Abstract
Subcellular compartmentalization of exoribonucleases (RNases) is an important control mechanism in the temporal and spatial regulation of RNA processing and decay. Despite much progress towards understanding RNase substrates and functions, we know little of how RNases are transported and assembled into functional, subcellularly-restricted complexes. To gain insight into this issue, we are studying the exosome-binding protein Dis3, a processive 3' to 5' exoribonuclease. Here, we examine the interactions and subcellular localization of the Drosophila melanogaster Dis3 (dDis3) protein. N-terminal domain mutants of dDis3 abolish associations with the `core' exosome yet only reduce binding to the `nuclear' exosome-associated factor dRrp6. We show that nuclear localization of dDis3 requires a C-terminal classic nuclear localization signal (NLS). Consistent with this, dDis3 specifically co-precipitates the NLS-binding protein importin-α3. Surprisingly, dDis3 constructs that lack or mutate the C-terminal NLS retain importin-α3 binding, suggesting that the interaction is indirect. Finally, we find that endogenous dDis3 and dRrp6 exhibit coordinated nuclear enrichment or exclusion, suggesting that dDis3, Rrp6, and importin-α3 interact in a complex independent of the core. We propose that the movement and deposition of this complex is important for the subcellular compartmentalization and regulation of the exosome core.
Keywords: Dis3, Rrp6, core exosome, importin-α3, RNase, PIN motif, OB fold, NLS, nucleocytoplasmic transport
Introduction
RNA-metabolizing (ribonucleometabolic) exosome complexes are fundamental players in the regulation of broad classes of RNAs in archaebacterial and eukaryotic cells (1, 2). These complexes consist of varying number of RNase PH components (Rrp41, 42, 43, 45, 46, and Mtr3) and S1 RNA binding proteins (Csl4, Rrp4, and Rrp40). In certain organisms, these complexes interact with Dis3 and Rrp6, 3' to 5' exoribonucleases and homologs of RNase R and RNase D, respectively (3). Despite great strides towards understanding the exosome-dependent mechanisms regulating RNA-specific processing, decay, or surveillance, there is a lack of knowledge regarding the cell biology and compartmentalization of these RNases and the associated exosome core (4, 5). This is a fundamental issue, as spatially- and temporally-controlled RNA processing and turnover events play critical roles in maintaining cell function as well as more generally in development, differentiation, and disease (6-9).
Defining the pathways and factors that regulate the subcellular distribution, interactions, and function of exoribonucleases is just beginning. The yeast exosome complex was originally proposed to exist in two forms, one nuclear and the other cytoplasmic (3). The nuclear complex was proposed to comprise ten `core' subunits plus a `nuclear' subunit Rrp6. Several biochemical lines of evidence suggest this idea requires revision. First, yeast proteomic studies indicate that exosome subunits, in addition to co-precipitating the expected core subunits, recover any number of accessory factors (10-12). Second, these proteomic studies and other work also revealed exosome subunits and exosome-associated factors in complexes distinct from the exosome (13-15). Third, exosome complexes isolated from human or trypanosome cells do not associate with Dis3 (16, 17). Fourth, an exosome complex purified from Drosophila cells lacks the core subunit dRrp45 (18). Fifth, fly exosome complexes do not recover Rrp43 (18, 19), indicating that they lack an architecturally critical component of the core (20).
Cytological evidence bolsters the argument derived from the biochemical findings. First, localization studies of fully functional GFP-tagged exosome subunits in yeast showed several subunits are enriched in the nucleolus whereas others localize primarily in the cytoplasm (21). Second, the human homolog of Rrp6, hPM/Scl-100, localizes in both the nucleoplasm and cytoplasm of HeLa cells whereas it is excluded from the nucleoplasm, yet in the nucleolus and/or the cytoplasm, in 293T cells (22). Third, tagged human subunits expressed from a heterologous promoter were exclusively nucleolar (23, 24). Fourth, endogenous and epitope-tagged Drosophila exosome subunits had distinct localization profiles between one another as well as from cell to cell (5). These conflicting results have unfortunately created a muddled picture of exoribonuclease and core exosome compartmentalization. Thus, a better understanding of the mechanisms and signals regulating subcellular localization of these proteins is needed to help clarify form and function of exosome subunit complexes in vivo.
The proper temporal and spatial distribution of macromolecules in cells depends on several highly regulated and compartmentally-restricted interactions, post-translational modifications, and enzymatic activities. A clear and well-defined example of this is found in nucleocytoplasmic transport (25, 26), the movement of protein cargoes in and out of the nucleus though the nuclear pore complex (NPC). Proteins fated for nuclear import commonly contain a set of basic amino acids called a nuclear localization sequence (NLS). In the cytoplasm, the NLS is bound by a protein called a karyopherin, importin-α. In yeasts, there is one importin-α molecule (Srp1) and in metazoans there are three or more importins (in Drosophila they are α1, α2, and α3 (27)). A cytoplasmic cargo/importin-α complex then associates with importin-β, and this cargo/α/β complex is imported into the nucleus. Once in the nucleus, this complex is dissociated by the action of the small GTPase Ran through a sophisticated series of biochemical interactions (28). The directionality of nucleocytoplasmic transport is thought to be maintained by the high concentration of Ran-GTP in the nucleus. This so-called `Ran-GTP gradient' is regulated by the compartmentalization of the Ran GEF (guanine nucleotide exchange factor), Rcc1, to the nucleus, and the Ran GAP (GTPase activating protein; induces conversion of Ran-GTP to Ran-GDP) to the cytoplasmic face of the NPC (25, 26, 28, 29). Whether the subcellular distribution of RNases and the core requires interaction with, and regulation by, the nucleocytoplasmic transport machinery has not been formally addressed.
Curiously, Dis3 has physical and genetic interactions with Ran in S. pombe and S.cerevisiae (13, 14, 30). We previously observed that dDis3 localizes, in a non-overlapping fashion, to the nucleus, the nuclear rim, or the cytoplasm of Drosophila melanogaster S2 cells (5). Given these links between dDis3 and nucleocytoplasmic transport, we sought to investigate the relationship between dDis3 localization and interactions and subcellular distributions of other RNases and the core exosome. Our results suggest that dDis3, dRrp6 and exosome subunits use a dedicated importin-α3-dependent pathway for nuclear targeting. These results allow us to present an inchoate model for mechanisms underlying RNase and core exosome subcellular compartmentalization.
RESULTS
Bioinformatic analysis of dDis3 reveals new motifs
An alignment of dDis3 with two possible eubacterial homologs, RNase II and RNase R, is shown in Figure 1A. Based upon sequence alignments and the fact that yeast Dis3 can digest RNA with secondary structure (20), as can RNase R (31), Dis3 appears to be the eukaryotic homolog of RNase R. This alignment also showed the existence of an N-terminal extension of ~210 amino acids in dDis3.
Figure 1. dDis3 N-terminal sequences are required for interactions with the core exosome but not with dRrp6.
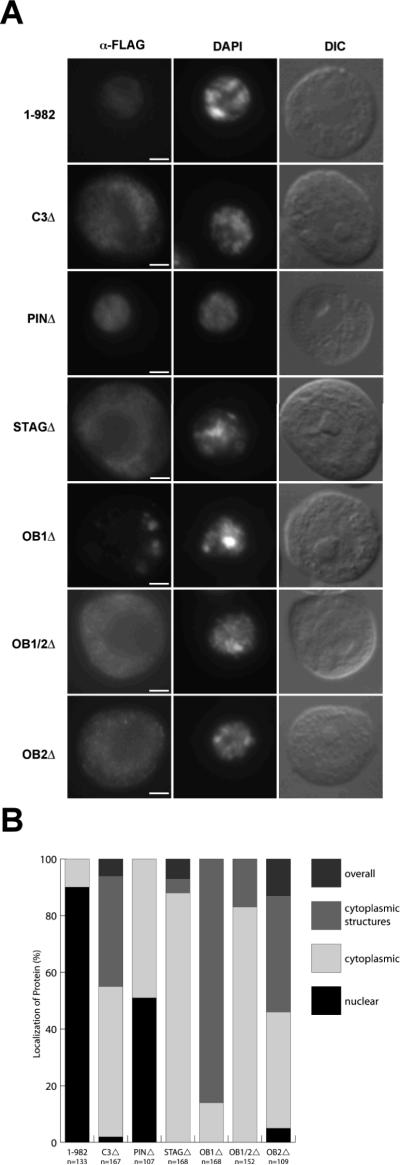
(A) Schematic representation and domain comparisons of Escherichia coli RNase II, RNase R, and Drosophila melanogaster Dis3 (dDis3). In the case of Dis3, only the RNB domain has been shown to have a defined activity. The bioinformatic identification of domains, putative functions, and conservation are discussed in the text. (B) Removal of the dDis3 PIN domain ablates interaction between dDis3 and core exosome subunits but only reduces the interaction efficiency between dDis3 and dRrp6. Antibodies used to detect exosome subunits alone or on a single blot (dRrp4, dRrp46, dCsl4, and dRrp47) are designated on the left side. Dis3F (F, FLAG) fusions is a blot with the anti-FLAG antibody. 1-982 is full-length dDis3. Asterisk, background IgG bands that are only present in the immunoprecipitate (IP) lanes. Input, 2.5%. (C) dDis3 N-terminal domain mutants used in this study. (D) dDis3 N-terminal domain mutants interact with dRrp6 but not core exosome subunits. Blots to detect both the FLAG tag (Dis3F fusions) and both endogenous and tagged dDis3 (dDis3) are shown to determine expression level of tagged relative to endogenous dDis3. Note that there is little or no effect on the steady-state levels of endogenous dDis3 or of other exosome subunits. Likewise notable is that C3Δ, PINΔ, and STAGΔ mutants have minor but observable N-terminal degradation products and that the individual OB deletion mutants migrate anomalously given their expected molecular weights. Asterisk, background IgG band. Input, 2.5%.
Bioinformatic inspection of the Dis3 N-terminal extension revealed several interesting features of the polypeptide that are not found in its eubacterial homologs (Figure 1A). The PIN (PilT N-terminus) domain, originally considered a motif with phosphodiesterase activity (32), has been shown to have DNA flap endonuclease (33) and ssRNA endonuclease activity (34) in other proteins. Three N-terminal cysteine residues (CX4CX2C, where X is any amino acid; referred to henceforth as C3), conserved in all Dis3 homologs, bear a close resemblance to those found in the iron-sulfur cluster binding motif (CX4CX2C20-40C) of ferredoxin family members (35). However, Dis3 homologs have no additional sequence homology with ferredoxins and lack the critical fourth cysteine. Finally, a dDis3 N-terminal sequence (amino acids 205-222) has similarity to the Pfam entry STAG, a domain found in the mitotic cohesin Scc3/STAG (Figure S1; (36)). Despite the presence of these three conserved Dis3 regions, the functional significance of these amino acid motifs to Dis3 exoribonuclease function remains undetermined.
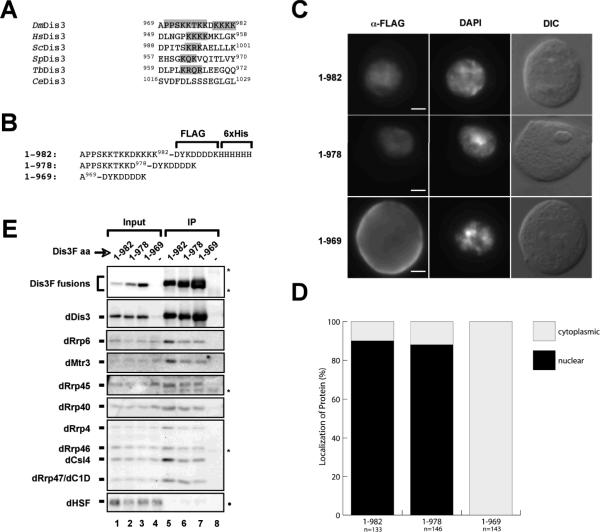
Sequence alignments of RNase R with Dis3 suggests the presence of two tandem OB (oligonucleotide-binding) folds (called here OB1 and OB2), domains involved in ssRNA or ssDNA binding (37). These have been confirmed in the recent crystal structure of the yeast homolog of Dis3, Rrp44 (38). The so-called RNB domain is the only functionally-validated domain in Dis3, as point mutations in critical residues in this domain disrupt 3' to 5' exoribonuclease activity (39, 40). The C-terminal tail of dDis3 shares an S1 RNA-binding domain with both its prokaryotic counterparts. Finally, the tip of the dDis3 C-terminus contains two consensus classic nuclear localization sequences (NLS), neither one of which have been shown to function as such. The number and C-terminal positioning of basic amino acids comprising the putative NLSs vary widely in animalia, fungi, plantae, and protista. Oddly, only C. elegans and C. briggsae lack these putative C-terminal NLSs (Figure 3A and not shown).
Figure 3. The dDis3 C-terminus harbors an nuclear localization sequence that is conserved and necessary for nuclear targeting.
(A) Sequence alignment of Dis3 C-termini from a number of model eukaryotes. Note that dDis3 (shown here as DmDis3 for continuity) is the only member of this family to have two putative NLSs. Dm, Drosophila melanogaster; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Tb, Trypanosoma brucei; Ce, Caenorhabditis elegans. (B) Truncation mutants designed to determine whether either or both NLSs are required for dDis3 nuclear import. These are referred to here as NLS1 and NLS2. dDis31-982 is the full-length protein, dDis31-978 removes only NLS2, and dDis31-969 removes both NLS1 and NLS2. (C) Loss of NLS1 and NLS2 but not NLS2 alone abrogates dDis3 import. Stably transfected S2 cells were analyzed as described in Figure 2. Bar, 2 microns. (D) Quantitative distribution analysis of full-length or NLS mutant dDis3 polypeptides. (E) C-terminal mutant polypeptides assemble into exosome subunit complexes in whole cell extracts. Asterisk, background IgG bands; Dot, cross-reactivity of dHSF antibody with co-migrating dDis3 NLS mutant polypeptides. Input, 2.5%
dDis3 N-terminal domain mutants lose interactions with the core exosome and reduce, but do not eliminate, binding to dRrp6
The observation that Dis3 is not found in all exosome core complexes led us to investigate how Dis3 associates with the core and how this relates to its subcellular distribution. Our previous results in Drosophila melanogaster S2 cells showed that purification of an epitope-tagged form of dDis3 from whole cell extracts recovers the full complement of exosome subunits (5). Thus, using this system, we addressed the domain requirements for Dis3-exosome interactions.
Since eubacterial cells do not have core exosome subunits, we reasoned that the Dis3 N-terminus evolved in eukaryotic cells to facilitate Dis3-exosome interaction(s). The PIN domain occupies the largest sequence space in the dDis3 N-terminus, spanning ~130 amino acids, 61% of the N-terminal extension. We engineered a mutant dDis3 polypeptide that lacks the PIN domain (PINΔ; Figure 1C; Table S1). We purified the wild-type full-length and PINΔ proteins from stably transformed Schneider cells and analyzed the co-precipitating exosome subunits. As seen previously (5), the full-length FLAG-tagged dDis3 co-precipitated the core exosome subunits dCsl4, dMtr3, dRrp4, dRrp40, dRrp42, dRrp45, and dRrp46, as well as dRrp6 and the nuclear cofactor dRrp47 (also referred to as C1D; Figure 1B, lane 4; data not shown; (41, 42)). As a control for specificity, the heat shock transcription factor dHSF showed little or no background binding to the immunoprecipitated dDis3. Moreover, there was no background resin binding in immunoprecipitations with cell extracts lacking a tagged protein (Figure 1B, lane 6). Although PINΔ is expressed and recovered efficiently, it does not co-precipitate any of the core exosome subunits (Figure 1B, lane 5). However, PINΔ retains interactions with dRrp6, although to a reduced degree (see also Figure 1D). These data indicate that the PIN domain is necessary for dDis3-core exosome interaction and important, but not essential for dDis3-dRrp6 interaction. Our data also show that dDis3 interacts with dRrp6 independently of the core exosome.
The removal of such a large domain from the dDis3 N-terminus could have a significant impact on the architecture of the entire N-terminus, thereby affecting dDis3-exosome interactions. Alternatively, the PIN domain alone may be the direct physical link between dDis3 and the core exosome. To address these alternatives, we constructed a set of internal domain mutants that singularly removed the predicted motifs that flank the PIN domain. A C3Δ mutant was engineered that removed the three conserved cysteines and C-terminal amino acids abutting the PIN domain. We also created a mutant dDis3 that deleted the STAG homology. Both of these mutants remove ~10% of the total amino acid content of the N-terminal extension and ~3% of the full-length dDis3 (Figure 1C, Table S1). Removal of these amino acids in the C3 and STAG regions does not disrupt PIN domain residues. We extended our analysis to other regions of dDis3 by engineering mutations that removed most of the predicted OB1 (OB1Δ), OB1 and the N-terminal part of OB2 (OB1/2Δ), and OB2 (OB2Δ) (Figure 1C and Table S1). A deletion of the first OB in RNase II (OB1 of dDis3) in the context of the full-length protein does not affect its RNase activity (43), suggesting that the corresponding dDis3 mutant should be enzymatically active.
Each of these dDis3 deletion constructs were stably transfected into S2 cells and induced with copper, then whole cell extracts were prepared and dDis3 complexes isolated by anti-FLAG immunoaffinity chromatography. Using an antibody to dDis3, we are able to show that the mutant polypeptides are expressed at the same level as or 2- to 4-fold more than endogenous dDis3 (Figure 1D, dDis3 blot). Expression of these mutant polypeptides had no observable effect on the level of endogenous dDis3, core exosome subunits, dRrp6, or dRrp47 (Figure 1D, lanes 2-7). Remarkably, every internal deletion mutant lost the ability to bind to the core exosome (Figure 1D, lanes 10-15). In contrast, dRrp6 was still recovered with these mutant polypeptides above background (Figure 1D, dRrp6 blot, compare lanes 10-15 to lane 16). Finally, the control protein dHSF was not recovered in any of the immunoprecipitates. These results show that dDis3 requires an intact N-terminus to interact with the core exosome and confirm that it has core exosome-independent interactions with dRrp6.
The results above indicate that the dDis3 N-terminus is necessary for interaction with the core exosome. To determine whether it was sufficient for this interaction, we tested whether a dDis3 polypeptide containing the first 479 amino acids (up to, but not including the RNB domain) could immunoprecipitate core exosome subunits. Indeed, dDis3 1-479 recovered core subunits dRrp40, dRrp46, dRrp40, and the nuclear exosome cofactor dRrp47 (Figure S2). Thus, the dDis3 N-terminus is necessary and sufficient for interaction with the core exosome.
dDis3 N-terminal mutants that retain binding to dRrp6 fail to localize properly to the nucleus
We had previously observed that both epitope-tagged dDis3 and dRrp6 predominantly localize to S2 cell nuclei and nucleoli, respectively, whereas most other subunits were cytoplasmic (5). We hypothesized that the inability of dDis3 mutants to interact with the core may be due to mislocalization. Thus, we examined the subcellular distribution of the dDis3 N-terminal mutants.
S2 cells expressing the wild-type dDis3 or N-terminal mutants were fixed and stained with the anti-FLAG antibody to detect the polypeptides and with a DNA dye to mark the nucleus. The full-length dDis3 polypeptide localizes predominantly to the nucleus but is sometimes found in the cytoplasm and at the nuclear rim ((5); Figure 2A, Table 1). In contrast, the C3Δ, STAGΔ, OB1Δ, OB1/2Δ, and OB2Δ polypeptides localized to the cytoplasm or throughout the cell, with very little or no exclusive staining of the nucleus (Figures 2A and 2B, Table 1). Several of these mutant proteins, in particular OB1Δ, accumulated in foci or structures in the cytoplasm, as shown previously for exosome subunits (5). Finally, although these mutants are mostly cytoplasmic, they retain the ability to bind to the `nuclear' dRrp6 (Figure 1D). The observation that dDis3 N-terminal domain mutants lost interaction with the core exosome yet associated with dRrp6 was intriguing. We will return to this issue in a later section.
Figure 2. dDis3 N-terminal mutants lose proper nucleocytoplasmic distribution.
(A) S2 cells stably transfected with dDis3 N-terminal mutants were fixed and stained with the FLAG antibody to detect the epitope-tagged dDis3 and with DAPI to visualize nuclear DNA. The differential interference contrast (DIC) images show that expression of these mutant dDis3 constructs does not have a severe impact either on cell shape or cytoplasmic, nucleolar, or nuclear structure. Representative images of major localization patterns are presented. Bar, 2 microns. (B) Quantitative analysis and grouping of localization patterns of dDis3 N-terminal mutants. A more comprehensive, quantitative analysis can be found in Table 1.
Notably, unlike the other N-terminal domain mutants, PINΔ is still targeted to the nucleus and exhibits a restricted nuclear or cytoplasmic staining (Figure 2A and 2B). However, the localization profile is shifted from ~9:1 nuclear-to-cytoplasmic distribution to an almost 1:1 ratio (Figure 2B and Table 1). These observations suggest that PIN domain (and/or surrounding, interacting domains) has an activity or an interaction that is critical for maintaining proper dDis3 nucleocytoplasmic balance. It is quite surprising that this and other dDis3 mutants contain two putative NLSs (Figure 3A), yet all of these mutants mislocalize compared to the wild-type polypeptide. Either these NLSs are either non-functional or are insufficient to target the dDis3 N-terminal domain mutants to the nucleus.
dDis3 C-terminus contains an NLS that is necessary for dDis3 nuclear targeting
An alignment of the dDis3 C-terminus with those of several other homologs demonstrates the presence of basic amino acids in most Dis3 molecules (Figure 3A). The C-terminus of dDis3 is unlike other homologs in that it has two positively-charged amino acid patches, separated by an aspartic acid. To address whether either or both of these charge patches are nuclear localization signals (tentatively called NLS1 and NLS2), we engineered two truncation mutants. The first mutant, 1-978, removed NLS2, and the second mutant, 1-969, removed both NLS1 and NLS2; both were appended with the FLAG epitope for localization and interaction studies (Figure 3B, Table S1). dDis31-978 localized similarly to the full-length dDis31-982, with approximately a 9:1 nuclear to cytoplasmic ratio. In contrast, dDis31-969 was throughout the cell or exclusively cytoplasmic, sometimes accumulating at the cell cortex (Figures 3C and 3D). This mislocalization to the cytoplasm was not a consequence of loss of interaction with the exosome, as dDis31-969 still recovered exosome subunits from extracts (Figure 3E). This indicates that the dDis3 C-terminus is necessary for nuclear targeting.
In order to determine whether either NLS1 alone or NLS2 alone is functional for dDis3 nuclear import, we engineered point mutations into them individually (Figure 4A); these mutants were then examined as described above. The three NLS2 mutants all properly localized to the nucleus and showed a ~9:1 nuclear to cytoplasmic ratio. In contrast, the dDis3 NLS1 mutant polypeptide was exclusively cytoplasmic (Figures 4B and 4C, Table 1). All of these mutants are expressed at approximately equal levels, do not affect the stability of exosome subunits, and recover subunits from cell extracts (Figure 4D). Thus, NLS1, not NLS2, is necessary for dDis3 nuclear accumulation.
Figure 4. dDis3 NLS1, not NLS2 is functionally important for nuclear import.
(A) Sequence comparison of wild-type dDis3 C-terminus with point mutations designed to disrupt either NLS1 alone or NLS2 alone in the context of the full-length polypeptide. (B) Localization of dDis3 NLS point mutants. Bar, 2 microns. (C) Quantitative analysis of dDis3 NLS point mutants. (D) Immunoprecipitation of exosome subunits with dDis3 NLS point mutants. Dot as described in Figure 3E. Input, 2.5%
dDis3 interaction with importin-α3 is specific but independent of dDis3 C-terminal NLSs and of its interaction with the core exosome
Because NLS1 is necessary (Figures 3 and 4) but insufficient (Figure 2) for dDis3 nuclear targeting, we thought that dDis3-interacting proteins other than dRrp6 and the core control its subcellular distribution. To identify such proteins, full-length dDis3 was purified from S2 cell extracts by tandem affinity chromatography and co-precipitating proteins were examined. Protein bands that were specifically and reproducibly recovered were excised and subjected to mass spectrometric analysis. One band corresponded to importin-α3, a karyopherin protein critical for nucleocytoplasmic transport of cargoes containing a canonical NLS; perhaps dDis3 nucleocytoplasmic distribution is controlled by importin-α3. However, because dDis3 interacts with dRrp6 and the core exosome in this preparation (Figure 5A, data not shown), we needed to determine whether the importin-α3 interaction was specific and direct.
Figure 5. dDis3 specifically interacts with importin-α3 in an NLS- and core exosome-independent manner.
(A) Immunoaffinity chromatographic purification of dDis3 from S2 whole cell extracts recovers importin-α3. Exosome subunit identity (dRrp4, dMtr3, and dRrp42 shown here) confirmed by diagnostic complex pattern and by western blotting. -, immunoprecipitated material from extracts prepared from control cells stably transfected with the Mtn vector. Asterisk, dDis3FH degradation products. (B) dDis3 interaction with importin-α3 is specific but independent of its C-terminal NLSs. (C) dDis3 NLS point mutants interact with importin-α3. Shown is a anti-dDis3 blot of these mutants to show their expression relative to endogenous dDis3 (shown in lane 8). (D) dDis3 N-terminal mutants interact with importin-α3. For all panels, dot and asterisk as described in Figure 3E; Input, 2.5%
Therefore, we directly tested whether the interaction between dDis3 and importin-α3 was specific. Whereas dDis3FH co-precipitates importin-α3 in a single-step immunoprecipitation reaction, importin-α1, importin-α2, or importin-β were not (Figure 5B, lane 5). Moreover, we do not observe any physical interaction between dDis3FH with the small GTPase Ran nor with a high molecular weight nuclear pore complex protein. Since importin-α proteins bind directly to NLSs, and NLS1 is important for dDis3 nuclear targeting, retention, or both, we predicted that importin-α3 would interact with the dDis3 C-terminus. We first examined interactions with the C-terminal truncation mutants. As expected, the dDis31-978 mutant, retaining NLS1, still effectively co-precipitated importin-α3 (Figure 5B, lane 6). Surprisingly, however, dDis31-969 also bound to importin-α3 (Figure 5B, lane 7). Moreover, the dDis3 polypeptides with point mutations in NLS1 or NLS2 all recover importin-α3 (Figure 5C, lanes 12-15). Recovery of importin-α3 in all cases was above background binding to the resin (Figure 5B, lane 8 and 5C, lane 16). This suggests that either dDis3 binds importin-α3 through a sequence other than its C-terminal NLSs or importin-α3 may bind another NLS-containing protein co-precipitating with dDis3. To examine these ideas, we tested whether the mis-localized dDis3 N-terminal deletion polypeptides (Figure 2) co-precipitated importin-α3. They did (Figure 5D, lanes 10-15). We provide here additional evidence that dRrp6, but not the core subunit dRrp42, interacts with these mutants, as initially shown in Figure 1. Importantly, in all of these immunoprecipitation experiments, there was little or no recovery of the non-specific protein dHSF. Since dDis3 stably interacts with importin-α3 without its NLSs, without core exosome interactions, and regardless of its subcellular localization, we surmise that this interaction may be indirect.
Interdependent localization and stability of dDis3 with dRrp6 and the core exosome subunit dRrp40
We considered the possibility that dRrp6 could be the direct target of importin-α3 binding because dRrp6 has a putative NLS (functionally validated in yeast Rrp6 (44)) and the interaction profile of dDis3 and importin-α3 was similar to that of dDis3 and dRrp6. To explore this idea, we investigated the cell biological relationship between dDis3 and dRrp6. We first performed a rigorous quantitative analysis of the distribution of endogenous dRrp6. We find that although dRrp6 is highly enriched in the nucleus in some cells and in a punctate stain throughout the cytoplasm, in other cells the nuclear signal is quite weak or absent (Figure 6A). In previous work (5), we had assumed that this lack of signal was due to uneven staining by the antibody. However, this cell-to-cell difference in dRrp6 staining may reflect mechanisms controlling dRrp6 subcellular compartmentalization and stability.
Figure 6. Subcellular coincidence of dDis3 and dRrp6.
(A) Endogenous dRrp6 is either enriched in the nucleus or distributed throughout the cell (scored here as cytoplasm to emphasize the fact that dRrp6 is not enriched in the nucleus). The graph at right represents the sum total of 457 cells from three independent experiments. (B) Specificity of dDis3 antibody. The antibody recognizes the full-length dDis3 by western blotting of whole S2 cells. (C) Endogenous dDis3 exhibits a distribution profile similar to that of dRrp6. A total of 334 cells were scored in three independent experiments. (D) dDis3 and dRrp6 are coincident in all cells examined. The scoring in the graph is derived from two independent experiments, with a total of 210 cells scored. In the first experiment, dDis3 was scored first and then dRrp6, with the converse in the second experiment. Bar, 10 microns.
Given that the dRrp6 distribution pattern and core-independent interaction with dDis3, we hypothesized that the nuclear compartmentalization of these two proteins would be highly coordinated. To test this, we generated a polyclonal antibody against full-length dDis3 and determined its specificity. A major reactive band at the appropriate molecular weight was observed by western blotting of whole S2 cells (Figure 6B). Immunofluoresence analysis of endogenous dDis3 with this antibody revealed the polypeptide enriched in the nucleus in only about one-third of the cells (Figure 6C). Given the similar quantitative and qualitative distribution patterns of dRrp6 and dDis3, we performed co-immunofluorescence experiments with the two antibodies. A striking correlation between dDis3 and dRrp6 nucleocytoplasmic distribution was observed (Figure 6D).
To test whether this co-compartmentalization could be disrupted by changing protein concentration, we examined the localization of endogenous dDis3 in cells expressing an FLAG-tagged form of dRrp6 (dRrp6FH) from a copper-inducible promoter. Because dRrp6FH is targeted to the nucleolus, whereas dDis3 is not (5), we could determine whether dRrp6FH expression induced dDis3 recruitment or retention to the nucleolus. Although dDis3 did not localize to the nucleolus in cells harboring dRrp6FH, it was, remarkably, enriched in the nucleus of these cells (Figure 7A); tautologically, dDis3 was never not found in the nucleoplasm of cells harboring dRrp6FH. In light of these findings, we also determined whether overexpression of dDis3FH could affect endogenous dRrp6 distribution. Indeed, we find a high, almost direct, correlation of the subcellular distribution of these two polypeptides, with an approximately 9:1 nuclear-to-cytoplasmic ratio (Figure 7B, top; cf. Figure 2B). Remarkably, in cells expressing the dDis3 PINΔ mutant polypeptide, endogenous dRrp6 mirrored the tagged protein and exhibited a 1:1 nuclear-to-cytoplasmic ratio (Figure 7B, bottom; cf. Figure 2B). Expression of the cytoplasmically-restricted dDis3 1-969, however, did not exclude dRrp6 from the nucleus (Figure S3A). The upshot is that dDis3 and dRrp6 localization is tightly coordinated.
Figure 7. dDis3-dRrp6 congruence revealed by overexpression studies.
(A) Nucleolar-and nuclear-targeted epitope-tagged dRrp6 exhibits exclusive nuclear enrichment of endogenous dDis3. Graphed data represents a total of 157 cells from two independent experiments. (B) Expression of dDis3 full-length (1-982, top) or mutant (PINΔ, bottom) polypeptides directly affects the subcellular distribution of endogenous dRrp6. Bar, 10 microns.
As a brief aside, the localization pattern of both the endogenous proteins and their FLAG-tagged counterparts are similar, but the distribution frequencies are different (cf. Figure 3A and Table 1 with Figure 6 and (5)). We interpret this to mean that overexpression causes additional copies of either dDis3 or dRrp6 accumulate in the nucleus and/or nucleolus. However, there is neither a change in the steady-state dRrp6 protein levels in response to overexpression of the dDis3 wild-type and mutant polypeptides nor the converse with the dRrp6 overexpression (Figure 1B, 1D, 5D, and 7A; data not shown; (5)).
Can nuclear-targeted dDis3 likewise regulate core exosome localization? To answer this question, we inspected and quantitated the subcellular distribution of the core subunit dRrp40 (Figure S3B). Notably, in cells expressing the nuclear dDis3 1-982, dRrp40 was found almost exclusively in the nucleus. As dRrp40 localization is observed predominantly cytoplasmic in untransfected cells (5), we find that expression of the cytoplasmic dDis3 1-969 did not change this distribution pattern. Thus, nuclear targeting of dDis3 dominantly affects both nuclear (dRrp6) and core (dRrp40) exosome components.
Depletion of dDis3 or dRrp6 affects protein stability and localization
We have shown a strong physical and cytological relationship between dRrp6 and dDis3. To extend upon this analysis, we performed RNA interference (RNAi) with double-stranded RNAs (dsRNA) direct towards dDis3, dRrp6, and GFP (as a non-specific control). We find that loss of dRrp6 does not affect endogenous dDis3 but that depleted dDis3 does reproducibly reduce endogenous dRrp6 levels from 10-30% (cf. Figure 8A and 8B, left panels). Consistent with this effect, dRrp6 depletion does not have a quantitiative effect on dDis3 nuclear accumulation but dDis3 depletion leads to an almost 3-fold reduction in dRrp6 nuclear occupancy (cf. Figure 8A and 8B, right panels). This suggests that dRrp6 stability and nuclear targeting is especially sensitive to dDis3 levels.
Figure 8. Depletion of dDis3 results in reduced levels of endogenous dRrp6 protein levels and reduced accumulation of dRrp6 in the nucleus.

(A) Western blot analysis showing that dDis3 levels are reduced in dsRNA to dDis3 (dsDis3), but not dsRrp6. Nuclear localization of dDis3 is not disrupted in dsRrp6-treated cells. (B) Western blot analysis showing that dRrp6 levels are reduced in both dsRrp6- and dsDis3-treated cells. Nuclear localization of dRrp6 is reduced in dsDis3 treated cells, as compared to dsGFP treated cells. Graphed data represents totals from two independent experiments.
To examine this relationship more thoroughly, we sought to determine whether depletion of dRrp6 would affect the localization or stability of dDis3 1-982, 1-969, or PINΔ (Figure S4). Although we observed no significant change in the subcellular distribution of these dDis3 polypeptides in the dRrp6-depleted cells, we did find that all of these polypeptides were reduced by 70-90% of levels observed in GFP dsRNA-treated cells. This finding is in sharp contrast with the previous finding (Figure 8A, left panel), showing that dRrp6 depletion does not co-deplete endogenous dDis3. We interpret these results to mean that dDis3 stability is sensitive to dRrp6 levels only when the molecular ratio of dRrp6:dDis3 exceeds a certain threshold. These studies also show that epitope-tagged dDis3 constructs do not require robust dRrp6 levels for targeting to or retention at subcellular locales. We conclude that molecular communication between dDis3 and dRrp6 and with the core exosome is extremely complex and highly regulated and therefore important for cellular ribonucleometabolic pathways.
DISCUSSION
In this study, we describe the interaction and distribution profiles of wild-type and mutant forms of the Dis3 3' to 5' exoribonuclease in Drosophila melanogaster S2 cells. We show that dDis3 interaction with the core exosome and localization to the nucleus is very sensitive to changes to its N-terminus. Furthermore, we present data showing that C-terminal NLS1 is necessary but, surprisingly, insufficient for dDis3 nuclear accumulation. We demonstrate that dDis3 interacts with both dRrp6 and importin-α3 and that dDis3 and dRrp6 nucleocytoplasmic distribution is coordinated.
Dis3: with or without the core exosome?
Our work suggests that dDis3 interacts with dRrp6 and the core subunits together as well as with dRrp6 in a core-independent manner. There are at least two possible explanations for why the dDis3 N-terminal domain mutants lose core interactions. On the one hand, there could be a single domain that bridges dDis3 and the core, and removal of these amino acids leads to loss of interaction. Proper folding or presentation of this hypothetically crucial domain requires other surrounding domains within the N-terminus. On the other hand, there could be multiple, cooperative contacts between dDis3 and the core. Loss of any one of these contacts is sufficient to see loss of dDis3-core interactions.
A recent electron microscopic (EM) study of the core associated with Dis3 (also called Rrp44) promulgates the latter of these two explanations (45). This study suggested that whereas Dis3 C-terminus (RNB domain) has conserved interactions with Rrp43 and Rrp45, the Dis3 N-terminus (PIN domain) associates with Rrp41. Our data is not wholly consistent with the interpretation of the EM results. For instance, we show that that the dDis3 N-terminus is necessary and sufficient to interact with the core. This suggests that the Dis3-core interaction is mediated in an important fashion by the N-terminus. One could argue, however, that the N-terminus is required for a nucleation step in the Dis3-core interaction but dispensable once Dis3 is anchored on the core. Alternatively, the N-terminal mutations could disrupt the fold of the entire Dis3 polypeptide, thus occluding potential RNB-exosome interaction interface.
Moreover, because Drosophila lacks Rrp43, a Dis3-Rrp43 interaction interface cannot be universal for all Dis3-core interactions. However, it is possible that another RNase PH contributes this interaction in the Drosophila core. Defining the exact physical interface between Dis3 and the core exosome should shed light on control of Dis3 exoribonuclease activity as well as why Dis3 associates with the core in only some eukaryotes.
Our demonstration that dDis3 associates with a small fraction of dRrp6 independent of the core exosome and that dDis3 and dRrp6 co-localize adds to mounting evidence linking these two proteins. Dis3 and Rrp6 are eubacterial (not archaebacterial (46)), mostly implicated in nuclear functions (1), found independent of the core in a Drosophila dosage compensation complex (15), linked genetically in yeast (47), and physically, genetically, and cytologically linked to mitotic progression and cell division (40, 48-50) (A. C. Graham, D. L. Kiss, and E. D. Andrulis, submitted). In short, Dis3 and Rrp6 appear to have conserved, core exosome-independent functions. Supporting this conclusion, Rrp6 functions independent of the core exosome in yeast (51).
Dis3: a karyopherin cargo or ferried through interactions with dRrp6?
A quintessential problem in cell biology involves how cargoes are transported to the appropriate subcellular location at the appropriate time. This question, when applied to understanding exosome core and subcomplex form and function, has remained largely unanswered. Our work addresses this matter and uncovers three connections between dDis3 and dRrp6 and nucleocytoplasmic transport, one that was anticipated and two others that were not.
The finding that dDis3 co-precipitates importin-α3 is not unexpected, for three reasons: (i) Tandem affinity purification of yeast Rrp4, Rrp40, Rrp45, Rrp46, Csl4, and Ski6 all co-precipitated Srp1, the sole yeast importin-α (10, 11); both Rrp6 and Dis3 were in these complexes. (ii) Another proteomic screen revealed that Srp1 co-precipitates Rrp4, Rrp43, and Rrp6 (52). (iii) A ~55 kDa band co-purifying with a nuclear Drosophila exosome complex (18) was importin-α3. The association between exosome subunits and Srp1 or importin-α3 is thus stable, and, in S2 cells, specific to this karyopherin class. However, the dDis3/a3 interaction is not mediated by NLSs and is not through the core. Although we cannot exclude the possibility that importin-α3 interacts with dDis3 in a direct, C-terminal NLS-independent manner, we propose an alternative explanation: dRrp6, not dDis3, is the stable and direct importin-α binding partner. In addition to reasons listed above, this is further supported by the observation that yeast Rrp6 nuclear localization requires functional NLSs (44).
The first of the two unanticipated findings is that dDis3 NLS1 is necessary for nuclear targeting of the full-length polypeptide but insufficient for nuclear targeting of the N-terminal internal deletion mutants. The second is that dDis3 mirrors the nucleocytoplasmic distribution of dRrp6. We deal with these issues in turn.
Given that dDis3 nuclear import requires NLS1, but that the dDis3-importin-α3 interaction does not require NLS1, then what binds NLS1? Perhaps dDis3 NLS1 interacts with another karyopherin (α1, α2, or β) in a manner that in unstable but direct. This proposal is weakened, however, by the inability of an intact NLS1 to confer nuclear retention or targeting to dDis3 N-terminal mutants that contain 92-98% of their coding sequences. Perhaps dRrp6 is the importin-α3 binding partner, and since we are able to isolate NLS1-lacking dDis3 in association dRrp6 and importin-α3, then this complex should ferry dDis3 into the nucleus. Paradoxically, it does not. Thus, either these dDis3 mutants mislocalize because they are not folding properly, are associating with a cytoplasmically-anchored binding partner or organelle, or are inactive for RNA binding or catalysis, or a combination thereof. Additional studies are needed to state unequivocally that the dDis3 NLS1 is functionally important, as we have not satisfied the four criteria required to make this claim (53). And what of NLS2? Although our data suggest that NLS2 is not important for dDis3 nuclear accumulation in interphase cells, we cannot exclude the possibility that it serves a specialized function at a particular stage of the cell cycle or for subnuclear targeting.
Returning to our other unexpected result: Why should dDis3 and dRrp6 interphase nucleocytoplasmic distribution be coordinated? A dDis3-dRrp6 complex, through associations with importin-α3, could ferry exosome subunits and complexes into the nucleus. Perhaps dDis3-dRrp6 complex activity is regulated by subcellular distribution. Or dDis3-dRrp6 localization could coordinate both interactions with the exosome, RNase activities, or both. Coordinated cell cycle-dependent proteolysis of these proteins could also be a mechanism of regulation. This seems possible, as the cell-to-cell difference in immunofluorescence would suggest.
Towards a general model of exosome and exosome-associated factor compartmentalization
It has been proposed that assembly and import of human exosome complexes proceeds in a step-wise manner, with assembly of the core in the cytoplasm followed by parallel but independent import of the core and hPM/Scl-100 into the nucleus, where they undergo further assembly, modification, and subnuclear targeting (4). Central to this model is the existence of an NLS in hRrp41 which is singularly responsible for core exosome complex import. However, the functional importance the hRrp41 NLS awaits confirmation and, notably, an NLS is not present in all Rrp41 homologs, including dRrp41. This realization suggests other mechanisms underlie exosome subunit and/or complex nuclear import. We propose that Dis3 and Rrp6 may work together or separately through importin-α3 to mediate exosome subunit/complex nucleocytoplasmic transport. This proposal is derived in part from the observation that nuclear-targeted dDis3 dominantly induces dRrp6 and dRrp40 nuclear localization.
Although human exosome subunits do not stably interact with hDis3, one cannot rule out the possibility of a transient, labile interaction among hDis3, hPM/Scl-100, and the core. Such a dynamic, unstable hDis3 interaction would be optimal from the standpoint of nucleocytoplasmic transport, where movement and deposition of cargoes relies on rapid and efficient assembly and disassembly of protein complexes (25). Incidentally, yeast Dis3 can be released from the core exosome under conditions that do not perturb the core (3, 39).
There have been several studies to investigate the subcellular distribution of exosome subunits and cofactors (3, 5, 21-24, 42, 44, 54, 55). Only a few of these have touched on the relationship among RNase and exosome subunit localization, protein-protein interactions, and ribonucleolytic function. Indeed, the most basic cell biological terrain of RNases is uncharted. In this study, we have begun to explore this area. In particular, we inquired how one exosome subunit, dDis3, gets to its intended subcellular destination and how this relates to its interactions with dRrp6, the core, and importin-α3. We sense this relationship important for understanding general mechanisms of compartmentalized exosome structure and function. Much remains to be uncovered about how dDis3 localization relates to its functions and interactions with dRrp6, with the core exosome, and the molecules and macromolecular structures involved in nucleocytoplasmic transport.
MATERIALS AND METHODS
Cell culture and stable cell line selection
Drosophila melanogaster embryonic Schneider (S2) cell lines were grown in HyQ-CCM3 (Hyclone) at 27°. Stable cell lines expressing dDis3 mutant polypeptides were prepared as described (5, 18), with a minor variation. After 4 weeks selecting and 2 weeks expanding cell lines in 300 μg/ml hygromycin B (Invitrogen), cells were carried in the absence of the drug. Comparing these conditions to those used in prior studies, we did not observe a change in either the protein expression level as detected by western blotting or the cell-to-cell expression frequency by immunofluorescence.
RNAi treatment
To prepare dsRNA specific for dRrp6 or GFP, PCR was carried out with primers carrying a 5' T7 RNA polymerase site. dsRNA was amplified by in vitro transcription reactions, purified with lithium chloride, and annealed at 70°C followed by slow cooling. Untransfected and stably transfected S2 cells were plated in 6-well plates at a density of 1×106 on Day 0, and treated with 30 or 45 μg dsRNA on Day 0, 1 and 3. Transiently transfected S2 cells were plated as stated above but only received dsRNA treatment on Day 0 and 1. They were transfected with 1μg of DNA using Cellfectin reagent (Invitrogen) on Day 2 and induced with copper for expression on Day 3. On Day 4, cells were fixed for immunofluorescence or lysed for western blot analysis.
Cloning and expression of dDis3 mutants
All dDis3 plasmids were made using basic molecular cloning techniques and PCR-based cloning strategies. Primers are shown in Table S2. The NLS mutants were made by introducing the truncation or point mutations into the 3' primer. The internal deletion mutants, however, required a two-step PCR mutagenesis using the full-length open-reading frame of dDIS3 as a template. The first step of PCR mutagenesis was performed using a 5' primer (with a unique BglII site) corresponding to the wild-type gene sequences in combination with a mutant 3' primer carrying an in-frame deletion as an overhanging fragment. The resulting PCR product was then used as a primer in a second PCR reaction with the wild-type 3' primer with an in-frame FLAG (DYKDDDK) tag and 6xHis epitope followed by a stop codon and a unique SalI site. This final PCR product was then digested with BglII and SalI and cloned into the BamHI and SalI sites of pRmHa3 to obtain epitope-tagged, domain-mutant dDIS3 genes downstream of the metallothionein (Mtn) promoter. Mtn-dDis3 constructs were transiently transfected using CELLFectin (Invitrogen) and tested for copper-inducible expression and then established as stable cell lines as described previously (5, 18).
Immunoprecipitations and mass spectrometric analysis
Whole cell extract preparation and immunoprecipitation of dDis3 polypeptides with anti-FLAG antibody agarose beads was performed in batch as described (5). To identify dDis3-interacting polypeptides, 8 liters of dDis3FH-expressing cells at a density of 1 × 107/ml were pelleted and washed; Mtn-expressing cells were processed in parallel as a control. Whole cell extracts were prepared and bound to anti-FLAG beads and eluted into fractions with 200 μg/ml FLAG peptide. Peak fractions were pooled, bound to nickel resin, washed, and eluted with imidazole. Peak fractions were pooled again, dialyzed, and analyzed by Coomassie staining and western blotting. Bands of interest were excised from the gel and subjected to mass spectrometric identification by the Proteomic Core in the Lerner Research Institute.
Antibodies and indirect immunofluorescence
Polyclonal antibodies were raised against recombinant MBP-dDis3 that was expressed in and purified from E. coli. MBP-dDis3 was injected into a guinea pig and sera recovered (Pocono Rabbit Farm and Lab, Inc.). Bleeds were all compared against pre-immune sera to determine specificity. Antibodies to dRrp6 were described previously (5, 18). The α-FLAG and α-Ran antibodies were purchased from Sigma. The nuclear pore complex antibody mAb414 was purchased from Covance. Importin-α3 antibodies were gifts of S. Cotterill (56) and C. Parker (57). Importin-α1 and -α2 antibodies were gifts of B. Mechler (58). Importin-β antibody was a gift of J. Szabad (59). Indirect immunofluorescence and microscopy were conducted as described (5), with the following exceptions. The antibody incubation buffer (PNS) contained 4%NDS and the rabbit anti-dRrp6 antibody was used at 1:1000. The guinea pig anti-dDis3 antibody was diluted 1:10 and then purified with Melon Gel (MG) IgG Spin Purification Kit (Pierce, Rockford, IL). The dilution used for this antibody was 1:25. Images were obtained using the Zeiss Axioplan 2 microscope.
Bioinformatic analyses
Sequence alignments were performed using BLAST at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). The dDis3 nuclear localization signals were identified by PSORT (http://psort.nibb.ac.jp/) and against a nuclear localization signals database (http://cubic.bioc.columbia.edu/db/NLSdb/). Both dDis3 OB1 and STAG (Pfam accession number PF08514) were identified by using dDis3 amino acids to search the EMBL-European Bioinformatic Institute (EBI) protein domain database (http://www.ebi.ac.uk/InterProScan/). Multiple sequence alignment was performed using ClustalW at EMBL-EBI (http://www.ebi.ac.uk/Tools/clustalw/).
ACKNOWLEDGEMENTS
The authors would like to thank Greg Matera and Alan Tartakoff for comments on the manuscript, Sue Cotterill, Paul Fisher, Bernard Mechler, Carl Parker, and János Szabad for antibodies, Piet de Boer for microscope use, and members of the Andrulis lab for discussions. This work is supported by grant GM072820 from the NIH (E.D.A). E.D.A. is a Mount Sinai Health Care Foundation Scholar.
REFERENCES
- 1.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 2.Lorentzen E, Conti E. The exosome and the proteasome: nano-compartments for degradation. Cell. 2006;125:651–4. doi: 10.1016/j.cell.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3' --> 5' exonucleases. Genes Dev. 1999;13:2148–58. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raijmakers R, Schilders G, Pruijn GJ. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur J Cell Biol. 2004;83:175–83. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- 5.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culbertson MR. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 7.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 8.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–37. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 9.Brewer G. Messenger RNA decay during aging and development. Ageing Res Rev. 2002;1:607–25. doi: 10.1016/s1568-1637(02)00023-5. [DOI] [PubMed] [Google Scholar]
- 10.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 11.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 12.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, Beattie BK, Lalev A, Zhang W, Davierwala AP, Mnaimneh S, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–39. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 13.Noguchi E, Hayashi N, Azuma Y, Seki T, Nakamura M, Nakashima N, Yanagida M, He X, Mueller U, Sazer S, Nishimoto T. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. Embo J. 1996;15:5595–605. [PMC free article] [PubMed] [Google Scholar]
- 14.Shiomi T, Fukushima K, Suzuki N, Nakashima N, Noguchi E, Nishimoto T. Human dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae dis3. J Biochem (Tokyo) 1998;123:883–90. doi: 10.1093/oxfordjournals.jbchem.a022020. [DOI] [PubMed] [Google Scholar]
- 15.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–23. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–64. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 17.Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–51. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- 18.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–41. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 19.Forler D, Kocher T, Rode M, Gentzel M, Izaurralde E, Wilm M. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–37. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 22.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–87. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 23.Raijmakers R, Noordman YE, van Venrooij WJ, Pruijn GJ. Protein-protein interactions of hCsl4p with other human exosome subunits. J Mol Biol. 2002;315:809–18. doi: 10.1006/jmbi.2001.5265. [DOI] [PubMed] [Google Scholar]
- 24.Brouwer R, Allmang C, Raijmakers R, van Aarssen Y, Egberts WV, Petfalski E, van Venrooij WJ, Tollervey D, Pruijn GJ. Three novel components of the human exosome. J Biol Chem. 2001;276:6177–84. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 25.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–94. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–51. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 27.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–14. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–44. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Clarke PR, Zhang C. Ran GTPase: a master regulator of nuclear structure and function during the eukaryotic cell division cycle? Trends Cell Biol. 2001;11:366–71. doi: 10.1016/s0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki N, Noguchi E, Nakashima N, Oki M, Ohba T, Tartakoff A, Ohishi M, Nishimoto T. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3' processing of 7S-to-5.8S rRNA and in degradation of the excised 5'-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics. 2001;158:613–25. doi: 10.1093/genetics/158.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 32.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–90. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 33.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004;279:16471–8. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 34.Glavan F, Behm Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. Embo J. 2006;25:5117–25. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lill R, Dutkiewicz R, Elsasser HP, Hausmann A, Netz DJ, Pierik AJ, Stehling O, Urzica E, Muhlenhoff U. Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim Biophys Acta. 2006;1763:652–67. doi: 10.1016/j.bbamcr.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 36.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 37.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–33. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the Active Subunit of the Yeast Exosome Core, Rrp44: Diverse Modes of Substrate Recruitment in the RNase II Nuclease Family. Mol Cell. 2008;29:717–28. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 40.Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease Activity of Dis3 Is Required for Mitotic Progression and Provides a Possible Link between Heterochromatin and Kinetochore Function. PLoS ONE. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3' processing of stable RNAs. Mol Cell Biol. 2003;23:6982–92. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in prerRNA processing. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amblar M, Barbas A, Fialho AM, Arraiano CM. Characterization of the functional domains of Escherichia coli RNase II. J Mol Biol. 2006;360:921–33. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 44.Phillips S, Butler JS. Contribution of domain structure to the RNA 3' end processing and degradation functions of the nuclear exosome subunit Rrp6p. Rna. 2003;9:1098–107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3' end processing. Proc Natl Acad Sci U S A. 2007;104:16844–9. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–52. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M. A novel plasmid-based microarray screen identifies suppressors of rrp6Delta in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:1044–55. doi: 10.1128/MCB.01299-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fomproix N, Hernandez-Verdun D. Effects of anti-PM-Scl 100 (Rrp6p exonuclease) antibodies on prenucleolar body dynamics at the end of mitosis. Exp Cell Res. 1999;251:452–64. doi: 10.1006/excr.1999.4578. [DOI] [PubMed] [Google Scholar]
- 49.Ohkura H, Adachi Y, Kinoshita N, Niwa O, Toda T, Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. Embo J. 1988;7:1465–73. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinoshita N, Goebl M, Yanagida M. The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol Cell Biol. 1991;11:5839–47. doi: 10.1128/mcb.11.12.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–55. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 53.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadowaki T, Schneiter R, Hitomi M, Tartakoff AM. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–10. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burkard KT, Butler JS. A nuclear 3'-5' exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol. 2000;20:604–16. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathe E, Bates H, Huikeshoven H, Deak P, Glover DM, Cotterill S. Importin-alpha3 is required at multiple stages of Drosophila development and has a role in the completion of oogenesis. Dev Biol. 2000;223:307–22. doi: 10.1006/dbio.2000.9743. [DOI] [PubMed] [Google Scholar]
- 57.Fang X, Chen T, Tran K, Parker CS. Developmental regulation of the heat shock response by nuclear transport factor karyopherin-alpha3. Development. 2001;128:3349–58. doi: 10.1242/dev.128.17.3349. [DOI] [PubMed] [Google Scholar]
- 58.Giarre M, Torok I, Schmitt R, Gorjanacz M, Kiss I, Mechler BM. Patterns of importin-alpha expression during Drosophila spermatogenesis. J Struct Biol. 2002;140:279–90. doi: 10.1016/s1047-8477(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 59.Timinszky G, Tirian L, Nagy FT, Toth G, Perczel A, Kiss-Laszlo Z, Boros I, Clarke PR, Szabad J. The importin-beta P446L dominant-negative mutant protein loses RanGTP binding ability and blocks the formation of intact nuclear envelope. J Cell Sci. 2002;115:1675–87. doi: 10.1242/jcs.115.8.1675. [DOI] [PubMed] [Google Scholar]