Abstract
The development of molecular pathologic components in epidemiologic studies offers opportunities to relate etiologic factors to specific tumor types, which in turn may allow the development of better overall risk prediction and provide clues about mechanisms that mediate risk factors. In addition, this research may help identify or validate tissue biomarkers related to prognosis and prediction of treatment responses. In this mini-review, we highlight specific considerations related to incorporation of pathology in epidemiologic studies, using breast cancer research as a model. Issues related to ensuring the representativeness of cases for which research tissue is available and understanding limitations resulting from variable procedures for tissue collection, fixation and processing are discussed. The growing importance of molecular pathology in clinical medicine has led to increased emphasis on optimized tissue preparation, which should enhance this type of research. In addition, the availability of new technologies including tissue microarrays, image scanning and automated analysis to achieve high throughput standardized assessment of immunohistochemical markers, and potentially other assays, is enabling consistent scoring of a growing list of markers in large studies. Concurrently, methodological research to extend the range of assays that can be performed on fixed tissues is expanding possibilities for molecular pathologic studies in epidemiologic research.
Background
Large epidemiologic studies offer important opportunities to relate etiologic exposures to cancer subtypes defined by molecular pathologic characteristics. Such investigations may identify specific relationships between risk factors and tumor subtypes, enable improved overall risk prediction, reveal mechanisms that mediate risk factors and discover and validate biomarkers for early detection, prognosis or prediction of treatment responses1. Analyses of cancer incidence and mortality rates stratified by tumor subtypes may provide etiologic clues, highlight racial/ethnic disparities and improve surveillance2–4. The promise of this research is potentiated by a National Cancer Institute sponsored initiative to optimize tissue collection and storage through the Biorepository Research Network5. Despite enthusiasm for using molecular pathology in epidemiologic studies, this approach presents some specific challenges. In this mini-review, we discuss key principles related to adding pathology components to epidemiologic studies, drawing on examples from breast cancer research.
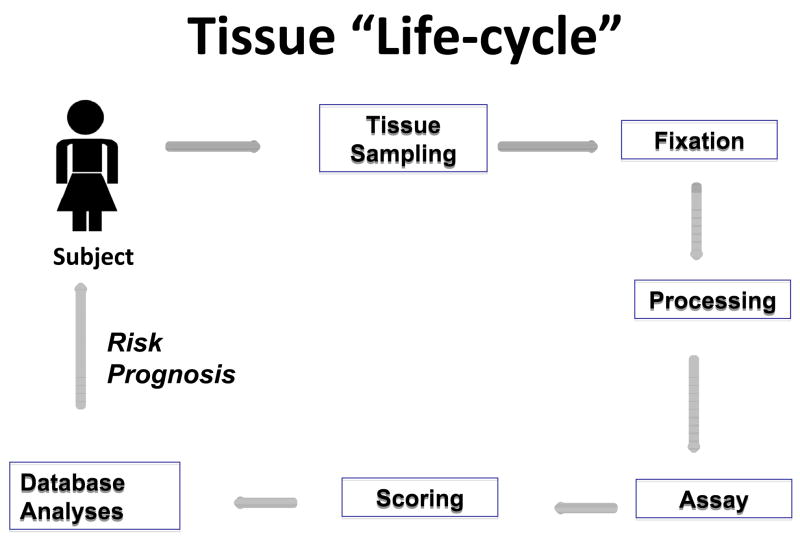
An overview of steps involved in developing the pathology components in epidemiologic studies is presented in figure 1. As described below, internal and external validity and quality assurance are important considerations throughout the process and technical improvements are anticipated to meet growing demands for use of molecular assays in clinical medicine.
Figure 1.
The patients for whom research tissues are available in epidemiologic studies are a function of multiple factors related to diagnosis and treatment. These practices may vary between populations, geographic regions and calendar period and by patient and tumor features. Practices of tissue sampling and preparation for diagnosis may vary with the type of procedure used to collect the sample, the clinical context and institutional protocols. All of the above may affect assay performance. In large studies, efforts to standardize, optimize and automate performance and scoring of assays are important to control costs, speed completion of studies and minimize variability. Computer infrastructure is important throughout the “cycle”, to provide tracking, data management and facilitate downstream statistical analysis.
Availability of Tissue: Ensuring Representativeness of Samples
Ensuring the representativeness of tissue specimens collected in epidemiologic investigations presents concerns that differ somewhat from those of biological specimens that researchers obtain directly from patients. Specifically, differential patterns of screening, diagnosis and treatment may be related to both patient and tumor characteristics and may affect the associations between the two. For example, in breast cancer, mammographically detected tumors differ from cancers that are either missed or found without screening6,7. Radiologically-guided biopsies performed in outpatient facilities may remove small tumors8, potentially skewing availability of tissues in hospital-based studies toward larger cancers. Digital mammography is more sensitive than film-based techniques among women with dense breasts9, and high density is related to younger age, specific exposures, higher growth fractions and interval cancers10. Similarly, use of magnetic resonance imaging for screening high-risk women may lead to diagnosis at younger ages and detection of smaller tumors, while paradoxically demonstrating more frequent multicentricity11.
Access to tissue is related to tumor size, which varies by population, risk factors and subtype. Use of neoadjuvant therapy either by necessity to render large tumors operable, or by preference, may limit availability of untreated specimens12, though providing opportunities to analyze risk factors for breast cancer phenotypes, defined by a combination of marker expression and therapeutic response. Some tumors detected by screening may be too small to use for research. Both patient characteristics (e.g. younger age, premenopausal status and obesity) and tumor pathology (e.g. higher grade and marker expression) may be related to larger tumor size, suggesting the need to consider pathologic factors in risk analyses13–15.
Impact of Study Design
Prospective studies may permit collection of research tissues according to protocols, but diagnostic requirements and batch processing for histology may be limiting. Tissue that is not needed for diagnosis, such as fat, normal or excess tumor may provide research opportunities, but require collaborations with pathology laboratories, especially if frozen tissue is sought. Developing a “virtual tumor bank” of fixed tissue may ensure later availability if prospective assembly is impossible. Retrospective studies may permit efficient targeted collection of tissues related to known outcomes, enabling the inclusion of pathology in “nested” retrospective case-control studies assessing questions such as risk of progression of cancer precursors16. The Surveillance, Epidemiology and End Results program has established Residual Tissue Repositories that can provide access to discarded blocks that are linked to registry data, which can potentially permit evaluation of population-based rates by molecular characteristics17; however, retrieval of representative tumors from a high percentage of cases is required to impute data for missing tumors. In retrospective studies, access, retrieval, variable routine processing, and degradation of tissues over time present challenges.
Tissue Preservation and Fixation
Optimized tissue preservation and fixation is critical for tissue-based research; unfortunately, factors that impact tissue quality such as prior biopsy, intra-operative hypoxia and ischemia; time to fixation; type, concentration and adequacy of fixative; and method of paraffin block preparation are poorly standardized and may introduce unanticipated variation18. Collection of fresh or frozen tissues averts some of these difficulties, but is expensive and infeasible in most epidemiologic studies.
Fortunately, optimizing tissue fixation to maximize its utility is a major area of research19–22; however, unless improved practices are widely adopted, such strategies will not be available for routine diagnostic specimens or archived tissues. Advances will allow improvements in immunohistochemistry and potentially wider application of proteomics, spectroscopy, DNA and RNA assays19–22. Given that histology laboratories employ batch processing, the pace of clinical advances may be rate limiting for epidemiologic research. Fixation in neutral buffered formalin is anticipated to remain as the gold standard because it provides morphologic appearances familiar to diagnosticians.
Assembling Tissues for Research
Most pathology reports specify blocks and their corresponding slides using designations linked to the diagnostic impression of the tissue at dissection. However, microscopic examination is needed to confirm the presence of histologic targets of interest and localize them within blocks. Off-site storage may increase time and costs for retrieval. Older tissues have declining value for some assays23 and are frequently discarded after a specified time period, whereas access to newer blocks may be limited by clinical demands. Applying bar code labels is helpful for tracking, but labels often adhere poorly to blocks, which has prompted some laboratories to embed radio tags into blocks.24
Tissue Microarrays for High Throughput Analysis
Tissue microarray (TMA) construction has been reviewed elsewhere. 24–28 Briefly, TMAs are constructed by removing microscopically identified tissue targets from routinely prepared tissue (“donor”) blocks and transferring them into a new (”recipient”) TMA block in pre-configured arrangements. Sections of the resulting TMA contain cores of many tumors, enabling rapid, cost-effective, high throughput performance of assays (figure 2). Selected, concerns specific to molecular epidemiology are discussed below.
Figure 2.
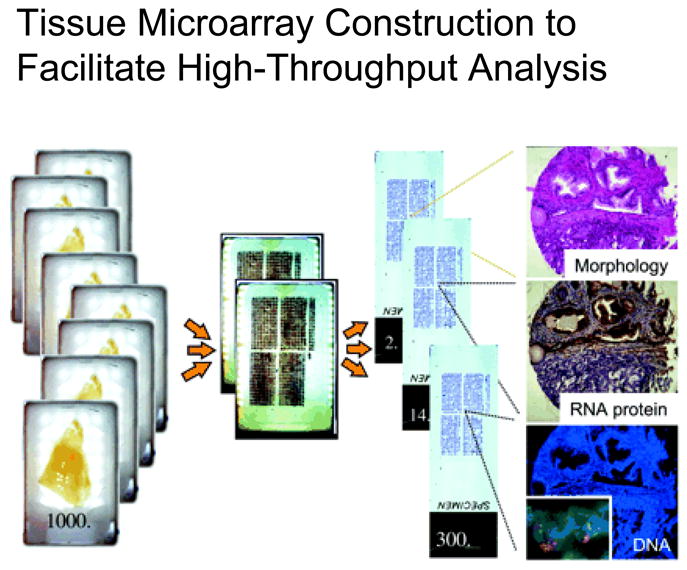
Tissue microarrays (TMAs) facilitate rapid, cost effective high throughput analysis of large tumor sets. Target areas within individual donor blocks are identified by examining corresponding hematoxylin and eosin stained tissues sections. Cylindrical cores are removed with semi-automated devices from each donor block and arrayed in a precise matrix in the recipient block to create the tissue microarray. Assays performed on single sections cut from TMA blocks enable the batched analysis of multiple tumors. Reprinted with permission: Kallioniemi O-P, et al, Tissue microarray technology for high-throughput molecular profiling of cancer, Human Molecular Genetics, 2001, volume 10, issue 7: 657–6227.
Designing Tissue Microarrays
Optimal TMA design varies by project aims and tumor site. Considerations include: 1) numbers of cores per target; 2) core diameter (typically 0.6 mm or 1.0 mm); 3) placement of replicate cores in single or multiple blocks; 4) total number of cores per block; 5) core arrangement and 6) segregation of cores from tumors with specified characteristics in dedicated blocks (Table 1).
Table 1.
Tissue Microarrays: Selected Design Considerations
| Design Consideration | Pros | Cons |
|---|---|---|
| Increased cores/sample | Reduces missing data Captures heterogeneity | Increases effort |
| Larger core diameter | Better target representation | Fewer cores/TMA |
| Replicate cores separated in different TMAs | Protects material if one TMA block lost | Need to ensure consistency |
| More cores/TMA | Reduces assay costs | Mapping more difficult |
| Grouping cores within TMA blocks by subject or tumor characteristics | Conserves material for relevant assays | Need to ensure consistency |
Redundant sampling reduces missing data per subject secondary to loss of cores, resulting from tumor depletion in deeper sections, non-adherence of sections or loss during vigorous antigen retrieval. However, high-level redundancy adds preparation time and cost of preparation and is impossible for small targets. Theoretically, replicate sampling should improve scoring accuracy by accounting for heterogeneity in marker expression. However, as has been shown for ER, there is probably more variation in expression between than within blocks because sampling more distant topographic regions is more likely to capture biological heterogeneity (e.g. hypoxic tumor center vs. growing edge).29 In the end, useful markers must be robust to sampling variability in research and clinical applications.
Using larger cores, facilitates histologic characterization and minimizes missing results, but as with multiplicity of cores, target size and efficiency are limiting. For tumors with low cellularity (e.g. lobular carcinoma), larger cores may improve representation, throughout the length of cores. TMA cores are often targeted to cellular tumor areas, which theoretically may bias marker levels (e.g. proliferation or cell cycling). Caution may be required when using larger needles to avoid damaging blocks.
In our experience, placing replicate cores per tumor in one TMA block may offer modest advantages for standardization, particularly for immunofluorescence assays30. For chromogenic assays, batch staining is probably sufficient, provided that sections are cut and stored consistently. Placing cores from the same specimen into different blocks prevents loss of all cores from a subject if a block is lost or damaged and ultimately permit more potential assays per tumor.
Many TMAs contain hundreds of cores, permitting representation of large collections in a few blocks. Preparation of high density TMAs raises the risk that damage or errors in preparation will impact many cases and poses greater challenges for mapping cores. Constructing TMA blocks by key characteristics (e.g. ER) may be advantageous, by allowing preservation of tissues for relevant assays only.
Increasingly, stained TMA sections are scanned as digital images that can be scored visually or by computer algorithms. Central placement of sections on slides is essential to ensure quality scanning and optimal staining when using automated immunostainers. Asymmetric TMA designs or placement of sentinel cores outside the main grid facilitate orientation. Use of a single scanner and maintaining a consistent orientation is also helpful.
Tissue Cores for Molecular Studies
Loose TMA cores provide an enriched source of target tissue for molecular extraction and banking. Punching blank paraffin in between targets is probably sufficient to avert cross-contamination for DNA assays and possibly others;31 however, we recommend saving and testing blank cores for testing to exclude carry over. The availability of disposable TMA needles presents the opportunity to receive loose tissue cores for molecular extraction without borrowing blocks. Techniques to build TMAs using loose cores as starting materials have not been widely applied.
Test Arrays and Cores
Preparing test arrays from representative specimens is useful for pilot testing. Inclusion of tissue cores from well-characterized tissues or cell lines in TMAs may allow evaluation of assay procedures, but does not address concerns about preservation or fixation of individual samples. The developmental of sentinel markers of tissue preservation/fixation for breast tissue akin to the suggested use of p27 for prostate specimens would be useful, allowing either comparison of each marker to an internal standard or discounting poor quality results.32
Sectioning and Staining TMAs
Minimizing the number of times a block is sectioned conserves tissue. However, given that cut sections suffer degradation, manifested as loss of antigenicity by immunohistochemistry during storage33–36, it is desirable to limit the number of sections to those needed for immediate use. Therefore cutting and staining in batches is recommended. The mechanism of degradation of cut sections remains unknown. Paraffin dipping, oxygen-free storage and cooling may slow degradation, but do not completely eliminate the effect for all tissues and markers.33–35
Cutting TMA blocks and mounting sections on slides is challenging; each core is potentially unique, so folds and irregularities result in lost data. Some laboratories favor use of tape transfer methods to maintain core alignment, whereas others prefer standard techniques, which limits background. Use of coated slides that enhance section adherence helps prevent loss of cores with antigen retrieval for immunohistochemistry and nucleic acid denaturation for in-situ hybridization. Standardized sectioning is important for accurate semi-quantitative or quantitative analyses. Given that typical sections are 4–5 uM thick, slight absolute differences in thickness translate into large percentage differences. All scoring methods reflect light penetration, which is impacted by tissue thickness.
Scoring
Microscopic scoring of TMAs is laborious and has largely been replaced by use of digitized images for visual or automated analysis. Technologies to improve the speed, automation, magnification and resolution of scanning methods and to increase image storage, enhance analysis and create flexible databases are under continuous development. In particular, the computer infrastructure required for this work is complex.
Quality assurance procedures to establish TMA orientation, core alignment and correct subject links are critical. Visual inspection of TMAs may reveal missing, misaligned or obscured cores. Computer algorithms to assess cores for area of tissue coverage or cell number may identify poor quality spots.
TMA data should be analyzed across rows and columns to exclude unequal staining. Highly disparate results for replicate cores per subject or differences in marker frequency distributions by study center should prompt review. Evaluation of expected associations is also useful. For example, in breast cancer, higher ER levels should be related to older patient age, postmenopausal status, lobular or tubular histology and low tumor grade. Similarly, a high frequency of ER/PR co-expression is expected. In general, percentage of cells and staining intensity are strongly related for most markers. Exploring results for correlative assays for cases tested by different methods (e.g. amplification of the human epidermal growth factor 2 (HER 2) gene and immunohistochemical analysis of protein), may substantiate quality. Similarly, highly correlated mRNA and immunohistochemical levels are encouraging, but this is only expected for some markers (e.g. ER, PR), reflecting both biological and technical factors. Demonstrating expected correlations between markers within a pathway also provides validation.
Options for scoring TMAs include visual assessment, semi-automated scoring and fully automated scoring. Software that displays images of cores for visual scoring matched to subject records for data entry offers organizational efficiency and reduced recording errors. However, availability of microscopic slides for review of individual cores as needed is desirable.
Automated assessment may permit rapid throughput, improved precision, especially compared with multiple readers, and theoretically, improved accuracy, although this has not been clearly established for most markers. Currently some staining patterns remain challenging for automated quantification. Human assessment has relative strengths related to recognition of tumor and artifacts, such as over-staining at edges of structures. Instrumentation has advantages for counting cells and assessing gradations of stain intensity. Semi-automated approaches in which a trained observer “segments” the region for computer analysis offer complementary strengths, but little efficiency. Automated scoring to triage cases for visual review may also have value for some markers. Specifically, an automated algorithm that achieves excellent negative predictive value but suboptimal positive predictive value for a marker may facilitate a strategy in which only cores presumptively scored positive by the computer require visual review.
Marker data should be formatted to reflect precision and biological effects. For markers such as HER2, for which biological effects seem linked to gene amplification, dichotomous scoring with an optimal threshold for distinguishing strong positive immunohistochemical staining may be best. For other markers, categorical or continuous analyses of stain intensity, percentage of cells positive or a combinatorial metric may be optimal. If multiple cores are scored, one can often use average or maximum values with similar results, but confirmation is required.
Future Directions
Recently, progress has been made towards developing and validating methods for methylation, mRNA and miRNA profiling on fixed tissues37–40. Many of these strategies compensate for the biochemical modifications and fragmentation that occur with fixation, and increasingly, newer technologies rely on sequencing strategies instead of detection using fixed probe sets. However, most carcinogenic mechanisms operate by affecting protein levels within normal, precancerous or cancerous cells, and their surrounding microenvironments. Therefore, application of robust marker specific assays in morphologically defined contexts may be required for marker validation, biological understanding and clinical translation.
Methods for relating immunohistochemical results to objective standards such as absolute numbers of molecules per cell could permit further advances. Development of robust assays for activated markers, such as phosphorylated proteins, may permit analysis of critical final steps in molecular pathways. Multiplexed labeling would increase the utility of individual tissue sections and enable characterization of marker co-expression within single cells. Finally, development of new statistical methods to concurrently model exposures and endpoints simultaneously, is important for analyzing tumor markers, many of which are highly associated.41
Relating epidemiologic exposures to molecular tumor markers is important for increasing biological understanding and clinical translation. Knowing a subject’s risk for cancer is a starting point, but knowing the type of tumor that is likely to develop, the threat it poses to health and the mechanisms that mediate its development are important. This detailed information may aid the implementation of prevention measures. For example, tamoxifen and raloxifene have established utility in preventing ER positive breast cancers, but our modest ability to predict risk for ER positive tumors specifically (similar to breast cancer overall), have limited implementation in practice42. As treatment for breast cancer and other tumors moves increasingly towards exploitation of molecular targets, risk modeling and prevention should pursue similar approaches in parallel. Incorporation of molecular pathology in epidemiologic studies will be required to realize this goal.
Acknowledgments
Supported in part by the Intramural Research Program of the National Cancer Institute, the Applied Molecular Pathology laboratory and the Tissue Array Research Program.
Bibliography
- 1.Garcia-Closas M, Vermeulen R, Sherman ME, Moore LE, Smith MT, Rothman N. Applicaton of Biomarkers in Cancer Epidemiology. In: Schottenfeld DFJJ, editor. Cancer Epidemiology and Prevention. Oxford, UK: Oxford University Press; 2006. pp. 70–88. [Google Scholar]
- 2.Ravdin PM, Cronin KA, Howlader N, et al. The decrease in breast-cancer incidence in 2003 in the United States. N Engl J Med. 2007;356:1670–4. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 3.Anderson WF, Matsuno RK, Sherman ME, et al. Estimating age-specific breast cancer risks: a descriptive tool to identify age interactions. Cancer Causes Control. 2007;18:439–47. doi: 10.1007/s10552-006-0092-9. [DOI] [PubMed] [Google Scholar]
- 4.Menashe I, Anderson WF, Jatoi I, et al. Underlying causes of the black-white racial disparity in breast cancer mortality: a population-based analysis. J Natl Cancer Inst. 2009;101:993–1000. doi: 10.1093/jnci/djp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore HM, Compton CC, Lim MD, et al. 2009 Biospecimen research network symposium: advancing cancer research through biospecimen science. Cancer Res. 2009;69:6770–2. doi: 10.1158/0008-5472.CAN-09-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smigal C, Jemal A, Ward E, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56:168–83. doi: 10.3322/canjclin.56.3.168. [DOI] [PubMed] [Google Scholar]
- 7.Gotzsche PC, Nielsen M. Screening for breast cancer with mammography. Cochrane Database Syst Rev. 2009:CD001877. doi: 10.1002/14651858.CD001877.pub3. [DOI] [PubMed] [Google Scholar]
- 8.Liberman L, Kaplan JB, Morris EA, et al. To excise or to sample the mammographic target: what is the goal of stereotactic 11-gauge vacuum-assisted breast biopsy? AJR Am J Roentgenol. 2002;179:679–83. doi: 10.2214/ajr.179.3.1790679. [DOI] [PubMed] [Google Scholar]
- 9.Pisano ED, Gatsonis C, Hendrick E, et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med. 2005;353:1773–83. doi: 10.1056/NEJMoa052911. [DOI] [PubMed] [Google Scholar]
- 10.Boyd NF, Martin LJ, Rommens JM, et al. Mammographic density: a heritable risk factor for breast cancer. Methods Mol Biol. 2009;472:343–60. doi: 10.1007/978-1-60327-492-0_15. [DOI] [PubMed] [Google Scholar]
- 11.Morris EA. Diagnostic breast MR imaging: current status and future directions. Radiol Clin North Am. 2007;45:863–80. vii. doi: 10.1016/j.rcl.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Kim R, Osaki A, Toge T. Current and future roles of neoadjuvant chemotherapy in operable breast cancer. Clin Breast Cancer. 2005;6:223–32. doi: 10.3816/CBC.2005.n.024. discussion 233–4. [DOI] [PubMed] [Google Scholar]
- 13.Althuis MD, Fergenbaum JH, Garcia-Closas M, et al. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol Biomarkers Prev. 2004;13:1558–68. [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Brinton LA, Lissowska J, et al. Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer. 2006;95:123–9. doi: 10.1038/sj.bjc.6603207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Bernstein L, Pike MC, et al. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast Cancer Res. 2006;8:R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacey JV, Jr, Mutter GL, Nucci MR, et al. Risk of subsequent endometrial carcinoma associated with endometrial intraepithelial neoplasia classification of endometrial biopsies. Cancer. 2008;113:2073–81. doi: 10.1002/cncr.23808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson WF, Luo S, Chatterjee N, et al. Human epidermal growth factor receptor-2 and estrogen receptor expression, a demonstration project using the residual tissue repository of the Surveillance, Epidemiology, and End Results (SEER) program. Breast Cancer Res Treat. 2009;113:189–96. doi: 10.1007/s10549-008-9918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leyland-Jones BR, Ambrosone CB, Bartlett J, et al. Recommendations for Collection and Handling of Specimens From Group Breast Cancer Clinical Trials. Journal of Clinical Oncology. 2008;26:5638–5644. doi: 10.1200/JCO.2007.15.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grizzle WE. Special symposium: fixation and tissue processing models. Biotech Histochem. 2009;84:185–93. doi: 10.3109/10520290903039052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt SM, Lewis FA, Cao Y, et al. Tissue Handling and Specimen Preparation in Surgical Pathology Issues Concerning the Recovery of Nucleic Acids From Formalin-Fixed, Paraffin-Embedded Tissue. Archives of Pathology & Laboratory Medicine. 2008;132:1929–1935. doi: 10.5858/132.12.1929. [DOI] [PubMed] [Google Scholar]
- 21.Paavilainen L, Edvinsson A, Asplund A, et al. The Impact of Tissue Fixatives on Morphology and Antibody-based Protein Profiling in Tissues and Cells. J Histochem Cytochem. 2009 doi: 10.1369/jhc.2009.954321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nirmalan NJ, Harnden P, Selby PJ, et al. Mining the archival formalin-fixed paraffin-embedded tissue proteome: opportunities and challenges. Molecular Biosystems. 2008;4:712–720. doi: 10.1039/b800098k. [DOI] [PubMed] [Google Scholar]
- 23.Cronin M, Pho M, Dutta D, et al. Measurement of gene expression in archival paraffin-embedded tissues - Development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. American Journal of Pathology. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26:5630–7. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- 25.Hewitt SM. The application of tissue microarrays in the validation of microarray results. Methods Enzymol. 2006;410:400–15. doi: 10.1016/S0076-6879(06)10020-8. [DOI] [PubMed] [Google Scholar]
- 26.Conway C, Dobson L, O’Grady A, et al. Virtual microscopy as an enabler of automated/quantitative assessment of protein expression in TMAs. Histochem Cell Biol. 2008;130:447–63. doi: 10.1007/s00418-008-0480-1. [DOI] [PubMed] [Google Scholar]
- 27.Kallioniemi OP, Wagner U, Kononen J, et al. Tissue microarray technology for high-throughput molecular profiling of cancer. Human Molecular Genetics. 2001;10:657–662. doi: 10.1093/hmg/10.7.657. [DOI] [PubMed] [Google Scholar]
- 28.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 29.Chung GG, Zerkowski MP, Ghosh S, et al. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest. 2007;87:662–9. doi: 10.1038/labinvest.3700543. [DOI] [PubMed] [Google Scholar]
- 30.Sherman ME, Rimm DL, Yang XR, et al. Variation in breast cancer hormone receptor and HER2 levels by etiologic factors: a population-based analysis. Int J Cancer. 2007;121:1079–85. doi: 10.1002/ijc.22812. [DOI] [PubMed] [Google Scholar]
- 31.Gaudet MM, Campan M, Figueroa JD, et al. DNA hypermethylation of ESR1 and PGR in breast cancer: pathologic and epidemiologic associations. Cancer Epidemiol Biomarkers Prev. 2009;18:3036–43. doi: 10.1158/1055-9965.EPI-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Marzo AM, Fedor HH, Gage WR, et al. Inadequate formalin fixation decreases reliability of p27 immunohistochemical staining: probing optimal fixation time using high-density tissue microarrays. Hum Pathol. 2002;33:756–60. doi: 10.1053/hupa.2002.126187. [DOI] [PubMed] [Google Scholar]
- 33.Fergenbaum JH, Garcia-Closas M, Hewitt SM, et al. Loss of antigenicity in stored sections of breast cancer tissue microarrays. Cancer Epidemiol Biomarkers Prev. 2004;13:667–72. [PubMed] [Google Scholar]
- 34.DiVito KA, Charette LA, Rimm DL, et al. Long-term preservation of antigenicity on tissue microarrays. Lab Invest. 2004;84:1071–8. doi: 10.1038/labinvest.3700131. [DOI] [PubMed] [Google Scholar]
- 35.Mirlacher M, Kasper M, Storz M, et al. Influence of slide aging on results of translational research studies using immunohistochemistry. Mod Pathol. 2004;17:1414–20. doi: 10.1038/modpathol.3800208. [DOI] [PubMed] [Google Scholar]
- 36.Wester K, Wahlund E, Sundstrom C, et al. Paraffin section storage and immunohistochemistry - Effects of time, temperature, fixation, and retrieval protocol with emphasis on p53 protein and MIB1 antigen. Applied Immunohistochemistry & Molecular Morphology. 2000;8:61–70. [PubMed] [Google Scholar]
- 37.Paik S, Kim CY, Song YK, et al. Technology insight: Application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat Clin Pract Oncol. 2005;2:246–54. doi: 10.1038/ncponc0171. [DOI] [PubMed] [Google Scholar]
- 38.Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 39.Beck AH, Weng Z, Witten DM, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Killian JK, Bilke S, Davis S, et al. Large-scale profiling of archival lymph nodes reveals pervasive remodeling of the follicular lymphoma methylome. Cancer Res. 2009;69:758–64. doi: 10.1158/0008-5472.CAN-08-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chatterjee N. A two-stage regression model for epidemiological studies with multivariate disease classification data. JASA. 2004:127–38. [Google Scholar]
- 42.Waters EA, Cronin KA, Graubard BI, Han PK, Freedman AN. Prevalence of tamoxifen use for breast cancer chemoprevention among U.S. women. Cancer Epidemiol Biomarkers Prev. 2010;19:443–6. doi: 10.1158/1055-9965.EPI-09-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]