the product-inhibited complex. The crystals belonged to space group P212121, with unit-cell parameters a = 53.0, b = 97.6, c = 109.5 Å and a = 51.2, b = 97.8, c = 110.1 Å, respectively, and both contained two molecules in the asymmetric unit. Complete data sets were collected to 1.5 and 1.4 Å resolution, respectively, from single crystals under cryogenic conditions using synchrotron radiation.
1. Introduction
Peptidoglycan is a specific dynamic structure which constitutes a vital component of the bacterial cell wall (Vollmer et al., 2008 ▶). The biosynthesis of peptidoglycan occurs in three distinct phases at different locations and stages of the cell cycle. The initial cytoplasmic stage sequentially assembles the peptidoglycan intermediates prior to their translocation across the cell membrane. At the bacterial exoplasmic surface subsequent transpeptidation and transglycosylation reactions occur to produce mature cross-linked chains of peptidoglycan. d-Alanyl-d-alanine ligase (Ddl) is a cytoplasmic enzyme specific to peptidoglycan synthesis; it provides the d-alanyl-d-alanine dipeptide substrate for MurF in the initial cytoplasmic phase of biosynthesis (Neuhaus, 1962 ▶). The d-alanyl-d-alanine dipeptide is a central component of the biosynthetic pathway and is produced by Ddl in an ATP-dependent reaction mechanism which proceeds in two steps (Parsons et al., 1988 ▶; Mullins et al., 1990 ▶). Although d-cycloserine has long been known to inhibit Ddls and alanine racemase, problems arising from the toxicity of the drug mean that its administration is restricted (Neuhaus & Lynch, 1962 ▶) apart from the treatment of Mycobacterium tuberculosis infections. Nevertheless, Ddls present an attractive target for chemotherapeutic intervention because of their essential and universal role in bacterial cell-wall formation.
The enterococcal homologue of EcDdlB, d-alanyl-d-alanine ligase (VanA), functions in the mechanism of vancomycin resistance and also represents a novel target in the reversal of vancomycin resistance (Bugg et al., 1991 ▶). Therefore, the study of these enzymes is of prime interest in the search for new antimicrobials, particularly by rational drug-design methods. In order to elucidate the structural and chemical basis of enzyme activity, the crystal structures of Ddl from Escherichia coli (Fan et al., 1994 ▶), Staphylococcus aureus (Liu et al., 2006 ▶), Thermus caldophilus (Lee et al., 2006 ▶), Helicobacter pylori (Wu et al., 2008 ▶) and T. thermophilus HB8 (Kitamura et al., 2009 ▶) have been reported. Additionally, the crystal structures of d-alanyl-d-lactate ligases from Enterococcus faecium (Roper et al., 2000 ▶) and Leuconostoc mesenteroides (Kuzin et al., 2000 ▶) have been reported.
EcDdlB represents one of the two isoforms of Ddl that exist in E. coli and that have been kinetically characterized previously (Zawadzke et al., 1991 ▶). Currently, two wild-type structures of E. coli EcDdlB have been solved by X-ray crystallography in addition to a mutant form, Y216F EcDdlB. The first of these represented the first structure of a Ddl to be solved (Fan et al., 1994 ▶) and is in complex with ADP and a phosphophosphinate transition-state analogue. The structure identified several active-site residues and formed the basis for much of the subsequent work in the field of the Ddls (Shi & Walsh, 1995 ▶; Park et al., 1996 ▶; Lessard et al., 1999 ▶; Prevost et al., 2000 ▶). The subsequent structures of wild-type EcDdlB and Y216F EcDdlB with phosphonate and phosphinate intermediates revealed no major conformational changes in comparison to the first published structure (Fan et al., 1997 ▶): both are in complex with a transition-state intermediate and exist in closed enzyme conformations. Here, we report the crystallization and preliminary characterization of EcDdlB in complex with ATP and d-alanyl-d-alanine and in complex with ADP and d-alanyl-d-alanine. d-Alanyl-d-alanine acts as a substrate for the reverse reaction of ADP/Pi-dependent phosphorylysis, which occurs at a V max of less than 1% of the forward reaction (Mullins et al., 1990 ▶). EcDdlB has also previously been reported to display noncompetitive product inhibition (with a reported K i of 49 µM; Zawadzke et al., 1991 ▶). This suggests that the mode of binding of d-alanyl-d-alanine to the active site is different as a substrate and as an inhibitor, forming the rationale for cocrystallization studies of EcDdlB with ATP and d-alanyl-d-alanine. Furthermore, in contrast to previously reported transition-state mimic-containing EcDdlB structures, crystallographic studies of EcDdlB containing ADP and d-alanyl-d-alanine have been performed in order to determine the true ternary complex structure of EcDdlB. Further structural information regarding EcDdlB may contribute to a greater structural understanding of EcDdlB and may contribute to the design of effective ligase inhibitors.
2. Materials and methods
2.1. Cloning, overproduction and purification of E. coli EcDdlB
The gene for EcDdlB was amplified by PCR and cloned into the NcoI and XhoI sites of pProEXa (Invitrogen) to produce an amino-terminally histidine-tagged protein; the tag was not removed for crystallization. Freshly transformed E. coli BL21(λDE3)Star-pRosetta cells harbouring pProEX::EcDdlB were grown in 1 l Luria broth medium containing 50 µg ml−1 ampicillin and 30 µg ml−1 chloramphenicol. The culture was grown at 310 K to an OD600 of 0.5, at which point isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM. After 3 h of induction at 310 K, cells were harvested by centrifugation, resuspended in 30 ml buffer A (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole) and sonicated. Cellular debris was removed by centrifugation at 20 000g for 30 min and the supernatant was applied onto a 5 ml HisTrap Fast Flow column (GE Healthcare) equilibrated with buffer A. EcDdlB was eluted with a gradient of 10–250 mM imidazole over 60 ml.
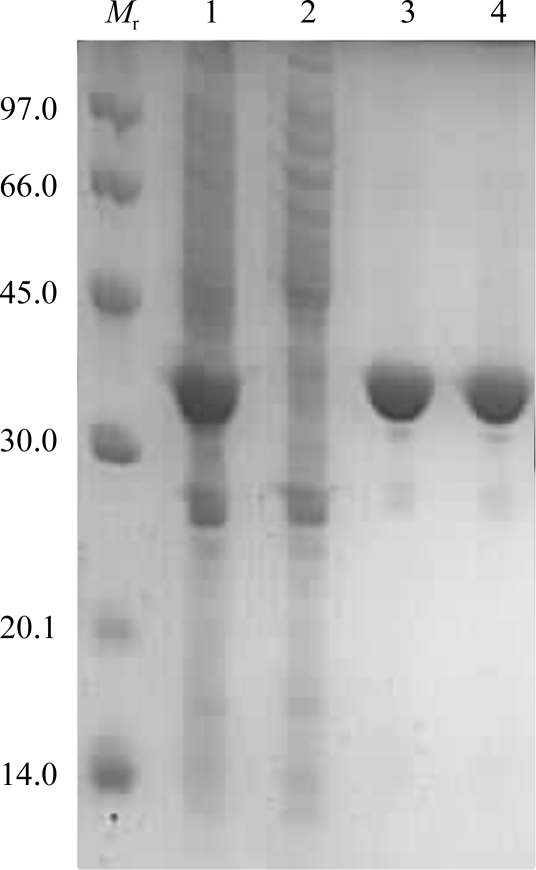
The fractions containing EcDdlB were pooled, concentrated with a Centripep-30 concentrator and applied onto a Superdex 200 column equilibrated with 150 mM KCl, 50 mM HEPES pH 7.5 and 0.5 mM EDTA. EcDdlB was eluted with approximately 180 ml buffer. The fractions containing recombinant EcDdlB were analysed by SDS–PAGE (Fig. 1 ▶), pooled and concentrated to 12 mg ml−1 using a Centriprep-30 concentrator. Enzyme activity was determined using a continuous spectrometric assay monitoring ADP release as described previously (Daub et al., 1988 ▶).
Figure 1.
12% SDS–PAGE analysis of the purification of EcDdlB. Lane M r, molecular-weight standards (kDa); lane 1, crude cell lysate; lane 2, flowthrough from immobilized metal-affinity chromatography; lane 3, peak from immobilized metal-affinity chromatography; lane 4, peak from size-exclusion chromatography.
2.2. Crystallization
All crystallization experiments were performed using vapour diffusion. Initial crystallization was performed using the sitting-drop vapour-diffusion method. The protein concentration used for crystallization was 12 mg ml−1 in 5 mM ATP or 5 mM ADP and 50 mM d-alanyl-d-alanine and a ratio of one volume of protein solution to one volume of precipitant solution was used throughout. Crystals were obtained in conditions from a commercially available crystal screen (Molecular Dimensions). Refinement of the Tris–HCl conditions from pH 8.5 to pH 8 using the hanging-drop vapour-diffusion method gave crystals of sufficient quality for data collection.
2.3. Data collection
Single crystals of the EcDdlB complexes were flash-cooled in liquid nitrogen using reservoir solution containing 30% glycerol as a cryoprotectant. X-ray diffraction data were collected on beamline I04 at Diamond (UK). Diffraction images were measured using an ADSC Quantum 315 CCD detector.
3. Results and discussion
The E. coli EcDdlB protein was purified by immobilized metal-affinity chromatography and size-exclusion chromatography to yield 10 mg purified protein from 1 l culture. SDS–PAGE analysis revealed that the preparation was free of major and minor contaminating species. The subunit molecular mass on SDS–PAGE appeared to be consistent with the calculated molecular mass of 34 533 Da. The catalytic efficiency for d-alanyl-d-alanine synthesis was found to be 477.4 ± 0.7 min−1 mM −1, a value that is comparable to that obtained in a previous kinetic analysis of EcDdlB (440 min−1 mM −1) in a non-histidine-tagged form (Park et al., 1996 ▶).
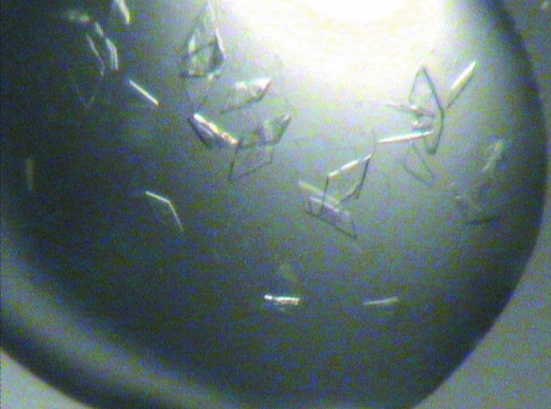
The best crystals were obtained at 291 K using the hanging-drop method by mixing 1 µl protein solution containing 5 mM ATP or 50 mM ADP and 50 mM d-alanyl-d-alanine with 1 µl crystallization solution containing 0.2 M MgCl2, 25% PEG 3350, 0.1 M Tris–HCl pH 8.0 and equilibrating against a 500 µl reservoir. Crystals appeared after 2 d as plates of dimensions of 0.1 × 0.35 mm (Figs. 2 ▶ and 3 ▶). We are confident these crystals contained the ligands that were cocrystallized since EcDdlB did not crystallize under the same conditions in the apo form or with ATP, ADP or d-alanyl-d-alanine alone.
Figure 2.
Photograph of EcDdlB crystals obtained in the presence of ATP and d-alanyl-d-alanine grown in 0.2 M MgCl2, 25% PEG 3350, 0.1 M Tris–HCl pH 8.0. The approximate dimensions of the individual crystals are 0.1 × 0.35 × 0.02 mm.
Figure 3.
Photograph of EcDdlB crystals obtained in the presence of ADP and d-alanyl-d-alanine grown in 0.2 M MgCl2, 25% PEG 3350, 0.1 M Tris–HCl pH 8.0. The approximate dimensions of the individual crystals are 0.1 × 0.4 × 0.02 mm.
Initial diffraction experiments were carried out at the ESRF, but complete data sets were collected to resolutions of 1.4 Å for EcDdlB–ATP–d-alanyl-d-alanine and 1.5 Å for EcDdlB–ADP–d-alanyl-d-alanine on the I04 beamline at the Diamond synchrotron (UK) using an ADSC Q315 CCD detector with an oscillation angle of 1° and a crystal-to-detector distance of 160 mm. All data were indexed, integrated and scaled using the HKL suite of programs (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are shown in Table 1 ▶. For each data set, Matthews probability calculations suggested the presence of two molecules in the asymmetric unit, although a self-rotation function did not reveal the presence of a noncrystallographic rotational axis (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). A native Patterson map suggested the presence of pseudo-translation, with a high peak in the (u, v, ½) Harker section (94σ and 31σ, corresponding to 40% and 13% of the origin-peak height, for the two data sets, respectively). Molecular-replacement solution using Phaser (McCoy et al., 2007 ▶) with the coordinates of the Y216F EcDdlB mutant (PDB code 1i0w; Fan et al., 1997 ▶) as a model subsequently revealed two molecules in the crystallographic asymmetric unit which are related to each other by a twofold rotation parallel to the z crystallographic axis for both data sets.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| EcDdlB–ATP–D-alanyl-D-alanine | EcDdlB–ADP–D-alanyl-D-alanine | |
|---|---|---|
| Beamline | I04, Diamond | I04, Diamond |
| Detector | ADSC Q315 CCD | ADSC Q315 CCD |
| Wavelength (Å) | 0.9763 | 0.9763 |
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 51.23 | 53.00 |
| b (Å) | 97.80 | 97.62 |
| c (Å) | 110.12 | 109.49 |
| Molecules per ASU | 2 | 2 |
| Matthews coefficient (Å3 Da−1) | 2.09 | 2.14 |
| Solvent convent (%) | 41 | 43 |
| Resolution range (Å) | 49–1.4 (1.45–1.4) | 48–1.5 (1.55–1.5) |
| Total observations | 648917 | 601481 |
| Unique reflections | 106197 | 91443 |
| Average I/σ(I) | 22.4 (2.1) | 18.9 (2.1) |
| Rmerge† (%) | 0.069 (0.577) | 0.093 (0.578) |
| Completeness (%) | 97.1 (94.6) | 99.9 (99.1) |
R
merge = 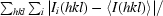
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the mean intensity of that reflection
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the mean intensity of that reflection
Models of the EcDdlB–ATP–d-alanyl-d-alanine complex and the EcDdlB–ADP-d-alanyl-d-alanine complex are currently being built and refined. We are also seeking to crystallize the protein in the apo form and in complex with inhibitors. Ultimately, these studies may contribute to further understanding of the individual reaction steps of the mechanism, the activity-associated conformational changes and the rational design of specific inhibitors against Ddls.
Acknowledgments
This work was supported by a University of Warwick PhD studentship to SB and in part by MRC research grants G500643 and G0600801 and Wellcome Trust equipment grants 071998 and 068598. Equipment used in this research was obtained through the Birmingham–Warwick Science City Translational Medicine: Experimental Medicine Network of Excellence project, with support from Advantage West Midlands (AWM). Crystallographic data were collected at beamlines IO4 at Diamond Light Source, UK and at ID14.1 at the ESRF, France, and we acknowledge the support of beamline scientists Ralf Flaig (Diamond) and Matthew Bowler (ESRF).
References
- Bugg, T. D., Dutka-Malen, S., Arthur, M., Courvalin, P. & Walsh, C. T. (1991). Biochemistry, 30, 2017–2021. [DOI] [PubMed]
- Daub, E., Zawadzke, L. E., Botstein, D. & Walsh, C. T. (1988). Biochemistry, 27, 3701–3708. [DOI] [PubMed]
- Fan, C., Moews, P. C., Walsh, C. T. & Knox, J. R. (1994). Science, 266, 439–443. [DOI] [PubMed]
- Fan, C., Park, I. S., Walsh, C. T. & Knox, J. R. (1997). Biochemistry, 36, 2531–2538. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Kitamura, Y., Ebihara, A., Agari, Y., Shinkai, A., Hirotsu, K. & Kuramitsu, S. (2009). Acta Cryst. D65, 1098–1106. [DOI] [PMC free article] [PubMed]
- Kuzin, A. P., Sun, T., Jorczak-Baillass, J., Healy, V. L., Walsh, C. T. & Knox, J. R. (2000). Structure, 8, 463–470. [DOI] [PubMed]
- Lee, J. H., Na, Y., Song, H. E., Kim, D., Park, B. H., Rho, S. H., Im, Y. J., Kim, M. K., Kang, G. B., Lee, D. S. & Eom, S. H. (2006). Proteins, 64, 1078–1082. [DOI] [PubMed]
- Lessard, I. A., Healy, V. L., Park, I. S. & Walsh, C. T. (1999). Biochemistry, 38, 14006–14022. [DOI] [PubMed]
- Liu, S., Chang, J. S., Herberg, J. T., Horng, M. M., Tomich, P. K., Lin, A. H. & Marotti, K. R. (2006). Proc. Natl Acad. Sci. USA, 103, 15178–15183. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Mullins, L. S., Zawadzke, L. E., Walsh, C. T. & Raushel, F. M. (1990). J. Biol. Chem.265, 8993–8998. [PubMed]
- Neuhaus, F. C. (1962). J. Biol. Chem.237, 778–786. [PubMed]
- Neuhaus, F. C. & Lynch, J. L. (1962). Biochem. Biophys. Res. Commun.8, 377–382. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Park, I. S., Lin, C. H. & Walsh, C. T. (1996). Biochemistry, 35, 10464–10471. [DOI] [PubMed]
- Parsons, W. H., Patchett, A. A., Bull, H. G., Schoen, W. R., Taub, D., Davidson, J., Combs, P. L., Springer, J. P., Gadebusch, H., Weissberger, B., Valiant, M. E., Mellin, T. N. & Busch, R. D. (1988). J. Med. Chem.31, 1772–1778. [DOI] [PubMed]
- Prevost, M., Van Belle, D., Tulkens, P. M., Courvalin, P. & Van Bambeke, F. (2000). J. Mol. Microbiol. Biotechnol.2, 321–330. [PubMed]
- Roper, D. I., Huyton, T., Vagin, A. & Dodson, G. (2000). Proc. Natl Acad. Sci. USA, 97, 8921–8925. [DOI] [PMC free article] [PubMed]
- Shi, Y. & Walsh, C. T. (1995). Biochemistry, 34, 2768–2776. [DOI] [PubMed]
- Vollmer, W., Blanot, D. & de Pedro, M. A. (2008). FEMS Microbiol. Rev.32, 149–167. [DOI] [PubMed]
- Wu, D., Zhang, L., Kong, Y., Du, J., Chen, S., Chen, J., Ding, J., Jiang, H. & Shen, X. (2008). Proteins, 72, 1148–1160. [DOI] [PubMed]
- Zawadzke, L. E., Bugg, T. D. & Walsh, C. T. (1991). Biochemistry, 30, 1673–1682. [DOI] [PubMed]