SUMMARY
The 2.9 Å crystal structure of the core SRPK1:ASF/SF2 complex reveals that the N-terminal half of the basic RS domain of ASF/SF2, which is destined to be phosphorylated, is bound to an acidic docking groove of SRPK1 distal to the active site. Phosphorylation of ASF/SF2 at a single site in the C-terminal end of the RS domain generates a primed phosphoserine that binds to a basic site in the kinase. Biochemical experiments support a directional sliding of the RS peptide through the docking groove to the active site during phosphorylation, which ends with the unfolding of a β strand of the RRM domain and binding of the unfolded region to the docking groove. We further suggest that the priming of the first serine facilitates directional substrate translocation and efficient phosphorylation.
INTRODUCTION
The SR proteins represent a family of splicing factors that are essential for both constitutive and alternative splicing and are highly conserved in metazoans and plants (Birney et al., 1993; Black, 2003; Blencowe, 2000). All SR proteins share a common structural organization; the N terminus consists of one or two RNA-recognition motifs (RRMs), whereas the C terminus contains arginine/serine (RS) dipeptide repeats and is referred to as the RS domain (Fu, 1995; Graveley, 2000). SR proteins are extensively phosphorylated at their RS domains and can exist in differentially phosphorylated (hypo- and hyperphosphorylated) states (Fu, 1995; Graveley, 2000). For proper execution of splicing activity, cycles of phosphorylation and dephosphorylation of SR proteins are important (Misteli et al., 1997; Shi et al., 2006a; Xiao and Manley, 1997, 1998). Phosphorylation of SR proteins also dictates their nuclear import (Lai et al., 2001), and some SR proteins have been shown to serve additional roles in mRNA export and translation regulation (Huang et al., 2003, 2004; Sanford et al., 2004, 2005).
Several protein kinases have been reported to phosphorylate the RS domains of SR proteins (Blaustein et al., 2005; Colwill et al., 1996; Gui et al., 1994a; Rossi et al., 1996). The SR protein kinase (SRPK) and Clk/Sty family of protein kinases predominantly phosphorylate SR proteins in vivo. SRPKs are highly RS-specific kinases and only phosphorylate serine (not threonine) residues that are located adjacent to arginines (not lysines) in their substrates (Gui et al., 1994b; Wang et al., 1998). One of the well-studied SR proteins is ASF/SF2. There are 20 serines within the RS domain of ASF/SF2, of which SRPK1 processively phosphorylates nearly 12 series present within the N-terminal half of the RS domain (RS1 motif) (Aubol et al., 2003; Velazquez-Dones et al., 2005). Genetic and biochemical experiments have demonstrated critical functions of ASF/SF2 in heart development, genomic stability, and mRNA surveillance (Ghigna et al., 2005; Li and Manley, 2005; Xu et al., 2005; Zhang and Krainer, 2004). A recent report has also demonstrated oncogenic transformation activity of overexpressed ASF/SF2. Aberrant alternative splicing of target genes critical for cell proliferation, growth, and apoptosis might be responsible for cellular transformation (Karni et al., 2007).
Although all protein kinases adopt the common kinase fold, they are highly specific and can recognize and phosphorylate a unique subset of the cellular proteins. Several kinases have been identified to rely on direct docking interactions with substrates, using sites distinct from the phosphor-acceptor sequences to achieve substrate specificity (Biondi and Nebreda, 2003; Pellicena and Kuriyan, 2006; Remenyi et al., 2006; Shi et al., 2006b). The docking groove of SRPK1 is constituted in part by an insert found in CMGC kinases known as the MAP kinase insert (Zhang et al., 1994). This docking groove binds to an ASF/SF2 docking motif, shown to be present immediately N-terminal to the RS domain. We showed that the docking interaction between SRPK1 and ASF/SF2 is important for processive and restrictive phosphorylation and promotes assembly of ASF/SF2 in nuclear speckles. Further phosphorylation (hyperphosphorylation) of the RS domain by Clk/Sty results in its dissociation from the speckles (Ngo et al., 2005).
In this study, we present the X-ray crystal structure of an active fragment of SRPK1 bound to ASF/SF2. Our structure shows that three discontinuous segments of ASF/SF2 interact with SRPK1. The second RRM (RRM2) of ASF/SF2 interacts with both the small and large lobes of SRPK1, the N-terminal part of the RS domain interacts with the distal kinase-docking groove, and a phosphoserine from the C-terminal part of the RS domain contacts a basic site located near the P+1 loop of the kinase. Extensive contacts seen in our structure explain the high affinity of this complex, which is unusual for a kinase-substrate pair. Our structure also suggests a possible role for the RRM2 domain of ASF/SF2 in regulation of the phosphorylation mechanism by controlling the relative movement of the lobes of the kinase. The binding of the other two segments of ASF/SF2 to SRPK1 presents features that allow us to propose how the kinase sequentially and processively phosphorylates its substrate by mechanism that involves priming, translocation, and unfolding of the substrate.
RESULTS AND DISCUSSION
Isolation of the Crystallizable Complex of SRPK1 and ASF/SF2
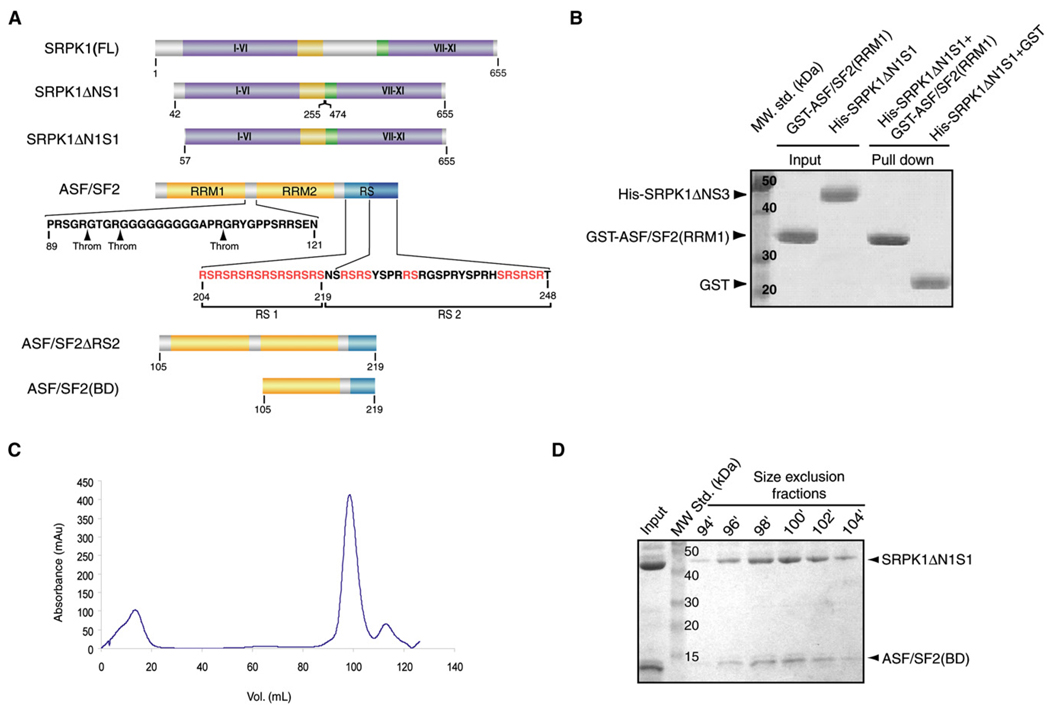
We previously showed that SRPK1 and ASF/SF2 form a high-affinity complex of apparent Kd ~50 nM. We also demonstrated that the insert region of SRPK1 did not play a role in the binding interaction with ASF/SF2 (Aubol et al., 2003). Furthermore, the X-ray crystal structure of a truncated SRPK1, SRPK1ΔNS1, which lacks residues 256–473 in the insert and residues 1–56 at the N terminus was previously determined in our laboratory (Figure 1A) (Ngo et al., 2005, 2007). Therefore, our initial approach was to crystallize the complex of ASF/SF2 and SRPK1ΔNS1. Both proteins were expressed independently in E. coli as polyhistidine fusion proteins. ASF/SF2 was purified under denaturing condition and mixed with pure SRPK1 followed by isolation of the complex by removing the denaturant through dialysis. This complex did not crystallize, which led us to generate several other deletion complexes using similar unfolding-refolding strategy. Diffraction-quality crystals were obtained from one of these complexes that formed between SRPK1ΔN1S1 (15 residues of the N terminus is removed in the context of SRPK1ΔNS1) and ASF/SF2ΔRS2 (residues 1–219) (Figure 1A). Analysis of these crystals by SDS-PAGE surprisingly revealed that RRM1 of ASF/SF2 was removed during the crystallization process (see Figure S1 available online). Detail sequence analysis of ASF/SF2 revealed that putative thrombin cleavage sites are present within the linker region between the two RRMs (Figure 1A). We reasoned that a trace amount of thrombin was retained in the complex that led to the removal of RRM1. These observations also suggested that the RRM1 does not interact with SRPK1 and thus is dispensable for the complex formation. To confirm this, we performed GST pull-down assays by incubating GST-RRM1 with WT and different truncated mutants of SRPK1. At no condition did we observe any interaction between SRPK1 and RRM1 of ASF/SF2 (Figure 1B). Based on these observations, we generated a construct of ASF/SF2 that contains only the RRM2 and RS1 domains (residues 105–219, termed ASF/SF2[BD]) and formed complex with SRPK1ΔN1S1 (hereafter we will refer to SRPK1ΔN1S1 as SRPK1) (Figures 1A, 1C, and 1D). The structure was solved by molecular replacement and refined to 2.9 Å resolution (Table 1).
Figure 1. Isolation of the Core of the SRPK1:ASF/SF2 Complex.
(A) Domain organization of SRPK1 and ASF/SF2 and the fragments used for the complex formation and crystallization. I–XI denotes the substructures in the two kinase lobes as defined by Hanks and Quinn (1991). The putative thrombin cleavage sites in ASF/SF2 are shown by arrows.
(B) RRM1 does not interact with SRPK1. GST pull-down assay shows no interaction between SRPK1ΔN1S1 and RRM1 of ASF/SF2.
(C) Gel filtration elution profile of the complex. Size-exclusion chromatography shows that the purified complex of SRPK1ΔN1S1 and ASF/SF2(BD) is homogeneous in solution.
(D) SDS-PAGE analysis of peak fractions from size-exclusion chromatography.
Table 1.
Data Collection and Refinement Statistics
| Crystal | SRPK1ΔN1S1:ASF/ SF2(BD):AMP-PNP |
|---|---|
| Data collection | |
| Data collection source | APS ID-23 |
| Wavelength (Å) | 0.9793 |
| Resolution (Å) | ∞–2.9 |
| Number of measured reflections (unique) | 79,879 (14,976) |
| Completeness (%)a | 93.0 (62.2) |
| I/σa | 19.9 (2.54) |
| Rsym (%)a,b | 5.8 (44.3) |
| Refinement | |
| Resolution (Å) a | 50–2.90 (2.976–2.901) |
| Rcrys (%)a,c | 23.66 (41.5) |
| Rfree (%)a,d | 29.80 (45.7) |
| Rmsd | |
| Bond lengths (Å) | 0.012 |
| Bond angles | 1.493 |
| Ramachandran Distribution (Number of Residues/Percent of Total Residues)e | |
| Favorable regions | 306 (80.1%) |
| Additional regions | 66 (17.3%) |
| Generously allowed regions | 8 (2.1%) |
| Disallowed regions | 2 (0.5%) |
Values in parentheses indicate the specific values for the outermost-resolution shell.
Rsym = Σ|I − < I > |/ΣI/.
Rcrys = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree was calculated with 5% of the data excluded from the refinement calculation.
Ramachandran analysis was determined using PROCHECK (Laskowski et al., 1993).
An Overview of the Complex
The overall structure of the complex is shown in Figure 2A. The model of the SRPK1:ASF/SF2 includes residues 69–235 and 475–655, which comprise the kinase core of SRPK1; residues 121–196 and 201–210 of ASF/SF2, which make up the RRM2 and part of the RS1 domains; and, intriguingly, a 2-mer peptide containing a phosphoserine that interacts with a basic site in the kinase (Figure 2A). One molecule of AMP-PNP binds at the typical ATP-binding cleft between the kinase lobes. Within the ternary complex, SRPK1 adopts a conformation similar to that observed in the apo structure, with an rmsd of 0.857 Å over 348 residues of the kinase core. One exception is the glycine (Gly)-rich loop of the substrate-bound kinase, which shows a downward movement toward the large lobe when compared to the apo enzyme. This is not surprising, since the Gly-rich loop forms part of the binding interface between the kinase and substrate.
Figure 2. Overview of the SRPK1:ASF/SF2 Complex Structure.
(A) Overall structure of the complex shown in two orientations. The kinase (SRPK1) and substrate (ASF/SF2) are represented in gray and gold, respectively. The dotted line marks the lack of electron density of four residues connecting the RRM and RS domains of the substrate. A molecule of AMP-PNP lies between a cleft between the small and large lobes of SRPK1. The phosphoserine (P-Ser) shown in gold is located near the P+1 loop of the kinase.
(B) Ribbon diagram of the RRM2 domain of ASF/SF2 (left panel). The right panel shows a canonical RRM fold from the RNP domain of U1 A protein (Nagai et al., 1990). Note that the strand βN is absent in the canonical RRM fold.
(C) Amino acid sequence of the ASF/SF2 fragment used in structure determination and multiple sequence alignment of this fragment with other SR proteins containing two RRMs. The secondary structural elements seen in the structure are assigned to the sequence of the RRM2 domain of ASF/SF2. Residues that are disordered are colored gray. The unique RRM2 SWQDLKD sequence is shaded by a yellow box. The kinase-contacting residues within the RRM2 domain are colored blue. The cysteine residue denoted by green makes a disulfide bond with a symmetry-related substrate molecule. The previously identified docking motif is highlighted by a red box.
The electron density of the substrate is discontinuous at the junction between the RRM2 and RS domains, thus revealing a bipartite mode of interaction. The observed electron density of RRM2 of ASF/SF2 starts at residue 121 and ends at residue 196. Although the RRM2 seems to adopt the canonical RRM fold where two α helices are packed against an antiparallel β sheet, residues 177–182 of ASF/SF2 incorporate into an extra β strand (βN) antiparallel to β4 (Figure 2B, left panel). This extra β strand lies between helix α2 and β4, but unlike other RRMs, where the same loop is folded into a small two-stranded antiparallel β sheet (β3′ and β3″), βN forms an antiparallel β strand with β4 and merges with the β sheet to form a five-strand antiparallel β sheet instead of the four-strand β sheet seen in canonical RRM structures (Nagai et al., 1990) (Figure 2B, right panel). We conclude that RRM2 of ASF/SF2 adopts a topology of β1-α1-β2-β3-α2-βN-β4. The βN strand, along with the conserved SWQDLKD motif, appears to be a common feature of all SR proteins containing two RRMs, which may account for their distinct functional characteristics (Dauksaite and Akusjarvi, 2002, 2004; Tintaru et al., 2007) (Figure 2C).
We have previously described how a substrate-docking peptide binds to the docking groove of SRPK1. The current model of ASF/SF2 shows that our previously described docking motif of ASF/SF2 (residues 191–197), which plays an important role in SRPK1 interaction and the regulation of phosphorylation, actually constitutes the very C terminus of the RRM2 β4 strand (Figures 2B and 2C). The side chains of R191 and K193 that were previously shown to mediate docking interaction are solvent exposed and do not participate in SRPK1 binding in the current structure. Instead, residues 201–210 that correspond to the N-terminal region of the RS1 motif (N′-RS1) bind at the kinase-docking groove. The N′-RS1 peptide of ASF/SF2 adopts an extended conformation that is similar to that of the conformation of the peptide described in our previous SRPK1:peptide model. However, electron densities for the four residues (PRSP) that connect this region to the RRM2 are missing, resulting in a bipartite binding mode by the substrate. Residues 211–219 of the RS1 motif are also disordered in the structure. Nonetheless, the orientation of the N′-RS1 peptide and the distance between S210 to the active site of the kinase (~25 Å) suggest that the serines near the C terminus of the RS1 motif are most likely to occupy the P0 site. As we describe later, the substrate-docking motif is not a rigid element but is mobile. The β4 strand of ASF/SF2 unfolds during phosphorylation and occupies the kinase-docking groove during the late stage of substrate phosphorylation.
An intriguing feature of the current model of the complex is the presence of a dipeptide containing a phosphoserine residue at a basic site near the P+1 loop of the kinase (Figure 2A and Figures S2A and S2B). Since no ATP was introduced throughout the course of purification and crystallization, it raises the question as to how the phosphoserine is produced and from what component of the complex this peptide is derived. We reasoned that trace amounts of ATP, sufficient to partially phosphorylate the substrate, might be present in the AMP-PNP stock that was used for crystallization. Indeed, when we tested the AMP-PNP as a substrate in the kinase reaction containing ASF/SF2 and SRPK1, we noticed a small amount of ASF/SF2 undergoes phosphorylation, and the result was further confirmed by mass spectrometry (Figures S2C–S2E). The mass spectrometry results clearly showed that, under the crystallization condition, one serine of ASF/SF2 is phosphorylated on average. The linear distance between R210, the last substrate residue seen in our structure, and the positively charged surface where the phosphoserine binds (Figure S2B) suggests that the serine that underwent phosphorylation is located at the C terminus of our ASF/SF2 construct. The implications of this interaction will be discussed later.
Interactions between the Kinase and Substrate
AMP-PNP Binding
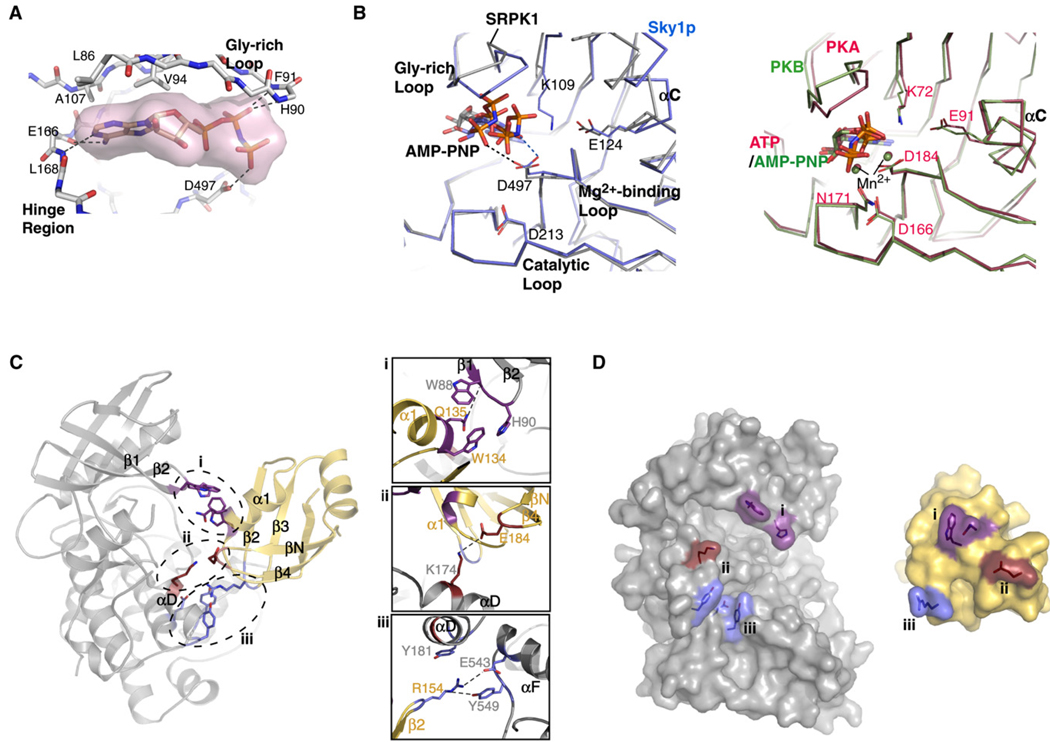
The AMP-PNP lies close to the small lobe of the kinase, contacting residues only from the upper cleft and hinge region of the nucleotide-binding pocket (Figure 3A). N1 of the adenine ring of AMP-PNP and the backbone amide of L165 from the hinge region of the kinase form a hydrogen bond, and three residues (L85, V93, and A106) of the small lobe make van der Waals contacts with the adenine ring. Interestingly, no Mg2+ ion at the nucleotide-binding pocket was observed, although magnesium chloride was included in the crystallization condition. The absence of Mg2+ results in a different orientation of the phosphates within the nucleotide-binding pocket, when compared to other ATP-bound or AMP-PNP-bound kinase structures (Figure 3B). The β-phosphate of the AMP-PNP forms hydrogen bonds with the backbone amides of H90 and F91 of the Gly-rich loop. The γ-phosphate hydrogen bonds to the side-chain carboxyl of D497 of the Mg2+-binding loop directly, instead of through a magnesium, as is typically observed in other kinase:nucleoside triphosphate structures. A similar phenomenon is seen in the structure of Sky1p bound to AMP-PNP (Nolen et al., 2003). In the case of Sky1p, Mg2+ is absent from the cofactor binding site and the side chain of D550 (equivalent to D497 in SRPK1) hydrogen bonds to the nitrogen bridging the βγ-phosphates of AMP-PNP (Figure 3B, left panel). These observations suggest that the SRPKs are sensitive to the structural differences of ATP and AMP-PNP and that their nucleotide-binding chemistry does not resemble that of other kinases (Figure 3B, compare left and right panels). The improper orientation of the phosphate groups appears to be responsible for the displacement of the K109 side chain, thus preventing the formation of the invariant ion pair (K109-E124) seen in all active kinases (Figure 3B, left panel).
Figure 3. Kinase Binding to AMP-PNP and RRM2.
(A) Nucleotide-binding cleft. AMP-PNP, represented by surface rendition, is bound to the cleft between the small and large lobes of SRPK1. Hydrogen bonds between the nucleotide and the kinase are depicted by dotted lines.
(B) Comparison of the orientations of critical residues in the active sites of SRPK1 and other kinases bound to nucleotides. (Left) The structures of SRPK1 (gray) and Sky1p (blue) bound to AMP-PNP. The side chain of residue K109 (part of the invariant K-E ion pair) of the current structure of SRPK1 is not visible, and D497 of the Mg2+-binding loop directly contacts a phosphate. In the case of Sky1p, no Mg2+ is observed, and the aspartate from the Mg2+-binding loop directly hydrogen bonds to the imido group of AMP-PNP. (Right) The structures of PKA bound to ATP (red, PDB ID 1ATP) and PKB bound to AMP-PNP (blue, PDB ID 1O6K). The phosphates of the nucleotides are positioned similarly through Mn2+ in both structures regardless of the type of nucleotide. Important residues at the active site are labeled based on the numbering in SRPK1 (left panel) and PKA (right panel).
(C) Interactions between RRM2 of ASF/SF2 and SRPK1. The three finger-like projections from RRM2 are marked by dotted ovals (left). Important residues involved in the interactions between each of the fingers and the kinase are shown in detail in the insets (right).
(D) Surface rendition of the binding interface between SRPK1 and RRM2 of ASF/SF2. Residues involved in the interactions between the kinase and substrate are colored correspondingly on the binding partner.
RRM2 Contacts Both Kinase Lobes Using Sporadic Contacts
ASF/SF2 projects three “fingers” that touch SRPK1: the α1/β1 loop, the hairpin loop connecting strands β2 and β3, and the loop connecting strands βN and β4. These three contact surfaces engage the kinase at both the small and large lobes from the same side (Figures 3C and 3D). The binding interface between RRM2 of ASF/SF2 and SRPK1 includes the Gly-rich loop, helices αD and αF, and the front of the nucleotide-binding pocket of the kinase and buries 1079 Å of solvent-accessible surface area. The RS1 domain of ASF/SF2 binds to the docking groove in SRPK1 and buries an additional 898 Å. Burial of such large surface area explains the overall high affinity of the complex and illustrates the importance of both RRM2 and RS1 domains in binding affinity. In particular, four residues of RRM2, W134, Q135, R154, and E184, directly contact the kinase (Figure 3C, insets). The conserved SWQDLKD motif is located at the C terminus of the helix α1 of ASF/SF2. This motif caps the helix and contacts the small lobe of SRPK1 at the tip of the Gly-rich loop. In particular, W134 is involved in a stacking interaction with H90 of SRPK1 and appears to stabilize the tip of the Gly-rich loop. Q135, which adopts a strained conformation (φ = 92.6°, ψ = −43.5°), is stabilized by stacking against W88, a unique residue in SRPKs, at the bottom of β1 of the Gly-rich loop and forms a hydrogen bond with its backbone carbonyl (Figure 3C, inset i, and Figure S3). These interactions are particularly intriguing, since the Gly-rich loop is known to be important for the proper orientation of the nucleotide for phosphoryl transfer. Our model suggests that this process could be further regulated through substrate binding. ASF/SF2 also engages the large lobe of the kinase at helix αF. R154 from the turn between strands β2 and β3 of ASF/SF2 projects into a pocket formed by residues from helices αD and αF of SRPK1 and is stabilized by hydrophobic and ionic interactions together with hydrogen bonds contributed by the side chains of Y181, E543, and Y549 from the kinase (Figure 3C, inset ii). Finally, the additional β strand (βN) in ASF/SF2 allows the formation of an extra hairpin loop, creating the third finger-like projection on the same surface of the molecule that serves as the binding surface for SRPK1. In particular, E184 from ASF/SF2 forms a salt bridge with K174 from the large lobe of the kinase (Figure 3C, inset iii). This binding mode between the substrate and kinase implies that relative motions of the lobes around the active site could be restricted by the substrate, an event that is counterintuitive to our current knowledge of substrate phosphorylation (Adams, 2001; Huse and Kuriyan, 2002). Importantly, despite the fact that the front of the nucleotide-binding pocket is covered by RRM2 upon substrate binding, an open channel is present between the spacer regions of SRPK1 and RRM2 (left side of the complex). This suggests that nucleotide binding by the kinase is not obstructed even when the active site is occupied by the substrate peptide, an important feature for a kinase that processively phosphorylates one molecule of substrate multiple times. In all, these observations suggest that the kinase-binding mode adapted by RRM2 of ASF/SF2 is specific and is likely to play an important role in determining the efficiency and processivity of phosphorylation.
The Docking Interaction Is Predominantly Electrostatic in Nature
N′-RS1, which spans residues 201–210 of ASF/SF2, binds SRPK1 in a manner almost identical to our previously described SRPK1:peptide structure (Figure 4A, compare left and right panels). The interactions between the N′-RS1 peptide and kinase are mostly electrostatic and are mediated by the alternate arginines on the peptide with acidic residues from the kinase. Specifically, the side chain of R204 of the N′-RS1 forms ion pairs with residues D564 and E571 from helix αG of SRPK1. The backbone amide of R206 of the substrate forms a hydrogen bond with E571, where its guanidinium group bonds to the backbone carbonyl of L550 and the side chain is additionally stabilized by van der Waals interactions with W606 of the kinase. The last arginine of the peptide (R210) forms an ion pair with D548, and hydrogen bonds to the backbone carbonyls of T546, Y181, and the side chain of N182 of SRPK1. Finally, K615 from the MAP kinase insert of SRPK1 donates a hydrogen bond to the backbone carbonyl of S207 to further stabilize the peptide backbone (Figure 4A, left panel). The most striking feature of this docking interaction is that the very residues that constitute this docking motif become substrates for phosphorylation at later stages of the reaction. This implies that this portion of the substrate moves from the docking groove to the active site. An extensive network of electrostatic interactions involving alternate arginines in the substrate and acidic residues in the kinase may facilitate the substrate movement, where similar functional groups are involved in the bond breakage and formation without any net change of energy. We suggest that these features are the hallmarks of processive phosphorylation.
Figure 4. RS Domain and Phosphoserine Binding to Kinase.
(A) The RS peptide from ASF/SF2 binds to the SRPK1 docking groove in an extended conformation. (Left) The docking peptide extends from residue 201 to 210. Residues R204, R206, S207, and R210 of the substrate and Y181, N182, T546, D548, L550, E571, D564, W606, and K615 of the kinase are involved in the binding interaction. The side chain of R208 is disordered, and only the main chain and Cβ atom are modeled. (Right) A 9-mer peptide, derived from the phosphorylation site of the yeast SR-like protein Npl3p, is involved in similar interactions with the kinase-docking groove, as described previously (Ngo et al., 2005). Hydrogen bonds are depicted by dotted lines.
(B) Comparison of phosphor-interacting sites in SRPK1 and other kinases. (Top) The phosphoserine (P-Ser) docks to a basic (P+2) pocket of SRPK1. The location of the pocket in the context of the entire kinase is shown on the left. Three basic residues from two different secondary structural elements (P+1 loop and helix αG) converge, forming a positively charged pocket (right). (Middle) Phosphothreonine (T197) of PKA stabilizes a basic cluster formed by R165, K189, and H87 and is important to the activation of the kinase (Knighton et al., 1991). (Bottom) A phosphate ion binds to a basic pocket at the activation segment of GSK3β and mimics the phosphate group of a primed phosphoserine in the substrate that facilitates substrate binding and activation of the kinase (ter Haar et al., 2001).
Substrate Binding at a Basic Site
As previously mentioned, the phosphate of a phosphoserine residue interacts with the basic site formed by residues R515, R518, and R561 of SRPK1 (Figure 4B, top panel, and Figure S2A). The same site was originally referred to as the P-2 pocket while describing the Sky1p and SRPK1 apo structures (Ngo et al., 2007; Nolen et al., 2001). In contrast to our previous assumption that phosphorylation occurs from the N′ to C′ direction of the substrate, our structure suggests that phosphorylation must occur in a C′ to N′ direction. We rename this site on the kinase the P+2 site, as phosphorylation of the RS substrate peptide in the C′ to N′ direction is likely to position the P+2 residue to this site. Similar highly positively charged pockets have been observed in other kinases, where they are involved in the interaction with a phosphorylated residue of the substrate, as in GSK3β, or a phosphorylated residue in the activation loop of kinase itself, as in PKA (Figure 4B, middle panel). A positively charged pocket in GSK3β interacts with the primed residue located at the P+4 position in the substrate to stabilize the active conformation of the kinase and thus facilitate phosphorylation (Figure 4B, bottom panel). We suggest that the phosphoserine in our structure represents a “primed residue” (Figure S2B). We also suggest that SRPK1 is responsible for priming its own substrate in contrast to substrate priming by different kinases, as observed in GSK3β binding (Dajani et al., 2001; ter Haar et al., 2001).
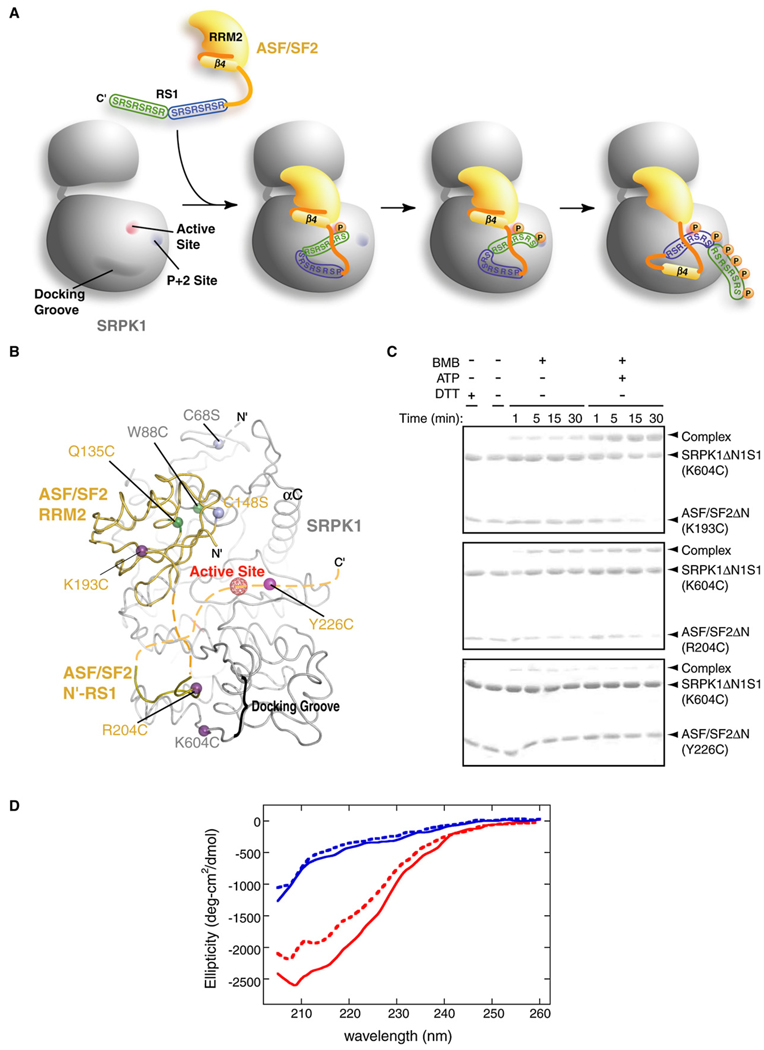
The Docking Motif of ASF/SF2 Slides during Phosphorylation
As mentioned above, interactions between the kinase-docking groove and N′-RS1 reveal an intriguing feature of substrate recognition by an enzyme, as this docked region must also be phosphorylated during the course of catalysis. Moreover, our previous experiments suggested that another docking site in the substrate is located N-terminal to the RS domain, which constitutes a β strand of RRM2. A plausible explanation to this conundrum is that the substrate translocates from the kinase-docking groove to the active site in a sequential manner during catalysis (Figure 5A).To examine if the substrate has a mobile docking motif that threads through the kinase-docking groove, we have carried out mutation-directed disulfide crosslinking experiments. First, the surface cysteine residues were mutated to serines (SRPK1[C68S] and ASF/SF2ΔN[C138S]) to prevent aberrant crosslinking of the complex (Figure 5B). No crosslinked product was observed when crosslinking experiments were carried out on this surface cysteine-free complex (Figure S4A). All cysteine mutations were then created in the background of this surface cysteine-free complex. A pair of proximal residues (SRPK1[W88C] and ASF/SF2ΔN[Q135C]) was mutated to cysteine, and crosslinking was done in the presence of the crosslinker BMB (1,4-bismaleimidobutane). As expected, crosslinked complex was observed in this positive control (Figure S4A). Cysteines were then placed at strategic sites in both the cysteine-free kinase and the substrate. Three independent cysteine mutations were created in ASF/SF2ΔN (at positions 193, 204, and 226) and one within the docking groove of SRPK1 (at position 604) (Figure 5B). All kinase and substrate mutants are functional in phosphorylation activity (Figures S4B and S4C). Residues 204 of ASF/SF2 and 604 of the kinase are in relatively close proximity and therefore they should crosslink in the bound state observed in our structure. Residue 193 of ASF/SF2 is part of the alternate docking motif identified in our earlier studies and lies at the end of β4 of RRM2, which is far apart from the kinase-docking groove in our structure. For this residue to crosslink with K604C of kinase, the β strand must unfold and move to the kinase-docking groove during processive phosphorylation (Figure 5A). Therefore, crosslinked product should be observed in the presence of ATP. Residue 226 is expected to be located near the active site, away from the kinase-docking groove. The cysteine at this position may only crosslink to the kinase, if at all, at low efficiency and in an ATP-independent manner. The crosslinker BMB was added in the presence and absence of ATP to all three purified complexes, SRPK1(K604C):ASF/SF2(K193C), SRPK1(K604C):ASF/SF2(R204C), and SRPK1(K604C):ASF/SF2(Y226C). The reactions were incubated for indicated lengths of time, followed by SDS-PAGE analysis (Figure 5C). Our results show that R204C and K193C crosslink to the kinase in the presence of ATP (Figure 5C, compare middle and top panels). R204C, but not K193, showed weak crosslinking in the absence of any nucleotide. To further test the phosphorylation-dependent crosslinking by K193C, we have carried out crosslinking reactions in the presence of ADP. As predicted, presence of ATP, but not ADP, was essential for crosslinking (Figure S4D). Finally, as expected, Y226C crosslinks to the kinase only minimally (Figure 5C, bottom panel). These results suggest that the previously identified docking motif (residues 191–196) docks onto the kinase-docking groove only at the end of the phosphorylation reaction, whereas parts of the RS1 motif bind the docking groove before initiation of the reaction.
Figure 5. Cysteine Crosslinking Suggests Unfolding of β4 of RRM2 during Phosphorylation.
(A) A model of substrate binding, priming, and processive phosphorylation as occurs during phosphorylation of ASF/SF2 by SRPK1. In order for the RS1 domain to be fully phosphorylated, the substrate needs to translocate from the docking site to the active site during reaction, which eventually leads to the unfolding of β4 from the core of ASF/SF2 RRM2.
(B) Locations of cysteine mutations. The locations of the three cysteine mutations in the substrate relative to K604C of SRPK1 located in the kinase-docking groove are indicated b1y purple balls (including the putative Y226C). Green balls indicate the residues mutated for the positive control. Pink balls represent the position of the background cysteine to serine mutations (see the Experimental Procedures).
(C) Disulfide crosslinking experiments. SDS-PAGE analysis of the crosslinking reactions between ASF/SF2 (K193C):SRPK1 (K604C) (top panel), ASF/SF2 (R204C):SRPK1 (K604C) (middle panel), and ASF/SF2 (Y226C):SRPK1 (K604C) (bottom panel). Reactions were carried out in the presence and absence of ATP to observe phosphorylation-dependent crosslinking. Efficient crosslinking between K193C from β4 of ASF/SF2 and K604C of the SRPK1 docking groove requires the presence of ATP.
(D) Phosphorylation impacts the secondary structure of ASF/SF2. ASF/SF2 (red) and the RS domain (188–248) (blue) are incubated with SRPK1 in the presence of AMP-PNP (solid lines) and ATP (dotted lines), and CD spectra are recorded between 205 and 260 nm.
To further confirm the unfolding event, we have carried out CD experiments to monitor structural transition of the complex. We observe a clear shift from a β sheet to a random coil in the presence of ATP, but not AMP-PNP, in the SRPK1:ASF/SF2 complex, while no such transition for the RS domain alone is observed (Figure 5D). Together, these observations suggest that β4 of RRM2 unfolds, thus allowing this segment to convert into a substrate-docking motif. It is consistent with a recent study that suggests regions of ASF/SF2 including strands βN and β4 are intrinsically disordered (Haynes and Iakoucheva, 2006). Unfolding of substrate has also been implicated for phosphorylation of eIF2α by the kinase PKR (Dar et al., 2005).
Mechanisms of Priming, Processive Phosphorylation, and Product Dissociation
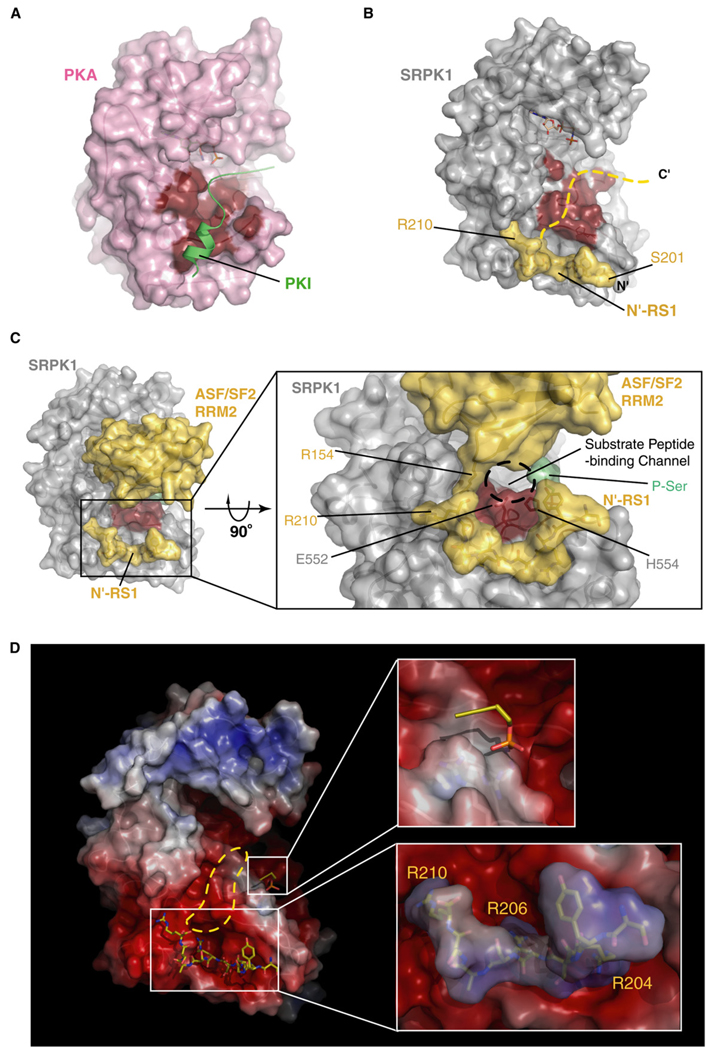
Based on the above observations, we propose a mechanism of substrate binding and phosphorylation by SRPK1. Comparison of the SRPK1:ASF/SF2 and PKA:PKI complexes reveals that the active-site peptide of the RS domain can be positioned along a path similar to that taken by PKI in the PKA:PKI complex (Figures 6A and 6B) (Knighton et al., 1991). Furthermore, despite the fact that the RRM2 of ASF/SF2 contacts SRPK1 at the front and covers a large accessible surface area, a hollow channel exists between the two proteins, which can accommodate the substrate peptide (Figures 6B and 6C). The linear distance between the active site and the Cα of the last residue of the docking peptide (Cα of residue 210 in the current structure) is nearly 25 Å, which can accommodate approximately seven residues. Together with the docking peptide, approximately 17 residues of ASF/SF2 interact with the kinase, spanning the beginning of the docking groove to the active site. A striking feature of this interaction is that the entire substrate-binding channel, extending from the docking groove to the active site, is highly negatively charged and can easily guide the binding of the entire basic peptide of the RS domain (Figure 6D). Based on these assumptions, we suggest that the entire RS1 domain can bind to the kinase and be phosphorylated through a threading manner, where the RS dipeptides at the C-terminal end occupy the active site and get phosphorylated first, while those at the N terminus occupy the docking groove. At a later stage of catalysis, the N-terminal RS dipeptides are displaced by the alternate docking motif (191–198) and move to the active site for the phosphoryl transfer (Figure 5A). Therefore, the complex in our structure represents an intermediate state of substrate phosphorylation, in which residues 201–219 occupy the expanded kinase groove (although we do not see the density of segment-spanning residues 211–219). In the case of full substrate, the phosphorylation initiation site is likely S225 or S227. The structural model is consistent with the recent observation from our biochemical experiments that the directionality of ASF/SF2 phosphorylation is from C′ to N′ (Ma et al., 2007).
Figure 6. Substrate Binding at the Active Site.
(A and B) Comparison of PKI-bound PKA and docking peptide-bound SRPK1. Surface renditions of the substrate-binding grooves in both kinases are highlighted in red. The dotted line in (B) is modeled based on the orientation of PKI upon binding to PKA and suggests the possible path of the substrate peptide of ASF/SF2 to the SRPK1 active site.
(C) Primed phosphoserine binding to the P+2 site forms a narrow substrate-binding channel. The complex is shown in surface rendition with the same view as in (B), except the entire complex is shown including the RRM2 of ASF/SF2. (Inset) The view from the bottom of the complex clearly shows the presence of the phosphoserine further constricts the narrow substrate-binding channel formed by the RRM2 of ASF/SF2 and E552 and H554 of SRPK1.
(D) SRPK1 solvent-accessible surfaces are colored by electrostatic potential (±7 kT/e) computed by APBS (Baker et al., 2001). (Upper inset) P-Ser interacts with a positively charged island (P+2 site) surrounded by a highly negatively charged environment. (Lower inset) Electrostatic potential of N′-RS1 is calculated independently and docked onto the docking groove in SRPK1. Highly negative potential within the docking groove (depicted by red) and positive potential of the N′-RS1 (depicted by blue) confirms that the docking interaction is predominantly electrostatic. Extension of the high negative charge potential to the active site implies that the rest of the highly basic peptide extends along this path.
The phosphoserine bound to the P+2 site on the kinase appears to be the result of the substrate “priming” by the kinase. We suggest that the first serine in the substrate after phosphorylation becomes a P+2 residue when the phosphoryl transfer takes place on the second serine. This phosphoserine (P+2) interacts strongly with the P+2 site of the kinase serving as a “prime site.” This priming event is likely to enhance the rate of the next phosphorylation step. After each round of phosphorylation, a new P+2 phosphoserine displaces the old one with enhanced catalytic efficiency. This mode of priming phosphorylation suggests that the rate-limiting step in the processive phosphorylation of ASF/SF2 might be the catalysis of the first serine. Moreover, the side chains of two residues, E552 and H554, unique in the SRPKs are projected into the substrate-binding channel and constrict the channel near the phosphoserine-binding site. The ionic interaction between the phosphoserine and the P+2 pocket might be responsible for fixing the orientation of these side chains and prevent backtracking of the phosphorylated product through the constricted narrow channel (Figure 6C). Consistent with our model, preliminary mutational studies show that alteration of the basic residues in the P+2 site changes the mode of phosphorylation from processive to distributive.
How does the substrate move toward the active site after each round of phosphorylation? Several contributing factors might facilitate the substrate movement. As mentioned earlier, the entire interaction path between the RS domain and the kinase is electrostatic in nature. It is possible that, during each step of catalysis, energy derived from ATP hydrolysis is utilized not only in bond formation but also to carry out the mechanical task in the movement of the substrate. It is also possible that the movement is linked to conformational changes at the active site during phosphorylation. Such conformational changes might link displacement and binding cycles at the P+2 site by phosphoserine and substrate translocation.
Dissociation of the fully phosphorylated product from the kinase also invokes an intriguing mechanism. In order for the phosphorylation of the last serines to occur (including S205 and neighboring ones), unfolding of the β4 strand from the rest of RRM2 of ASF/SF2 should occur for the last docking site (191–198) to interact with the kinase (Figure 5A). Unfolding, along with further stretching of the RRM2, may facilitate dissociation of the RRM2 from the kinase. Finally, increasing accumulation of negative charge is also expected to stimulate dissociation through charge repulsion. We suggest that the substrate loses affinity with progressive phosphorylation. In vivo, dissociation may also be favored by the presence of transportin-SR2, which binds the phosphorylated RS domain and imports ASF/SF2 to the nucleus (Lai et al., 2001). Recent studies have suggested direct involvement of the SWQDLKD motif in RNA recognition (Dauksaite and Akusjarvi, 2004; Tintaru et al., 2007). Therefore, competition for this recognition surface by both RNA and SRPK1 might be critical for p-ASF/SF2 release from the kinase.
In conclusion, the X-ray structure of the SRPK1:ASF/SF2 complex reveals new insight into the mechanisms of substrate selection, protection, and phosphorylation by a constitutively active kinase. The structure has helped us to identify three important functions of SRPK1 that are unique to a protein kinase: self priming of the substrate, substrate translocation, and substrate unfolding during phosphorylation. Interestingly, by interacting with a potential phosphorylation site, the kinase protects the substrate from being aberrantly and partially phosphorylated by other kinases. This substrate recognition and phosphorylation mechanism is apparently important for ASF/SF2 to carry out its diverse biological functions, which include splicing, mRNA export, translation, and maintaining genomic stability.
EXPERIMENTAL PROCEDURES
Recombinant SRPK1:ASF/SF2 Complex Formation
His-SRPK1ΔN1S1 and his-ASF/SF2(BD) were first purified separately as described previously (Aubol et al., 2003). SRPK1 (12 mg) and 16 mg of ASF/SF2 were mixed together in 100 ml buffer containing 500 mM KCl, 20 mM Tris-HCl (pH 7.5), 20% glycerol, 6 M urea, and 3 mM DTT. The mixture was mixed by stirring for 30 min at 4°C. The mixture was then dialyzed for 6 hr against 2 L of buffer containing 500 mM KCl, 20 mM Tris-HCl (pH 7.5), 15% glycerol, and 3 mM DTT followed by another 6 hr dialysis in buffer containing 500 mM KCl, 20 mM Tris-HCl (pH 7.5), 10% glycerol, and 3 mM DTT. A final dialysis was performed in buffer containing 500 mM KCl, 20 mM CAPSO (pH 9.5), 5% glycerol, and 1.5 mM DTT. The dialysate was then concentrated, and the His tag of the refolded protein was then removed by adding 0.2 NIH units (1 cleavage unit) of thrombin (Sigma-Aldrich) per milligram of complex. The reaction was allowed to proceed at room temperature for 16 hr and quenched by adding 1 mM EDTA. The untagged complex was then loaded on the Superdex 200 size-exclusion column equilibrated in 500 mM KCl, 20 mM CAPSO (pH 9.5), 5% glycerol, and 1 mM DTT. The pooled peak fractions were concentrated to ~11 mg/mL, aliquoted, and stored at −80°C.
Crystallization of SRPK1ΔN1S1:ASF/SF2(BD):AMP-PNP Complex
For crystallization, the SRPK1ΔN1S1:ASF/SF2(BD) complex was mixed at 6 mg/mL with 4 mM AMP-PNP and 8 mM MgCl2. Needle-shaped crystals were obtained by the hanging-drop vapor diffusion at 18°C using the Hampton Screen (Hampton Research). Optimal condition for crystallization was found to be 100 mM sodium citrate (pH 5.6), 200 mM sodium acetate, 5% MeOH or EtOH, and 8% PEG 5000MME in the reservoir. In order to improve the diffraction quality, the crystals were then dehydrated and cryoprotected by dialysis overnight against 100 mM sodium citrate (pH 5.6), 200 mM sodium acetate, 5% MeOH or EtOH, 15% PEG 5000MME, and 25% (V/V) ethylene glycol. All dialyzed crystals were flash frozen in liquid nitrogen prior to data collection.
Data Collection of SRPK1ΔN1S1:ASF/SF2(BD):AMP-PNP Complex Crystal
X-ray diffraction data of the ternary complex crystals were collected using a MAR CCD detector at GM/CA-CAT synchrotron beamline ID-23 of the Advanced Photon Source (APS) at Argonne National Laboratory. A complete data set of 180° was collected at 1° oscillations. The X-ray data was processed using program HKL2000 (Otwinowski, 1993). The complex crystals belong to orthorhombic space group I222 with unit cell dimensions a = 57.406 Å, b = 117.525 Å, c = 193.554 Å, a = b = c = 90°. Each asymmetric unit contains one complex molecule.
Structure Solution of SRPK1ΔN1S1:ASF/SF2(BD):AMP-PNP Complex
The structure of SRPK1ΔN1S1:ASF/SF2(BD):AMP-PNP ternary complex was solved by molecular replacement using the program AMoRe and the structure of apo SRPK1ΔNS1 as the search model, yielding clear rotation function and translation peaks (Navaza, 2001). After rigid-body refinement using CNS, Fo−Fc electron density map calculated with the model revealed large positive peaks and readily interpretable density for the ASF/SF2 and AMP-PNP. A polyalanine backbone of ASF/SF2 was manually built by using the NMR solution structure of ASF/SF2 (RRM2) (Protein Data Bank [PDB] accession code 1×4C) as a guide. The two lobes of the SRPK1 and the polyalanine backbone of ASF/SF2 were allowed to move independently during the first round of rigid-body refinement. Side chains of ASF/SF2 and a molecule of AMP-PNP were then fitted in, and the complex structure was refined with several cycles of manual refitting and refinements using REFMAC (Murshudov et al., 1997). The current model of the ternary complex includes residues 69–235 and 475–655 of the kinase, residues 121–196 and 201–210 of the ASF/SF2, a 2-mer peptide containing a phosphoserine, and one molecule of AMP-PNP.
GST Pull-Down
GST pull-down assays were done by incubating equal amounts of GST-RRM1 with wild-type and different truncated mutants of SRPK1 in a buffer containing 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, and 0.5% Triton X-100. Samples were washed extensively after pull-down and separated by SDS-PAGE and visualized by Coomassie blue staining.
Chemical Crosslinking
A surface-exposed cysteine in wild-type kinase (C68) was converted to serine to prevent aberrant crosslinking. Two independent cysteine mutations in the kinase (W88C and K604C) were created in the surface cysteine-free background. Similarly, ASF/SF2 contains a surface-exposed cysteine at position 148, which was mutated to a serine. All four cysteine mutations in the substrate (Q135C, K193C, 204C, and Y226C) were created in the C148S background. All mutants were made by using mutagenic oligonucleotides and confirmed by DNA sequencing. Mutant proteins were expressed and purified by using methods as described earlier for the wild-type complex. Each protein complex was diluted to 2 mg/ml using buffer containing 500 mM NaCl, 5 mM Tris (pH 7.5), 5% glycerol, and 10 mM DTT. After the complexes were incubated on ice for 1 hr, they were exchanged with the crosslinking buffer (500 mM NaCl, 5 mM Tris [pH 7.5], and 5% glycerol) using desalting spin columns (Pierce). The protein complexes were further diluted to 0.4 mg/ml with crosslinking buffer in order to be used for the following reactions: (1) 0.03 mM BMB (Pierce), (2) 0.03 mM BMB + 10 µM (or 50 µM) ATP or 50 µM ADP + 4 mM MgCl2, (3) 0.03 mM BMB + 4 mM MgCl2, (4) 10 mM DTT + DMSO (equal volume with the amount of BMB added in the reactions 1–3), and (5) DMSO. The reactions were carried out at 4°C. Equal-volume aliquots (reactions 1–4) were removed at each time point and quenched by 4× reducing SDS dye. The reaction 5 was quenched by 4× nonreducing SDS dye. The reactions were resolved by SDS-PAGE and analyzed by Coomassie blue staining.
Circular Dichroism Spectroscopy
ASF/SF2 (2 µM) and RS(188–248) (4.8 µM) were mixed with SRPK1 (90 nM) and a 12-fold molar excess of ATP (24 and 60 µM) in 5 mM acetate buffer (pH 5.5). After a 60 min reaction time, the samples were diluted in 5 mM acetate buffer (pH 5.5) and concentrated three times, using an Amicon Ultra 10K MWCO centrifugal filter. The circular dichroism(CD) spectra were recorded on an Aviv Circular Dichroism Spectrophotometer Model 202 purged with nitrogen using 2 mm thick quartz cells and 0.8 ml sample volume. Spectra were recorded from 205–260 nm, and appropriate baseline buffer spectra was collected and subtracted. Data points were converted from millidegrees to units of molar ellipticity (deg*cm2/dmole). Unphosporylated proteins were mock phosphorylated with AMP-PNP and processed in the same manner as the phosphorylated samples.
Mass Spectrometric Analysis of ASF/SF2 in the Presence of SRPK1 and AMP-PNP
In vitro phosphorylation experiments with the protein complex and AMP-PNP (Sigma) stocks were used for crystallization. Three distinct reaction conditions were studied. Buffer concentrations for all reactions were consistent with those used for crystallization, except that PEG 4000 was omitted. Reactions were incubated at room temperature for 1 hr, quenched with 5% acetic acid, and purified with C-18 Zip-Tips (Millipore). Mass spectrometric analysis of the reactions was carried out using high-resolution Fourier transform ion cyclotron resonance mass spectrometry (FC-ICRMS). Data were averaged with Qualbrowser and processed with Extract.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Tom Huxford and Simpson Joseph for carefully reading the manuscript and for suggestions. We also thank De-Bin Huang, Susan Taylor, Partho Ghosh, and Natarajan Kannan for suggestions. The work is supported by grants from the National Institutes of Health. J.C.K.N. was supported by the Cell Growth and Tumor Biology Training Grant.
Footnotes
ACCESSION NUMBERS
The coordinates of SRPK1-ASF/SF2 complex have been deposited in the Protein Data Bank under the accession number 3BEG.
SUPPLEMENTAL DATA
Supplemental Data include four figures and can be found with this article online at http://www.molecule.org/cgi/content/full/29/5/563/DC1/.
REFERENCES
- Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc. Natl. Acad. Sci. USA. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi RM, Nebreda AR. Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions. Biochem. J. 2003;372:1–13. doi: 10.1042/BJ20021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Dauksaite V, Akusjarvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- Dauksaite V, Akusjarvi G. The second RNA-binding domain of the human splicing factor ASF/SF2 is the critical domain controlling adenovirus E1A alternative 5′-splice site selection. Biochem. J. 2004;381:343–350. doi: 10.1042/BJ20040408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994a;369:678–683. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. USA. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM. Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thorton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu X-D, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J. Mol. Biol. 2007;376:55–68. doi: 10.1016/j.jmb.2007.08.029. Published online August 21, 2007. 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Gullingsrud J, Giang K, Yeh MJ, Fu XD, Adams JA, McCammon JA, Ghosh G. SR protein kinase 1 is resilient to inactivation. Structure. 2007;15:123–133. doi: 10.1016/j.str.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Nolen B, Yun CY, Wong CF, McCammon JA, Fu XD, Ghosh G. The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat. Struct. Biol. 2001;8:176–183. doi: 10.1038/84178. [DOI] [PubMed] [Google Scholar]
- Nolen B, Ngo J, Chakrabarti S, Vu D, Adams JA, Ghosh G. Nucleotide-induced conformational changes in the Saccharomyces cerevisiae SR protein kinase, Sky1p, revealed by X-ray crystallography. Biochemistry. 2003;42:9575–9585. doi: 10.1021/bi0344331. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. DENZO: An Oscillation Data Processing Program for Macromolecular Crystallography. New Haven, CT: Yale University Press; 1993. [Google Scholar]
- Pellicena P, Kuriyan J. Protein-protein interactions in the allosteric regulation of protein kinases. Curr. Opin. Struct. Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. USA. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol. Cell. 2006a;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Shi Z, Resing KA, Ahn NG. Networks for the allosteric control of protein kinases. Curr. Opin. Struct. Biol. 2006b;16:686–692. doi: 10.1016/j.sbi.2006.10.011. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, Wilson SA. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756–762. doi: 10.1038/sj.embor.7401031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKII-delta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith EJ. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.