The inverting trehalose phosphorylase from Thermoanaerobacter sp. was crystallized and its structure was determined by molecular replacement. Its purification, crystallization and preliminary X-ray diffraction analysis are presented.
Keywords: trehalose phosphorylase, GH65, disaccharide phosphorylases, inverting, crystal dehydration
Abstract
Disaccharide phosphorylases are attractive enzymatic platforms for tailor-made sugar synthesis owing to their ability to catalyze both the synthesis and the breakdown of disaccharides. Trehalose phosphorylase from Thermoanaerobacter sp. (TP) is a glycoside hydrolase family 65 enzyme which catalyzes the reversible breakdown of trehalose [d-glucopyranosyl-α(1,1)α-d-glucopyranose] to β-d-glucose 1-phosphate and d-glucose. Recombinant purified protein was produced in Escherichia coli and crystallized in space group P212121. Crystals of recombinant TP were obtained in their native form and were soaked with glucose, with n-octyl-β-d-glucoside and with trehalose. The crystals presented a number of challenges including an unusually large unit cell, with a c axis measuring 420 Å, and variable diffraction quality. Crystal-dehydration protocols led to improvements in diffraction quality that were often dramatic, typically from 7–8 to 3–4 Å resolution. The structure of recombinant TP was determined by molecular replacement to 2.8 Å resolution, thus establishing a starting point for investigating the structural and mechanistic determinants of the disaccharide phosphorylase activity. To the best of our knowledge, this is the first crystal structure determination of an inverting trehalose phosphorylase.
1. Introduction
Disaccharide phosphorylases (DSPs) catalyze the synthesis and degradation of disaccharides using phosphate-activated sugar donors (Kitaoka & Hayashi, 2002 ▶). DSPs can be found in both prokaryotic and eukaryotic organisms and can be either inverting or retaining, working on α-glycosidic or β-glycosidic bonds. Their enzymatic versatility is reflected in their classification: DSPs are found both in glycosyl hydrolase (GH) and glycosyl transferase (GT) families, e.g. chitobiose phosphorylase (GlcNAc-β1,4-GlcNAc, inverting, GH94; Honda et al., 2004 ▶), maltose phosphorylase (Glc-α1,4-Glc, inverting, GH65; Qian et al., 1994 ▶) and trehalose phosphorylase (Glc-α1,1α-Glc, retaining, GT4; Nidetzky & Eis, 2001 ▶). The ability of DSPs to catalyze reversible reactions is a consequence of the fact that the energy of a glycosyl-phosphate bond is lower than in other glycosyl adducts such as glycosyl nucleotides, which are the usual substrate of glycosyl transferases. This, together with their high substrate specificity, has led to the emergence of DSPs as attractive enzymes for the on-demand synthesis or breakdown of sugar chains. On the other hand, only a handful of DSPs are known to date and their narrow substrate specificity has proved to be a limiting factor in the development of a sugar-synthesis platform. The determination of the three-dimensional structure of these enzymes, especially in complex with their respective substrates, would therefore greatly contribute to our understanding of their reaction mechanism and the factors determining substrate specificity and promiscuity and would thus provide important insights towards the development of enzymes with altered specificities.
Trehalose phosphorylase (TP) catalyzes the reversible phosphorolysis of trehalose into d-glucose 1-phosphate and d-glucose. TPs have been found in various organisms such as Grifola frondosa (Saito et al., 1998 ▶), Schizophyllum commune (Eis & Nidetzky, 1999 ▶), Agaricus bisporus (Wannet et al., 1998 ▶), Catellatospora ferruginea (Aisaka & Masuda, 1995 ▶), Thermoanaerobacter brockii (Chaen et al., 1999 ▶) and Euglena gracilis (Belocopitow & Maréchal, 1970 ▶). The TPs above can be classified into two groups according to the anomeric specificity of d-glucose 1-phosphate. The first three enzymes (EC 2.4.1.231) from G. frondosa, S. commune and A. bisporus form α-d-glucose 1-phosphate as the product of phosphorolysis of trehalose and have been classified into glycoside transferase family 4. The latter three enzymes (EC 2.4.1.64) from C. ferruginea, T. brockii and E. gracilis produce β-d-glucose 1-phosphate. They have been classified into glycoside hydrolase family 65, together with maltose phosphorylase and kojibiose phosphorylase. All of these enzymes catalyze the reversible phosphorolysis of an α-glycosidic bond with inversion of the anomeric configuration. The reaction catalyzed by trehalose phosphorylase (EC 2.4.1.64) is
Here, we report the cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of trehalose phosphorylase from Thermoanaerobacter sp. (TP).
2. Materials and methods
2.1. Expression and purification
Thermoanaerobacter brockii subsp. brockii (DSMZ 1457, ATCC 33075) was purchased from DSMZ and was grown in anaerobically sealed 10–25 ml bottles at a temperature of 341 K in medium 144 as recommended by DSMZ. The complete coding sequence of the tp gene was amplified from genomic DNA by PCR using primers containing NdeI and BamHI restriction-enzyme cleavage sites (forward, 5′-GG GAA TTC CAT ATG GCC AAC AAA ACG AAG AAA CC-3′; reverse, 5′-GA CGG ATC CCT ACT TTA TTT CTC CTT TTT CAA G-3′). The PCR product was subcloned in the TOPO TA vector (Invitrogen) before final cloning into a pET-15b overexpression vector (Novagen). The pET-15b plasmid encodes an N-terminal His tag for ease of purification and an inducible T7 promotor. Digestion of pET-15b and the TOPO-TP construct with NdeI and BamHI and subsequent ligation yielded the final plasmid construct pXTP.
pXTP was then used to transform Escherichia coli BL21 (DE3) cells. A single colony was selected and grown in a small volume (50 ml) of LB broth medium containing 100 µg ml−1 carbenicillin (Cb). The overnight culture was used to inoculate 0.8 l LB/Cb broth medium at a ratio of 1:200; the culture was then grown at 310 K in a rotary shaker until an OD600 of 0.6–1.0 was reached. Protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cells were grown overnight after induction and harvested by centrifugation at 6000g for 30 min at 277 K.
Large amounts of recombinant protein were purified in four steps. Cell pellets were suspended in lysis buffer (50 mM Tris–HCl pH 8.4, 50 mM NaCl) and lysed by ultrasonication on ice (2 × 90 s, 30% amplitude) with a Branson 250-D sonicator. Cell debris was removed by two rounds of centrifugation (13 200 rev min−1, 277 K, 30 min). The supernatant was diluted in a 1:5 ratio with 50 mM Tris–HCl pH 8.4, 500 mM NaCl, 20 mM imidazole and loaded onto a 3 ml Ni-Sepharose column (GE Lifesciences) pre-equilibrated with 50 mM Tris–HCl pH 8.4, 500 mM NaCl, 20 mM imidazole. The protein was eluted with a stepwise imidazole gradient (50–500 mM). The fractions containing the protein of interest were resolved on SDS–PAGE, pooled and washed 3–4 times on a 30 kDa molecular-weight cutoff Centricon (Sartorius Stedim Biotech) with 20 mM Tris–HCl pH 8.4, 150 mM NaCl, 2.5 mM CaCl2. To remove the N-terminal His tag, an overnight digest with thrombin (Invitrogen) was carried out at room temperature (1 U enzyme per 2 mg TP). The digest was directly loaded onto a pre-equilibrated Ni-Sepharose column (50 mM Tris–HCl pH 8.4, 500 mM NaCl, 20 mM imidazole) and washed several times with 50 mM Tris–HCl pH 8.4, 500 mM NaCl, 20 mM imidazole to remove all nonbound protein. The flowthrough, which contained TP without the His tag, was pooled and concentrated in 50 mM Tris–HCl pH 8.4, 300 mM NaCl on a 30 kDa molecular-weight cutoff Centricon. As a final polishing step, the sample was loaded onto a Superdex 200 16/60 column (GE Lifesciences) equilibrated with 50 mM Tris–HCl pH 8.4, 300 mM NaCl. The fractions containing TP were pooled, concentrated in 20 mM Tris–HCl pH 8.4, 5 mM NaCl using a centrifugal filter device (30 kDa molecular-weight cutoff) and stored at 277 K. All purification steps were assessed on Coomassie-stained SDS–PAGE gels.
2.2. Production of the active-site mutant E498Q
An E498Q active-site mutant was constructed using the QuikChange XL Site-Directed Mutagenesis Kit (Qiagen) with 5′-GTCACTGGTCCGGATCAGTATACGGCATTAGTTG-3′ and 5′-CAACTAATGCCGTATACTGATCCGGACCAGTGAC-3′ as the forward and reverse primers, respectively. Sequencing confirmed the introduction of the E498Q mutation. The protocol developed for the wild-type enzyme (§2.1) was used for overexpression and purification.
2.3. Dynamic light scattering
Dynamic light-scattering studies (DLS) on recombinant TP were performed using a Zetasizer Nano S instrument (Malvern) equipped with a 633 nm He–Ne laser and a temperature-controlled measuring chamber. Samples of purified TP (13 mg ml−1 in 20 mM Tris–HCl pH 8.4, 5 mM NaCl) were clarified by centrifugation at 13 200 rev min−1 prior to all measurements.
2.4. Crystallization and crystal soaking
Initial crystallization screening was performed in-house based on the hanging-drop vapour-diffusion method and Crystal Screens 1, 2 and Lite from Hampton Research. 1 µl protein solution (13 mg ml−1 in 20 mM Tris pH 8.4, 5 mM NaCl) was mixed with an equal volume of reservoir solution on 18 mm siliconized round cover slides, sealed and equilibrated against 200 µl reservoir solution at room temperature (293 K). Additionally, cocrystallization with the natural substrates was attempted. 1 µl of 0.1–6%(w/v) trehalose, d-glucose or β-d-glucose 1-phosphate was mixed with 1 µl protein solution (13 mg ml−1 in 20 mM Tris–HCl pH 8.4, 5 mM NaCl) and 1 µl well solution. The drops were set up for hanging-drop vapour-diffusion crystallization as described above.
Promising leads were further optimized via additional crystallization grids in which the concentration of the precipitants was varied against a range of pH values. In parallel, the Additive Screen (Hampton Research) was employed in screens in which 1 µl protein solution (13 mg ml−1 in 20 mM Tris–HCl pH 8.4, 5 mM NaCl) was mixed with either 1 µl reservoir solution and 0.5 µl additive (in the case of nonvolatile additives) or 1 µl reservoir solution containing 10%(v/v) additive (in the case of volatile additives). The drops were set up for hanging-drop vapour-diffusion crystallization as described above.
In a second approach, protein–substrate complexes were obtained by soaking. Small 0.5 µl drops of mother liquor containing increasing concentrations of trehalose, d-glucose or β-d-glucose 1-phosphate were added directly to the crystallization droplet in 1–3% increments to a maximal final concentration of 1%(w/v). Soaking times varying between 45 min and overnight were employed. Alternatively, crystals were transferred directly with a nylon loop to a fresh drop containing 2–10%(w/v) substrate. However, in many cases this proved to be a rather harsh method and often resulted in crystal cracking.
2.5. Crystal dehydration
Four methods were applied in order to accomplish dehydration of the crystals (Heras & Martin, 2005 ▶). The first method was based on stepwise transfer of the cover slip carrying the crystallization droplets to reservoirs containing increasing concentrations of PEG 4000 [up to 18%(w/v)] using small increments of 2% PEG 4000 at a time. In a variation of this method 5% increments were used and 1 µl of well solution containing the higher PEG concentration was added to the crystallization droplet prior to sealing. In both methods the droplets were allowed to equilibrate for 8–24 h between transfers. In a third method the cover slides carrying the crystallization droplets were removed from the crystallization box and the droplets were exposed to air for short periods ranging between 5 and 30 min before adding cryoprotectant. Alternatively, the droplets were exposed to air by sealing them over an empty well. At 10 min intervals, 0.5 µl droplets of PEG 4000 were added to the crystallization droplets in 5% increments. On reaching the final concentration, the droplets were left to equilibrate for 1 h.
2.6. Cryoprotection
The presence of PEG 4000 in the crystallization conditions allowed us to exploit the possibility of using elevated concentrations of PEG 4000 as a cryoprotectant. Using a nylon loop, the crystals were transferred from the crystallization drop to drops with increasing concentrations of cryoprotectant. Crystals were left to equilibrate for 2–3 min between transfer steps. The crystals were then mounted in a nylon loop and flash-cooled by submerging them in liquid nitrogen. The poor reproducibility of this method prompted us to explore the use of other cryoprotective agents: sodium formate, lithium chloride, glucose, trehalose, glycerol, ethylene glycol, 2-methyl-2,4-pentanediol (MPD), paraffin oil and combinations thereof. The best results were obtained using MPD, whereby mother liquor containing increasing amounts of MPD was added to the crystallization drop in increments of 0.25–0.5 µl followed by equilibration over the reservoir for 10–15 min until a final concentration of around 10%(v/v) MPD was reached.
2.7. Data collection
Early batches of TP crystals were tested for diffraction on the beamlines of EMBL Hamburg (DESY, Hamburg, Germany). A total of five data sets were recorded.
Two data sets for wild-type TP were measured from single crystals on beamline X06SA at SLS (Swiss Light Source, Villigen, Switzerland). An initial crystal (TP_nat) was grown in 10%(v/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 1%(v/v) 2-propanol and cryoprotected by transfer to 25%(v/v) PEG 4000. A second data set (TP_OG) was recorded from a crystal grown in 10%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 1%(w/v) n-octyl-β-d-glucoside (OG). The crystal was cryoprotected by the gradual addition of MPD to a final concentration of 10%(v/v).
Two additional data sets for wild-type TP were recorded at BESSY (Berliner Elektronen-Speicherring Gesellschaft für Synchrotronstrahlung, Berlin, Germany). The first data set (TP_glc) was recorded from a crystal grown in 10%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 2%(v/v) 2-propanol. The crystal was soaked with glucose by direct transfer to 10%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 2%(w/v) glucose for approximately 2 h and cryoprotected with 20%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 10%(v/v) 2-propanol, 5%(w/v) glucose. A second crystal (TP_tre) was grown using 10%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 1%(v/v) 2-propanol and incubated with 0.4% trehalose (final concentration), which was gradually added to the crystallization droplet over a 15 min time course. MPD was the preferred cryoprotectant for these crystals.
Finally, a fifth data set (TP_mut) for the active-site mutant was recorded on the PROXIMA1 beamline at SOLEIL (Source Optimisée de Lumière d’Énergie Intermédiaire du LURE, Saint-Aubin, France). The crystal was grown from 12%(w/v) PEG 4000, 0.1 M Tris–HCl pH 7.5. Soaking and dehydration were performed simultaneously with the first method (§2.5) with 8 h between dehydration steps to give a final concentration of 22%(w/v) PEG 4000. In order to obtain a ternary complex, 0.5 µl 3%(w/v) trehalose was added to the droplet during the first and the last step of dehydration and 0.5 µl 150 mM phosphate was added during the next-to-last step, resulting in final concentrations of around 0.75%(w/v) trehalose and 19 mM phosphate. The total soaking times were thus 56 h for trehalose and 16 h for phosphate. MPD was used for cryoprotection.
Data-collection statistics are reported in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| TP_nat | TP_glc | TP_OG | TP_tre | TP_mut | |
|---|---|---|---|---|---|
| X-ray source | SLS X06SA | BESSY ID14.1 | SLS X06SA | BESSY ID14.2 | SOLEIL PROXIMA1 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 |
| Dehydration | No | No | No | No | Method 1 |
| Cryoprotectant | PEG 4000 | PEG 4000 | MPD | MPD | MPD |
| Ligand | None | n-Octyl-β-D-glucose | Glucose | Trehalose | Trehalose + Pi |
| Ligand concentration | — | 1%(w/v) | 2%(w/v) | 0.4%(w/v) | 0.75%(w/v) trehalose, 19 mM phosphate |
| Method for ligand complexes | — | Cocrystallization | Transfer soak (2 h) | Addition soak (30 min) | Addition soak (56 h) |
| Wavelength (Å) | 0.9998 | 0.9184 | 0.9999 | 0.9184 | 0.9184 |
| Data range (°) | 113 | 140 | 100 | 100 | 140 |
| Frame oscillation (°) | 0.125 | 0.2 | 0.2 | 0.2 | 0.2 |
| Data processing | XDS/XSCALE | XDS/XSCALE | XDS/XSCALE | XDS/XSCALE | XDS/XSCALE |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Resolution (Å) | 50–2.8 | 40–4.6 | 30–4.2 | 30–3.4 | 30–4.1 |
| Unit-cell parameters | |||||
| a (Å) | 97.84 | 102.4 | 103.8 | 98.39 | 101.2 |
| b (Å) | 104.28 | 105.2 | 105.4 | 103.92 | 107.4 |
| c (Å) | 415.71 | 423.1 | 424.0 | 420.6 | 422.0 |
| Molecules in asymmetric unit | 4 | 4 | 4 | 4 | 4 |
| Unique reflections | 100648 | 26162 | 34229 | 59496 | 36988 |
| Redundancy | 4.00 | 5.62 | 4.09 | 3.29 | 5.55 |
| Mosaicity (°) | 0.24 | 0.19 | 0.15 | 0.24 | 0.19 |
| Completeness (%) | 95.1 (96.1) | 98.3 (96.7) | 98.0 (96.2) | 98.4 (99.7) | 99.3 (99.3) |
| Rmeas† (%) | 11.0 (76.5) | 16.8 (93.2) | 9.8 (50.5) | 16.2 (73.6) | 7.4 (77.3) |
| 〈I/σ(I)〉 | 10.2 (2.4) | 9.08 (2.12) | 12.46 (3.05) | 8.55 (2.44) | 14.02 (2.55) |
| RFZ‡ | 6.1, 3.9, 3.7, 3.8 | ||||
| TFZ‡ | 4.4, 12.4, 19.3, 14.2 | ||||
| LLG‡ | 58, 1884, 381, 821 |
R
meas is the multiplicity-weighted R
merge, i.e.

 , where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the average value over multiple measurements (Diederichs & Karplus, 1997 ▶).
, where Ii(hkl) is the intensity of the ith measurement of reflection hkl and 〈I(hkl)〉 is the average value over multiple measurements (Diederichs & Karplus, 1997 ▶).
For monomers 1, 2, 3 and 4, respectively.
3. Results and discussion
The pXTP expression vector constructed to produce recombinant TP was sequenced to verify that no errors has been introduced during PCR and/or subsequent cloning steps. One mutation was found at base position 1082 (G→A, G361E) corresponding to the sequence of a glycoside hydrolase family 65 domain protein from T. pseudethanolicus annotated as a putative kojibiose phosphorylase (accession No. ABY95159). The protein from T. pseudethanolicus further differs from that of T. brockii subsp. brockii in that it contains an additional mutation at base position 2357 (G→A, E769K) and a deletion at base position 2370. However, these sites overlap with the primer used for amplification and thus are not present in the cloned construct.
Trehalose phosphorylase from Thermoanaerobacter sp. (TP) was recombinantly overexpressed in E. coli and purified to homogeneity. Furthermore, we produced the active-site mutant E498Q to enable additional structural and biochemical studies of the enzyme. The purified enzymes migrated as a single band on SDS–PAGE at a height of approximately 90 kDa, corresponding to the calculated molecular weight of 89 994.55 Da for the monomer. The elution profiles of native and mutant TP on a calibrated gel-filtration column, however, corresponded to a protein with a molecular weight of 180 kDa. This is consistent with the results from DLS, which gave a hydrodynamic radius of 11.8 nm, corresponding to a molecular weight of 214 kDa for a globular protein. This suggested that TP is a dimer in solution, as has been reported for the related maltose phosphorylase (Egloff et al., 2001 ▶).
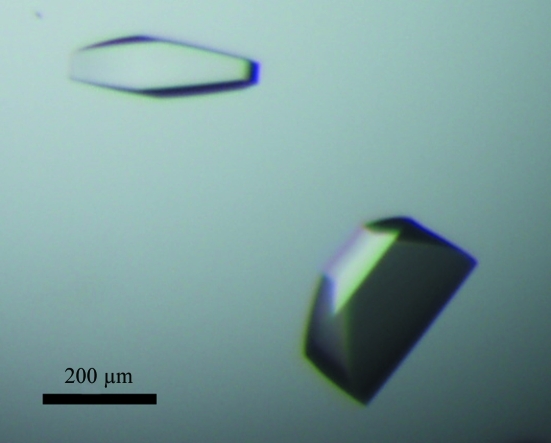
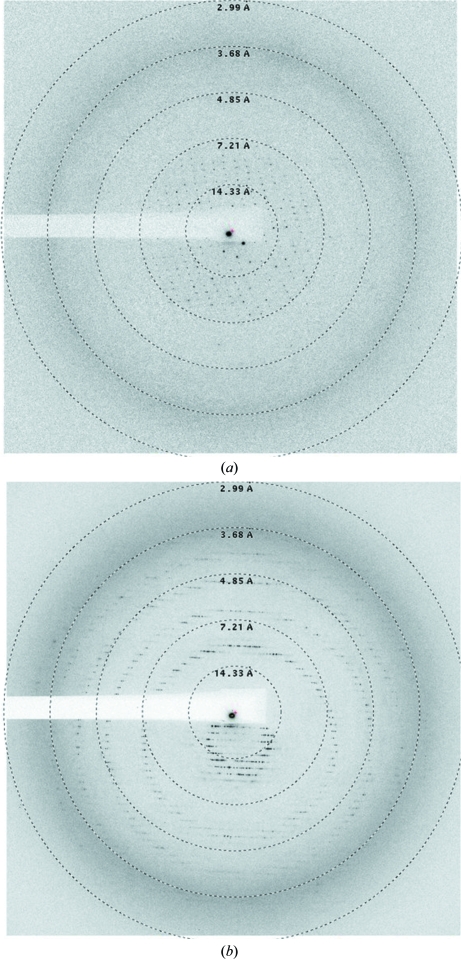
Initial crystallization screens on recombinant TP carrying an N-terminal His tag did not yield crystals of sufficient size and diffraction quality for data collection. Inspection of the structure of the related maltose phosphorylase from Lactobacillus brevis (33% sequence identity; PDB entry 1h54; Egloff et al., 2001 ▶) revealed that the N-terminus of the protein is conspicuously exposed at the surface of the protein. We pondered whether TP might be similar in this regard, so that a His tag at the N-terminus might compromise the formation of lattice interactions and thus affect crystal growth. We therefore initiated new crystallization trials of recombinant TP after proteolytic treatment with thrombin. This removes the His tag completely apart from a short Gly-Ser-His peptide preceding the initiating Met. Optimization led to hexagonal and trapezoidal crystals that were approximately 200–300 µm in the largest dimension on average (Fig. 1 ▶), although crystals of up to 500 µm could occasionally be grown. These crystals grew within a week from 5–15%(v/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 0–2%(v/v) 2-propanol. However, the diffraction quality of the crystals showed a large variability between protein batches. For poorly diffracting crystals, a substantial increase in diffraction quality could be achieved by crystal dehydration, by use of which the high-resolution limit could be dramatically improved from 7–8 to 3–4 Å (Fig. 2 ▶). This was particularly the case for native substrate-free crystals; a lesser degree of improvement was observed for soaked crystals. Nonetheless, the high susceptibility of the crystals to radiation damage, in combination with the need for fine-slicing (see below), resulted in a significant decrease in diffraction quality during data collection.
Figure 1.
Recombinant TP crystal grown from 10%(v/v) PEG 4000, 0.1 M Tris–HCl pH 7.5, 1% 2-propanol.
Figure 2.
(a) Diffraction image from a typical TP crystal without dehydration. (b) Diffraction image from the TP_mut crystal, which was dehydrated according to the first method described in §2.5.
We were able to record five complete data sets at the Swiss Light Source, Villigen, Switzerland (TP_nat, TP_OG), BESSY, Berlin, Germany (TP_glc, TP_tre) and SOLEIL, Saint-Aubin, France (TP_mut). The latter data set (TP_mut) was recorded from a crystal that was dehydrated using the first method (§2.5), while the other four data sets (TP_nat, TP_OG, TP_glc and TP_tre) were recorded from nondehydrated crystals (§2.7). All data sets could be indexed and processed using XDS (Kabsch, 1993 ▶) in the primitive orthorhombic space group P212121 (Table 1 ▶). The most remarkable feature of the unit cell was the very long c axis of 420 Å. Differences in the unit-cell dimensions were within 3% of the average values in all cases, the largest differences being observed along the a axis (Table 1 ▶). Analysis of crystal-packing densities suggested the presence of four monomers in the asymmetric unit, corresponding to an average solvent content of 60%. This rather high value, together with the large unit cell, may be part of the reason why the crystals only diffracted to moderate resolution despite their appreciable size (up to 500 µm). The long unit-cell c axis required special attention during data collection. To avoid reflection overlaps in reciprocal space, fine-slicing was used in combination with a finely collimated X-ray beam, while care was taken to avoid the c axis lying parallel to the X-ray beam.
For the first data set (TP_nat), the availability of the structure of the related maltose phosphorylase from L. brevis (MPLbre; PDB entry 1h54; Egloff et al., 2001 ▶) allowed us to solve the phase problem using maximum-likelihood molecular replacement as implemented in the program Phaser (McCoy et al., 2007 ▶). A semi-conservative monomeric search model was constructed with CHAINSAW (Stein, 2008 ▶) in which all nonconserved residues were truncated to the last common C atom. Molecular replacement with Phaser could place four monomers in the asymmetric unit with convincing Z scores and LLG values (Table 1 ▶). The solutions were confirmed by inspection of electron-density maps calculated with Fourier coefficients 2F o − F c,MR, αc,MR, which also showed extra density for missing structural elements and side chains. Additionally, the four monomers were found to form two dimers (Fig. 3 ▶).
Figure 3.

Cα-atom traces and packing of TP in the primitive orthorhombic cell. Subunits in dimers making up the asymmetric unit of each lattice are shown in blue and cyan and in pink and magenta, while symmetry-related molecules are shown in greyscale. This figure was prepared with the program PyMOL (DeLano, 2002 ▶).
For all other data sets, a partially refined TP_nat monomer was used for molecular replacement. In all cases, a convincing solution with four monomers in the asymmetric unit cell was found by Phaser and could be confirmed by inspection of the 2F o − F c,MR, αc,MR maps. Positive F o − F c,MR, αc,MR density could additionally be observed in the active sites of TP_OG, TP_glc, TP_tre and TP_mut.
TP_OG displayed clear positive difference density in all four active sites. This can be attributed to the binding of n-octyl-β-d-glucoside to the sugar-binding site, although no density could be observed for the octyl chain, probably owing to disorder. Similarly, TP_glc contained positive F o − F c,MR, αc,MR density in each active site indicative of the binding of glucose. In all four TP_tre monomers positive F o − F c,MR, αc,MR density was found in both sugar-binding subsites +1 and −1, suggesting the binding of trehalose. This was especially clear in chains A and B. However, no evidence for the binding of trehalose could be found in TP_mut, although positive F o − F c, MR, αc,MR density could be observed for phosphate in one chain.
Model building, refinement and structure description of trehalose phosphorylase from T. pseudethanolicus is under way and will be reported elsewhere.
Acknowledgments
We would like to thank the EMBL (Hamburg, Germany), BESSY (Berlin, Germany), the SLS (Villigen, Switzerland) and SOLEIL (Saint-Aubin, France) for beamtime allocation and technical support. This work was supported by grant IWT50191 from the ‘Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen’ (IWT Flanders, Belgium).
References
- Aisaka, K. & Masuda, T. (1995). FEMS Microbiol. Lett.131, 47–51. [DOI] [PubMed]
- Belocopitow, E. & Maréchal, L. R. (1970). Biochim. Biophys. Acta, 198, 151–154. [DOI] [PubMed]
- Chaen, H., Nakada, T., Nishimoto, T., Kuroda, N., Fukuda, S., Sugimoto, T., Kurimoto, M. & Tsujisaka, Y. (1999). J. Appl. Glycosci.46, 399–405.
- DeLano, W. L. (2002). The PyMOL Molecular Viewer. DeLano Scientific, Palo Alto, California, USA. http://www.pymol.org.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Egloff, M. P., Uppenberg, J., Haalck, L. & van Tilbeurgh, H. (2001). Structure, 9, 689–697. [DOI] [PubMed]
- Eis, C. & Nidetzky, B. (1999). Biochem. J.341, 385–393. [PMC free article] [PubMed]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed]
- Honda, Y., Kitaoka, M. & Hayashi, K. (2004). Biochem. J.377, 225–232. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kitaoka, M. & Hayashi, K. (2002). Trends Glycosci. Glycotechnol.14, 35–50.
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Nidetzky, B. & Eis, C. (2001). Biochem. J.360, 727–736. [DOI] [PMC free article] [PubMed]
- Qian, N., Stanley, G. A., Hahn-Hägerdal, B. & Radström, P. (1994). J. Bacteriol.176, 5304–5311. [DOI] [PMC free article] [PubMed]
- Saito, K., Yamazaki, H., Ohnishi, Y., Fujimoto, S., Takahashi, E. & Horinouchi, S. (1998). Appl. Microbiol. Biotechnol.50, 193–198. [DOI] [PubMed]
- Stein, N. (2008). J. Appl. Cryst.41, 641–643.
- Wannet, W. J., den Camp, H. J. O., Wisselink, H. W., van der Drift, C., Griensven, L. J. V. & Vogels, G. D. (1998). Biochim. Biophys. Acta, 1425, 177–188. [DOI] [PubMed]