Crystals of the Arabidopsis abscisic acid receptors PYL9/RCAR1, PYL5/RCAR8 and PYR1/RCAR11 have been obtained.
Keywords: plant hormone receptors, PYL9/RCAR1, PYL5/RCAR8, PYR1/RCAR11
Abstract
Abscisic acid (ABA) is a plant hormone that plays key regulatory roles in physiological pathways for the adaptation of vegetative tissues to abiotic stresses such as water stress in addition to events pertaining to plant growth and development. The Arabidopsis ABA receptor proteins PYR/PYLs/RCARs form a START family that contains 14 members which are classified into three subfamilies (I–III). Here, purification, crystallization and X-ray data collection are reported for a member of each of the subfamilies, PYL9/RCAR1 from subfamily I, PYL5/RCAR8 from subfamily II and PYR1/RCAR11 from subfamily III, in the presence of (+)-abscisic acid. The three proteins crystallize in space groups P3121/P3221, P2 and P1, respectively. X-ray intensity data were collected to 1.9–2.6 Å resolution.
1. Introduction
Recent advances and emerging trends in plant-hormone signalling are focused on understanding key proteins such as receptors at the molecular and structural levels (Santner & Estelle, 2009 ▶). Abscisic acid (ABA) is a plant hormone that plays key regulatory roles in physiological pathways related to plant growth and development, such as seed maturation and dormancy, and enables the adaptation of vegetative tissues to abiotic stresses such as water stress (Finkelstein et al., 2002 ▶; Yamaguchi-Shinozaki & Shinozaki, 2006 ▶). One conspicuous effect of ABA is the promotion of stomatal closure in guard cells, which is mediated by solute efflux. ABA regulates the expression of many genes, which in turn may regulate functions involving dehydration tolerance in both vegetative tissues and seeds. Since the discovery of ABA (Ohkuma et al., 1963 ▶; Eagles & Wareing, 1963 ▶), while advances have been made in the isolation and characterization of several key proteins that function in the downstream signalling network, little is known about the ABA-recognition mechanisms employed by receptor proteins. Although FCA, ABAR/CHLH/GUN5, GCR2, GTG1 and GTG2 have been reported to be ABA receptors, there is no evidence that the previously identified positive and negative regulators of ABA signalling, such as the PP2C protein phosphatases ABI1, ABI2 and HAB1, are regulated by these proposed receptors (Pennisi, 2009 ▶).
Recently, two reports describing a family of small soluble proteins that activate ABA signalling have drawn much attention from plant biologists (Park et al., 2009 ▶; Ma et al., 2009 ▶). These reports demonstrated that these proteins are able to recognize ABA and repress the key negative regulators in the ABA signalling pathways (ABI1, ABI2 and HAB1). Unexpectedly, this family contains 14 members in Arabidopsis: the regulatory components of ABA receptor 1–14 (RCAR1–14), also referred to as pyrabactin resistance 1 (PYR1) and PYR1-like 1–13 (PYL1–PYL13). These PYR/PYLs/RCARs belong to a protein family within the Bet v 1 superfamily that includes the birch pollen allergen Bet v 1a, class 10 pathogen-related proteins, polyketide cyclases and lipid-transfer proteins containing a steroidogenic acute regulatory protein-related lipid-transfer (START) domain (Radauer et al., 2008 ▶). Interestingly, the PYR/PYLs/RCARs have been classified into subfamilies I, II and III based on amino-acid sequence identity (Park et al., 2009 ▶). At present, whether each subfamily exerts different functions by binding to distinct downstream target proteins remains unknown. However, the presence of this many members in this family may suggest functional differentiation, as for other signalling proteins, and therefore it is important to clarify the structural differences in members of the three subfamilies. Here, we present preliminary crystallographic studies of PYL9/RCAR1 (subfamily I), PYL5/RCAR8 (subfamily II) and PYR1/RCAR11 (subfamily III).
2. Experimental
2.1. Protein expression
The nucleotides encoding Arabidopsis PYL9/RCAR1 (residues 1–187; NCBI accession No. AK227623), PYL5/RCAR8 (residues 1–203; AY052251) and PYRl/RCAR11 (residues 1–191; AY042890) were amplified by PCR using RIKEN clones pda18331, pda03718 and pda01246, respectively. The coding nucleotides were subcloned via SmaI/EcoRI sites into pET49b(+) plasmid (Novagen) containing an N-terminal glutathione S-transferase (GST) tag linked by an HRV3C protease site. Escherichia coli strain DH5α (DE3) cells were transformed with each recombinant DNA plasmid and selected using kanamycin. All plasmids extracted from the positive colonies were verified by DNA sequencing and transformed into E. coli strain Rosetta 2 (DE3) (Novagen) cells for protein expression.
An overnight culture (20 ml) of Rosetta 2 (DE3) cells transformed with each plasmid was inoculated into 2.5 l Luria–Bertani medium containing 30 µg ml−1 kanamycin and 20 µg ml−1 chloramphenicol. When the absorbance at 600 nm (OD600) of the cell culture reached 0.7–0.9, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce expression of each receptor gene to a final concentration of 400 µM (for PYL9/RCAR1), 10 µM (for PYL5/RCAR8) or 100 µM (for PYR1/RCAR11). Following IPTG induction, cells were grown for an additional 20–24 h at 289 K and then collected by centrifugation (Beckman Avanti J-HC).
2.2. Protein purification
Wet cells expressing each PYR/PYLs/RCARs protein were suspended in 20 mM Tris–HCl buffer pH 8.0 containing 150 mM NaCl (buffer A) and then disrupted by sonication on ice. The suspension was centrifuged at 140 500g for 30 min (Beckman Optima L-70) and the soluble portion of the cell extract was further purified using affinity column chromatography. The supernatant was applied onto a Glutathione Sepharose 4B column (GE Healthcare). After washing the column with buffer A, the GST-fusion protein was eluted with buffer A containing 20 mM glutathione.
The GST-fusion protein was cleaved using HRV3C protease. The expected peptide for the construct is located at a position following two extra residues (Gly-Pro) that are part of the HRV3C protease-recognition sequence. The cleaved protein was purified by HiTrap Q HP column (GE Healthcare) chromatography using a linear NaCl concentration gradient (0–0.5 M). Fractions containing the PYR/PYLs/RCARs protein were collected, concentrated and subjected to gel-filtration chromatography on a Superdex 75 prep-grade column (GE Healthcare) equilibrated and eluted with buffer A. Purified proteins were concentrated to 15–20 mg ml−1 in buffer A containing 2 mM DTT, divided into 20–50 µl aliquots in 0.5 ml Eppendorf tubes and then immediately frozen in liquid nitrogen. Frozen samples were stored at 193 K until use.
The identity of the purified protein was confirmed using matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI–TOF MS; Bruker Daltonics).
2.3. Crystallization
For crystallization, (+)-ABA (Tokyo Chemical Industry) was added to the purified protein solution to a final concentration of 2–3 mM. Initial crystallization conditions were screened employing the sitting-drop vapour-diffusion method using a Hydra II Plus One crystallization robot (Matrix Technology) with 96-well Intelli-Plates and the commercial crystallization-solution kits JCSG Core Suite I–IV and PACT Suite (Qiagen) at 293 K. Crystallization-solution droplets were prepared by mixing 0.2 µl protein solution with 0.3 µl of each reservoir solution and were equilibrated against 90 µl of each reservoir solution. The conditions obtained from the screening were optimized using the hanging-drop vapour-diffusion method with droplets prepared by mixing equal volumes (1.0–5.0 µl) of protein solution and reservoir solution and equilibrated against 200 µl reservoir solution. The crystals obtained were transferred stepwise into a cryoprotective solution containing 25%(v/v) glycerol or 15–20%(w/v) polyethylene glycol 400 (PEG 400) and flash-cooled at 100 K. Initial diffraction tests were performed using a home-source X-ray generator (Rigaku FR-E) equipped with a Rigaku R-AXIS VII detector at 100 K.
2.4. X-ray data collection
X-ray diffraction data were collected from native crystals of PYR/PYLs/RCARs proteins using a Rayonix MX225HE CCD detector installed on the BL41XU beamline at SPring-8. The camera was fixed at a distance of 260 mm with a wavelength of 1.0000 Å. Data collection for each native crystal was performed using an angular range of 180° (PYL9/RCAR1 and PYL5/RCAR8) or 360° (PYR1/RCAR11), an oscillation step of 1.0° (PYL9/RCAR1 and PYL5/RCAR8) or 2.0° (PYR1/RCAR11) and an exposure time of 1.0 s. All data were processed and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). Crystal data and intensity data-collection statistics using all data without I/σ(I) cutoff are summarized in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer resolution shell.
| PYL9/RCAR1–ABA | PYL5/RCAR8–ABA | PYL1/RCAR11–ABA | |
|---|---|---|---|
| Space group | P3121/P3221 | P2 | P1 |
| Unit-cell parameters | |||
| a (Å) | 111.4 | 190.8 | 60.6 |
| b (Å) | 111.4 | 146.3 | 71.2 |
| c (Å) | 40.5 | 191.9 | 49.4 |
| α (°) | 90 | 90 | 115.6 |
| β (°) | 90 | 119.8 | 113.2 |
| γ (°) | 120 | 90 | 100.2 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 |
| Resolution | 50–1.9 (1.97–1.90) | 50–2.6 (2.69–2.60) | 50–2.00 (2.07–2.00) |
| Rmerge† (%) | 5.0 (57.0) | 12.4 (43.0) | 5.5 (32.9) |
| I/σ(I) | 60.8 (4.7) | 18.8 (3.2) | 31.66 (2.49) |
| Mosaicity | 0.51–0.80 | 0.27 | 0.69 |
| Completeness (%) | 99.8 (100) | 98.5 (97.3) | 96.5 (96.4) |
| Redundancy | 10.7 (10.4) | 3.7 (3.4) | 3.2 (3.2) |
R
merge = 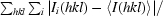
 , where I
i(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
, where I
i(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
3. Results and discussion
Each of the PYR/PYLs/RCARs proteins was successfully purified to homogeneity suitable for crystallization. MALDI–TOF MS analysis of the purified samples yielded a single peak (21 055.5, 22 815.1 or 21 728.1 Da) corresponding to the calculated molecular mass (21 055, 22 816 or 21 729 Da) of PYL9/RCAR1, PYL5/RCAR8 or PYRl/RCAR11, respectively.
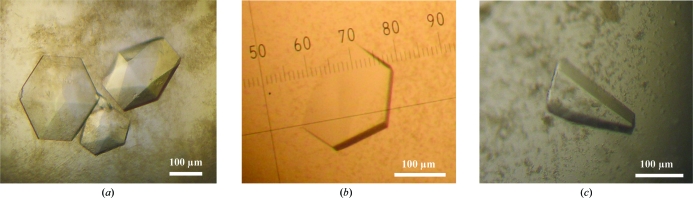
PYL9/RCAR1 protein solution was concentrated to 15 mg ml−1 (∼0.7 mM) with buffer A containing 2 mM (+)-ABA for crystallization. Hexagonal PYL9/RCAR1 crystals were obtained from solutions with a pH of ∼7.0 in the presence of PEGs. A microseeding method was then employed in an effort to obtain larger crystals suitable for X-ray diffraction. The best crystals were obtained from a solution consisting of 7.5 mg ml−1 protein, 1.5 mM (+)-ABA and 10 mM Tris–HCl buffer pH 8.0 containing 75 mM NaCl with 40 mM malic acid–MES–Tris (MMT) buffer pH 7.0 and 10%(w/v) PEG 1500 equilibrated against 16.5%(w/v) PEG 1500 in 100 mM MMT buffer pH 7.0. The crystals (Fig. 1 ▶ a) were found to belong to space group P3121/P3221, with unit-cell parameters a = b = 111.4, c = 40.5 Å, and diffracted to a resolution of 1.9 Å (Table 1 ▶).
Figure 1.
PYR1/PYLs/RCARs crystals. (a) PYL9/RCAR1 crystals. (b) A plate-like crystal of PYL5/RCAR8. (c) A chunky PYR1/RCAR11 crystal.
The PYL5/RCAR8 and PYRl/RCAR11 protein solutions were concentrated to 20 mg ml−1 (∼1 mM) in buffer A containing 3 mM (+)-ABA for crystallization. Thin plate-like crystals of PYL5/RCAR8 were obtained from solutions with pH 6.5–8.0 in the presence of PEG 3000, ammonium hydrogen phosphate or magnesium sulfate as precipitant. These crystals were relatively small and several trials using additive screening and the microseeding technique were required in order to obtain larger crystals. The best crystals were obtained from a solution consisting of 12.0 mg ml−1 protein, 4.0 mM (+)-ABA and 8 mM Tris–HCl buffer pH 8.0 containing 60 mM NaCl with 3.6%(w/v) PEG 3000 and 60 mM nondetergent sulfobetaine (NDSB) 201, 120 mM MgCl2 and 60 mM HEPES buffer pH 7.5 equilibrated against 6.0%(w/v) PEG 3000, 200 mM NDSB 201 and 200 mM MgCl2 in 100 mM HEPES buffer pH 7.5. The crystals (Fig. 1 ▶ b) were found to belong to space group P2, with unit-cell parameters a = 190.8, b = 146.3, c = 191.9 Å, and diffracted to a resolution of 2.6 Å.
Plate-like PYRl/RCAR11 crystals were obtained from a solution consisting of 14.6 mg ml−1 (∼0.7 mM) protein, 1.3 mM (+)-ABA and 13.3 mM Tris–HCl buffer pH 8.0 containing 100 mM NaCl with 67 mM MES buffer pH 6.5 and 5.8%(w/v) PEG 6000 equilibrated against 17.5%(w/v) PEG 6000, 200 mM MgCl2 and 100 mM MES buffer pH 6.5. Although the crystals were found to diffract to a resolution of 2.5 Å, these crystals were found to be comprised of clusters or cracked crystals despite appearing to be single crystals. The crystals (Fig. 1 ▶ c) were found to belong to space group P1, with unit-cell parameters a = 60.6, b = 71.2, c = 49.4 Å, α = 115.6, β = 113.2, γ = 100.2°, and diffracted to a resolution of 2.0 Å.
During the course of this research, structures of PYR1, PYL1 and PYL2 have been reported by five groups (Nishimura et al., 2009 ▶; Melcher et al., 2009 ▶; Miyazono et al., 2009 ▶; Santiago, Rodrigues et al., 2009 ▶; Yin et al., 2009 ▶). These studies have shown that the ABA-recognition mechanism is distinct from those of the recently elucidated plant hormone-recognition mechanisms employed by the auxin receptor TIR1 (Tan et al., 2007 ▶) and the gibberellin GID1 receptors (Murase et al., 2008 ▶; Shimada et al., 2008 ▶). However, all of the structurally elucidated ABA receptors (PYR1, PYL1 and PYL2) belong to subfamily III and no insight has been provided concerning how members of the three subfamilies differ with respect to structure and function. Indeed, the affinity for ABA may differ for members of different subfamilies, which could provide the molecular basis for a variety of responses of plants to different levels of ABA (Finkelstein et al., 2002 ▶; Yamaguchi-Shinozaki & Shinozaki, 2006 ▶). Interestingly, recently reported quantitative data suggest that PYR9/RCAR1 and PYR5/RCAR8 display a strong affinity for ABA with small values of the dissociation constant K d (∼1 µM; Ma et al., 2009 ▶; Santiago, Dupeux et al., 2009 ▶) that are 50-fold or 90-fold smaller than the K d values reported for PYL1/RCAR12 and PYL2/RCAR14 (Yin et al., 2009 ▶). Our structural studies, especially those concerning PYL9/RCAR1 and PYL5/RCAR8, could reveal the structural basis for these differences in affinity and clarify the modified binding modes to downstream target proteins. Structure determinations employing the molecular-replacement method are currently in progress.
Acknowledgments
We would like to thank J. Tsukamoto for technical support in performing the MALDI–TOF MS analysis. This work was supported by a Grant-in-Aid for Scientific Research (A) and a Grant-in-Aid for Scientific Research on Priority Areas, Protein Degradation, Macromolecular Assembly and Cancer from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, a research grant in the natural sciences from the Mitsubishi Foundation and a research grant in the life sciences from the Takeda Science Foundation (to TH). YH was supported by a research grant from a Grant-in-Aid for the Global COE Research from MEXT. We acknowledge Drs N. Shimizu, M. Kawamoto, M. Yamamoto and I. Ohki at SPring-8 for help with data collection at synchrotron beamline BL41XU.
References
- Eagles, C. F. & Wareing, P. F. (1963). Nature (London), 199, 874–875.
- Finkelstein, R. R., Gampala, S. S. & Rock, C. D. (2002). Plant Cell, 14, S15–S45. [DOI] [PMC free article] [PubMed]
- Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A. & Grill, E. (2009). Science, 324, 1064–1068. [DOI] [PubMed]
- Melcher, K. et al. (2009). Nature (London), 462, 602–608.
- Miyazono, K. I., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H. J., Asano, A., Miyauchi, Y., Takahashi, M., Zhi, Y., Fujita, Y., Yoshida, T., Kodaira, K., Yamaguchi-Shinozaki, K. & Tanokura, M. (2009). Nature (London), 462, 609–614. [DOI] [PubMed]
- Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. (2008). Nature (London), 456, 459–463. [DOI] [PubMed]
- Nishimura, N., Hitomi, K., Arvai, A. S., Rambo, R. P., Hitomi, C., Cutler, S. R., Schroeder, J. I. & Getzoff, E. D. (2009). Science, 326, 1373–1379. [DOI] [PMC free article] [PubMed]
- Ohkuma, K., Lyon, J. L., Addicott, F. T. & Smith, O. E. (1963). Science, 142, 1592–1593. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Park, S.-Y. et al. (2009). Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed]
- Pennisi, E. (2009). Science, 324, 1012–1013. [DOI] [PubMed]
- Radauer, C., Lackner, P. & Breiteneder, H. (2008). BMC Evol. Biol.8, 286–304. [DOI] [PMC free article] [PubMed]
- Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S.-Y., Jamin, M., Cutler, S. R., Rodriguez, P. L. & Márquez, J. A. (2009). Nature (London), 462, 665–668. [DOI] [PubMed]
- Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., Park, S. Y. & Márquez, J. A., Cutler, S. R. & Rodriguez, P. L. (2009). Plant J.60, 575–588. [DOI] [PubMed]
- Santner, A. & Estelle, M. (2009). Nature (London), 459, 1071–1078. [DOI] [PubMed]
- Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T., Nakajima, M., Naoe, Y., Ohmiya, H., Kato, H. & Matsuoka, M. (2008). Nature (London), 456, 520–523. [DOI] [PubMed]
- Tan, X., Calderon-Villalobos, L. I., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M. & Zheng, N. (2007). Nature (London), 446, 640–645. [DOI] [PubMed]
- Yamaguchi-Shinozaki, K. & Shinozaki, K. (2006). Annu. Rev. Plant Biol.57, 781–803. [DOI] [PubMed]
- Yin, P., Fan, H., Hao, Q., Yuan, X., Wu, D., Pang, Y., Yan, C., Li, W., Wang, J. & Yan, N. (2009). Nature Struct. Mol. Biol.16, 1230–1236. [DOI] [PubMed]