Very-long-chain acyl-CoA dehydrogenase from Caenorhabditis elegans (cVLCAD) has been crystallized in space group C2 and its X-ray diffraction data set has been collected to 1.6 Å resolution. Unlike other VLCADs that were reported to form dimers, the purified cVLCAD was found as a homotetrameric protein according to static light-scattering measurements.
Keywords: very-long-chain acyl-CoA dehydrogenases, fatty-acid β-oxidation, Caenorhabditis elegans, SEC-LS/UV/RI
Abstract
Acyl-CoA dehydrogenase [acyl-CoA:(acceptor) 2,3-oxidoreductase; EC 1.3.99.3] catalyzes the first reaction step in mitochondrial fatty-acid β-oxidation. Here, the very-long-chain acyl-CoA dehydrogenase from Caenorhabditis elegans (cVLCAD) has been cloned and overexpressed in Escherichia coli strain BL21 (DE3). Interestingly, unlike other very-long-chain acyl-CoA dehydrogenases, cVLCAD was found to form a tetramer by size-exclusion chromatography coupled with in-line static light-scattering, refractive-index and ultraviolet measurements. Purified cVLCAD (12 mg ml−1) was successfully crystallized by the hanging-drop vapour-diffusion method under conditions containing 100 mM Tris–HCl pH 8.0, 150 mM sodium chloride, 200 mM magnesium formate and 13% PEG 3350. The crystal has a tetragonal form and a complete diffraction data set was collected and processed to 1.8 Å resolution. The crystal belonged to space group C2, with unit-cell parameters a = 138.6, b = 116.7, c = 115.3 Å, α = γ = 90.0, β = 124.0°. A self-rotation function indicated the existence of one noncrystallographic twofold axis. A preliminary molecular-replacement solution further confirmed the presence of two molecules in one asymmetric unit, which yields a Matthews coefficient V M of 2.76 Å3 Da−1 and a solvent content of 55%.
1. Introduction
The acyl-CoA dehydrogenases (ACADs) constitute a family of FAD-dependent flavoproteins that catalyze the α,β-dehydrogenation of thioester substrates (Beinert, 1963 ▶). There are nine members of this family, five of which are involved in the mitochondrial fatty-acid β-oxidation process: short-chain, medium-chain, long-chain and very-long-chain acyl-CoA dehydrogenases (SCAD, MCAD, LCAD and VLCAD-1, respectively) and the recently reported very-long-chain acyl-CoA dehydrogenase ACAD-9 (VLCAD-2) (Zhang et al., 2002 ▶; Ensenauer et al., 2005 ▶; Kim & Miura, 2004 ▶). The other four members are involved in the amino-acid oxidation pathway and comprise iso(3)valeryl-CoA dehydrogenase (I3VD) for leucine, iso(2)valeryl-CoA dehydrogenase (I2VD) for isoleucine, isobutyryl-CoA dehydrogenase (IBD) for valine and glutaryl-CoA dehydrogenase (GD) for lysine and tryptophan (Kim & Miura, 2004 ▶).
As enzymes involved in the initial step of the mitochondrial β-oxidation spiral, ACADs catalyze the dehydrogenation of acyl-CoAs with the formation of a trans double bond between the Cα and Cβ atoms (Thorpe & Kim, 1995 ▶). ACADs have distinct substrate specificities: SCAD, MCAD, LCAD and VLCAD have optimal activity towards acyl-CoAs with four-carbon, eight-carbon, 14-carbon and 16-carbon chain lengths, respectively (Ikeda et al., 1985 ▶; Izai et al., 1992 ▶). However, they exhibit similar catalytic mechanisms that involve base-catalyzed α-proton abstraction from the substrate followed by the transfer of the β-hydrogen as a hydride to the N-5 position of the isoalloxazine ring of the enzyme-bound FAD (Engel, 1992 ▶; Ghisla et al., 1984 ▶, 1994 ▶; Thorpe & Kim, 1995 ▶). Finally, the electrons are transferred to the mitochondrial respiratory chain via a sequential electron-transfer flavoprotein (ETF) that is the physiological electron acceptor of ACADs (Crane & Beinert, 1956 ▶) and then to a membrane-bound ETF-ubiquinone oxidoreductase (Ruzicka & Beinert, 1977 ▶).
Very-long-chain acyl-CoA dehydrogenase (VLCAD) is a special member of the ACAD family. Unlike the other ACADs, which are soluble homotetramers with ∼45 Da subunits, VLCADs have been found to be homodimers with ∼45 kDa subunits and are associated with the inner mitochondrial membrane (Souri et al., 1998 ▶). To date, a number of ACAD structures have been solved, including those of MCAD (Kim et al., 1993 ▶; Lee et al., 1996 ▶; Satoh et al., 2003 ▶), SCAD (Battaile et al., 2002 ▶), I3VD (Tiffany et al., 1997 ▶) and GD (Fu et al., 2004 ▶). All of these soluble tetrameric ACADs exhibit ‘dimer-of-dimer’ structures and all their subunits possess a similar fold consisting of an N-terminal α-helical domain, a middle β-sheet domain and a C-terminal α-helical domain. The crystal structure of human VLCAD (hVLCAD) complexed with myristoyl-CoA was first determined at 1.91 Å resolution and the overall fold of its N-terminal ∼400 residues is similar to those of other soluble ACADs; its additional C-terminal 180 residues are proposed to be involved in binding to the membrane (McAndrew et al., 2008 ▶).
ACDH-11 (WormBase Protein ID WP:CE19166) is a VLCAD from Caenorhabditis elegans that shares 26% sequence identity with hVLCAD. Although C. elegans is a good animal model for studying fatty-acid metabolism regulation, which is important for obesity therapy, its β-oxidation system has not been well characterized to date. As a project on C. elegans mitochondrial β-oxidation regulation, cVLCAD was cloned from the cDNA library of C. elegans and expressed in Escherichia coli for biochemical and structural studies. The purified cVLCAD was found to form a tetramer according to size-exclusion chromatography (SEC) experiments, unlike hVLCAD and rat VLCAD (Izai et al., 1992 ▶), which have been observed to be dimers. The cVLCAD protein was successfully crystallized and its crystal structure was solved by molecular replacement. The crystal structure of cVLCAD confirms its tetrameric assembly mode and reveals a novel substrate-binding pocket compared with hVLCAD (to be published elsewhere). Here, we report the cloning, expression and preliminary crystallographic analysis of cVLCAD.
2. Materials and methods
2.1. Molecular cloning, protein expression and purification
The coding sequence of cVLCAD was amplified from the cDNA library of C. elegans using the forward primer 5′-GGTATACCATATGCATCGAATTGGCAACGCTGTCAG-3′ and the reverse primer 5′-GGAATCTCGAGGAAATTTTGCTCTTCTCAAGCGCAG-3′. The amplified product was inserted into the vector pEXS-DH, which contains coding sequences for 8×His tags at both the 5′ and 3′ ends of its multiple cloning site and which was developed in our laboratory on the basis of the pET-22b(+) vector (Novagen). The resulting plasmid encodes the recombinant fusion cVLCAD protein with a C-terminal 8×His tag. The recombinant plasmid was sequenced to verify the accuracy of the PCR amplification and was transformed into E. coli BL21 (DE3) cells.
Cells were grown in LB medium with 100 µg ml−1 ampicillin and were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at an OD600nm of 0.7. After 8 h induction at 289 K, the cells were harvested and resuspended in buffer A (50 mM Tris–HCl pH 8.0, 300 mM sodium chloride, 10% glycerol) and lysed by sonication. The lysate was centrifuged at 27 000g for 30 min at 277 K and the cell debris was discarded. The supernatant was loaded onto an Ni Sepharose 6 Fast Flow column (GE Healthcare) equilibrated with buffer A and the column was washed with 20 mM imidazole. The target protein was eluted using 200 mM imidazole and concentrated by ultrafiltration. After desalting, the protein was loaded onto a RESOURCE S column (GE Healthcare) pre-equilibrated with buffer B (20 mM MES pH 6.5, 10% glycerol). The target protein was eluted with a linear gradient of sodium chloride from 0 to 500 mM in buffer B. The yellow cVLCAD target protein was collected, pooled, concentrated and further purified by gel filtration using a Superdex 200 100/300 GL column (GE Healthcare) with buffer C (20 mM Tris–HCl pH 8.0, 150 mM sodium chloride). The cVLCAD fractions were pooled and concentrated to 42 mg ml−1.
2.2. Size-exclusion chromatography coupled with in-line static light-scattering, refractive-index and ultraviolet measurements (SEC-LS/UV/RI)
The instrumental setup used for the SEC-LS/UV/RI experiment consisted of an Agilent 1100 HPLC system (Agilent Technologies) connected in series to a DAWN HELEOS II light-scattering detector (Wyatt Technology) and an Optilab rEX interferometric refractometer detector (Wyatt Technology). Analytical size-exclusion chromatography was performed at 289 K using a Shodex Protein 803 KW column (Shoko America) equilibrated with a mobile phase consisting of 20 mM Tris–HCl pH 8.0, 150 mM sodium chloride. 50 µl purified cVLCAD sample at 5.0 mg ml−1 was injected onto the column and eluted at a flow rate of 0.5 ml min−1. The column effluent was monitored in-line with three detectors that simultaneously monitored ultraviolet absorption (UV), light scattering (LS) and refractive index (RI), respectively. The three resulting chromatograms were aligned with that of the LS output after corrections for the inter-detector volume delays between the UV and LS detectors and between the RI and LS detectors. Detector outputs were digitized and acquired by a computer running the ASTRA V software (Wyatt Technology). The oligomeric molecular weight of cVLCAD was calculated using the parameters from the LS and RI detectors using the ASTRA software (Wyatt Technology).
2.3. Crystallization and optimization
Initial screening was performed by the hanging-drop vapour-diffusion method using Index, Crystal Screen I and Crystal Screen II kits (Hampton Research) at 289 K. 2 µl protein solution (7 mg ml−1) buffered in 20 mM Tris–HCl pH 8.0 and 150 mM sodium chloride was mixed with an equal volume of reservoir solution and equilibrated against 200 ml reservoir solution. After 3 d, crystal-like spherical species appeared from condition No. 92 of the Index kit (100 mM magnesium formate, 15% PEG 3350). After optimizing these crystallization conditions by adding 100 mM Tris–HCl pH 8.0, yellow rhombic crystals appeared. Further optimizations were performed by varying the concentrations of PEG 3350, protein and magnesium formate. Good large single crystals were obtained using the conditions 12 mg ml−1 protein (buffered in 20 mM Tris–HCl pH 8.0, 150 mM sodium chloride), 100 mM Tris–HCl pH 8.0, 200 mM magnesium formate and 13% PEG 3350. Most large crystals diffracted to 2.6 Å resolution in-house.
2.4. Data collection and processing
Immediately prior to data collection, the cVLCAD crystal was quickly soaked in cryoprotectant solution (13% PEG 3350 and 20% glycerol) and flash-cooled at 100 K in a stream of nitrogen gas. An initial diffraction data set was collected to 2.6 Å resolution in-house using an FR-E SuperBright copper rotating-anode X-ray generator (Rigaku, Japan) operating at 45 kV and 45 mA and equipped with a 300 mm R-AXIS IV++ image-plate detector (Rigaku, Japan). The high-resolution diffraction data set was collected on beamline BL5A of the Photon Factory (KEK, Japan) using radiation of wavelength 1.0 Å. A Quantum 315 CCD detector with an active area of 315 × 315 mm was used and the crystal-to-detector distance was set to reach a corner resolution of 1.42 Å and an edge resolution of 1.6 Å. All diffraction images were processed using the HKL-2000 package (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C121 |
| Unit-cell parameters (Å, °) | a = 138.6, b = 116.7, c = 115.3, α = γ = 90.0, β = 124.0 |
| Wavelength (Å) | 1.00000 |
| Resolution range (Å) | 50–1.80 (1.83–1.80) |
| Completeness (%) | 97.5 (92.4) |
| Redundancy | 5.2 (3.0) |
| Rmerge† (%) | 4.9 (29.7) |
| Average I/σ(I) | 33.1 (2.2) |
| Total reflections | 705157 |
| Unique reflections | 136815 |
| Molecules per asymmetric unit | 2 |
R
merge = 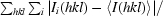
 , where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of reflections.
, where Ii(hkl) is the intensity of the ith observation and 〈I(hkl)〉 is the mean intensity of reflections.
2.5. Self-rotation function calculation
A self-rotation function was calculated by MOLREP (Vagin & Teplyakov, 1997 ▶) in the resolution range 50.0–6 Å; the integration radius was set to 60 Å.
3. Results and discussion
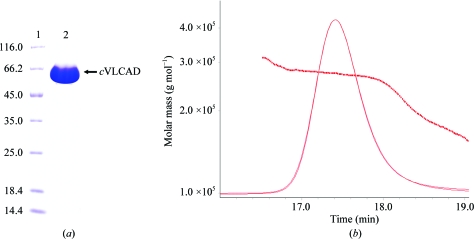
After affinity, ion-exchange and gel-filtration chromatography, the recombinant fusion protein cVLCAD was purified intensively to high purity (∼99%). Its apparent molecular weight was about 66 kDa (Fig. 1 ▶ a); the theoretical molecular weight of cVLCAD fused with a C-terminal 8×His tag is 70 kDa. However, cVLCAD was found to form oligomers in solution according to SEC-LS/UV/RI experiments and the molecular weight of the cVLCAD oligomer was determined to be ∼264 kDa (Fig. 1 ▶ b). The results suggest that cVLCAD exists as a tetramer in solution, which differs from the commonly held view that VLCAD assembles into a dimer.
Figure 1.
(a) SDS–PAGE of purified cVLCAD. Lane 1, protein molecular-weight markers (Fermentas; labelled in kDa); lane 2, purified cVLCAD. SDS–PAGE was performed on a 12%(w/v) gel; the gel was stained using Coomassie Brilliant Blue. (b) The profile of the SEC-LS/UV/RI experiment. The parameters monitored by the light-scattering (LS) and refractive-index (RI) detectors are plotted as curves and the molecular weights calculated according to LS and RI measurements at each time point are plotted as dots.
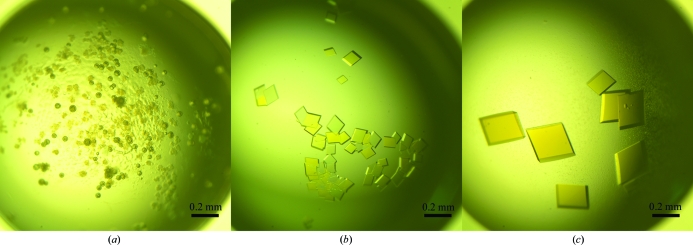
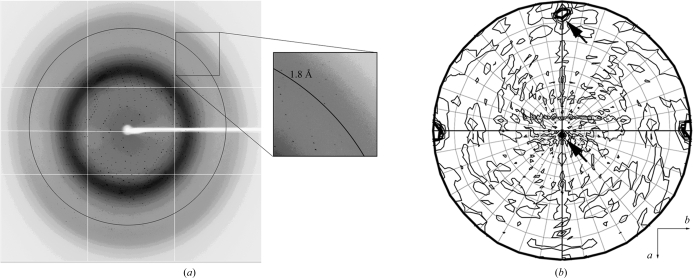
The purified cVLCAD protein was crystallized successfully to give crystals with optimal dimensions of ∼0.3 mm (Fig. 2 ▶). Good single crystals diffracted to better than 1.8 Å resolution (Fig. 3 ▶ a) on beamline BL5A at Photon Factory and a diffraction data set was collected. Because the average I/σ(I) was less than 2.0 when the resolution was higher than 1.8 Å, the diffraction data were finally processed to 1.8 Å resolution. The crystal belonged to space group C2, with unit-cell parameters a = 138.6, b = 116.7, c = 115.3 Å, α = γ = 90.0, β = 124.0° (Table 1 ▶). A self-rotation function indicated the existence of noncrystallographic twofold symmetry (Fig. 3 ▶ b), suggesting that there are two identical copies in the asymmetric unit. Furthermore, assuming the presence of two cVLCAD molecules in the asymmetric unit yields a rational Matthews coefficient of 2.76 Å3 Da−1 (Matthews, 1968 ▶), with a solvent content of 55%. These data indicate that there are most likely to be two molecules in the asymmetric unit.
Figure 2.
(a) Crystals of cVLCAD grown under condition No. 92 of the Index kit (100 mM magnesium formate, 15% PEG 3350). 5 mg ml−1 protein buffered in 20 mM Tris–HCl pH 8.0, 150 mM sodium chloride was used. (b) Crystals of cVLCAD grown using 10 mg ml−1 protein solution (buffered in 20 mM Tris–HCl pH 8.0, 150 mM sodium chloride), 100 mM Tris–HCl pH 8.0, 100 mM magnesium formate, 15% PEG 3350. (c) Crystals of cVLCAD grown using 12 mg ml−1 protein solution (buffered in 20 mM Tris–HCl pH 8.0, 150 mM sodium chloride), 100 mM Tris–HCl pH 8.0, 200 mM magnesium formate, 13% PEG 3350. cVLCAD has a yellow colour.
Figure 3.
(a) Diffraction pattern of a cVLCAD crystal on a Quantum 315 CCD detector. The crystal diffracted to better than 1.8 Å resolution. The black ring indicates 1.8 Å resolution. (b) A self-rotation function map of cVLCAD diffraction data plotted at χ = 180°. Latitude (θ angle) and longitude (ϕ angle) grid lines are drawn at 10° intervals. The related noncrystallographic twofold axis is indicated by an arrow.
Molecular replacement was used to determine the crystal structure of cVLCAD using the structure of E. coli AidB (PDB code 3djl; Bowles et al., 2008 ▶), which has 30% sequence identity to cVLCAD, as a search model. One unique solution with two monomers was obtained with a likelihood score of 310.8 and a Z score of 16.7 using the program Phaser (McCoy et al., 2007 ▶; Collaborative Computational Project, 1994 ▶). The two monomers contact each other closely and form a dimer in the asymmetric unit, which is correlated by the noncrystallographic twofold axis that was detected in the self-rotation function (Fig. 3 ▶ b). Crystal-packing analysis further indicated that the two dimers in two neighbouring asymmetric units that are related by the twofold crystallographic axis could form a compact tetramer with a large interface. This observation is consistent with the SEC-LS/UV/RI experiments, which showed that cVLCAD is a homotetrameric protein.
The structure determination and analysis of cVLCAD will be published elsewhere. With a tetrameric assembly mode that differs from the common dimeric mode of other VLCADs, the high-resolution structure of cVLCAD should provide new information on the structure and function of the ACAD family.
Acknowledgments
We thank Professor Weimin Gong for the gift of the C. elegans cDNA library and we also appreciate the help of Kai Zhang (from the group of FS) with the self-rotation function calculation. This work was supported by the National Natural Science Foundation of China (No. 30721003) and the ‘973’ program of the Chinese Ministry of Science and Technology (Nos. 2006CB911001 and 2006CB806506).
References
- Battaile, K. P., Molin-Case, J., Paschke, R., Wang, M., Bennett, D., Vockley, J. & Kim, J. J. (2002). J. Biol. Chem.277, 12200–12207. [DOI] [PubMed]
- Beinert, H. (1963). The Enzymes, 2nd ed., edited by P. D. Boyer, H. Lardy & K. Myrback, Vol. 7, pp. 447–466. New York: Academic Press.
- Bowles, T., Metz, A. H., O’Quin, J., Wawrzak, Z. & Eichman, B. F. (2008). Proc. Natl Acad. Sci. USA, 105, 15299–15304. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Crane, F. L. & Beinert, H. (1956). J. Biol. Chem.218, 717–731. [PubMed]
- Engel, P. C. (1992). Chemistry and Biochemistry of Flavoenzymes, edited by F. Müller, pp. 597–655. Boca Raton: CRC Press.
- Ensenauer, R., He, M., Willard, J. M., Goetzman, E. S., Corydon, T. J., Vandahl, B. B., Mohsen, A. W., Isaya, G. & Vockley, J. (2005). J. Biol. Chem.280, 32309–32316. [DOI] [PubMed]
- Fu, Z., Wang, M., Paschke, R., Rao, K. S., Frerman, F. E. & Kim, J. J. (2004). Biochemistry, 43, 9674–9684. [DOI] [PubMed]
- Ghisla, S., Engst, S., Vock, P., Kieweg, V., Bross, P., Nandy, A., Rasched, I. & Strauss, A. W. (1994). Flavins and Flavoproteins, edited by K. Yagi, pp. 283–292. University of Tokyo Press.
- Ghisla, S., Thorpe, C. & Massey, V. (1984). Biochemistry, 23, 3154–3161. [DOI] [PubMed]
- Ikeda, Y., Okamura-Ikeda, K. & Tanaka, K. (1985). J. Biol. Chem.260, 1311–1325. [PubMed]
- Izai, K., Uchida, Y., Orii, T., Yamamoto, S. & Hashimoto, T. (1992). J. Biol. Chem.267, 1027–1033. [PubMed]
- Kim, J. J. & Miura, R. (2004). Eur. J. Biochem.271, 483–493. [DOI] [PubMed]
- Kim, J. J., Wang, M. & Paschke, R. (1993). Proc. Natl Acad. Sci. USA, 90, 7523–7527. [DOI] [PMC free article] [PubMed]
- Lee, H. J., Wang, M., Paschke, R., Nandy, A., Ghisla, S. & Kim, J. J. (1996). Biochemistry, 35, 12412–12420. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McAndrew, R. P., Wang, Y., Mohsen, A. W., He, M., Vockley, J. & Kim, J. J. (2008). J. Biol. Chem.283, 9435–9443. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ruzicka, F. J. & Beinert, H. (1977). J. Biol. Chem.252, 8440–8445. [PubMed]
- Satoh, A., Nakajima, Y., Miyahara, I., Hirotsu, K., Tanaka, T., Nishina, Y., Shiga, K., Tamaoki, H., Setoyama, C. & Miura, R. (2003). J. Biochem.134, 297–304. [DOI] [PubMed]
- Souri, M., Aoyama, T., Hoganson, G. & Hashimoto, T. (1998). FEBS Lett.426, 187–190. [DOI] [PubMed]
- Thorpe, C. & Kim, J. J. (1995). FASEB J.9, 718–725. [DOI] [PubMed]
- Tiffany, K. A., Roberts, D. L., Wang, M., Paschke, R., Mohsen, A. W., Vockley, J. & Kim, J. J. (1997). Biochemistry, 36, 8455–8464. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Zhang, J., Zhang, W., Zou, D., Chen, G., Wan, T., Zhang, M. & Cao, X. (2002). Biochem. Biophys. Res. Commun.297, 1033–1042. [DOI] [PubMed]