Abstract
Brain-derived neurotrophic factor (BDNF) is important to brain functions such as plasticity and repair. A single nucleotide polymorphism for this growth factor, val66met, is common and associated with decreased activity-dependent BDNF release. The current study evaluated the effects of this polymorphism in relation to human brain motor system function, short-term plasticity, and learning. Functional magnetic resonance imaging (fMRI) scanning during right index finger movement (n = 24) identified activation in a broad sensorimotor network. However, subjects with the polymorphism showed smaller activation volume within several brain regions as compared with subjects without the polymorphism. Repeat fMRI after 25 min of right index finger training found that the 2 genotype groups modulated brain activation differently. In several brain regions, subjects with the polymorphism showed greater activation volume reduction, whereas subjects without the polymorphism showed greater activation volume expansion. On a driving-based motor learning task (independent cohort, n = 29), subjects with the polymorphism showed greater error during short-term learning and poorer retention over 4 days, relative to subjects without the polymorphism. The presence of this BDNF polymorphism is associated with differences in brain motor system function, altered short-term plasticity, and greater error in short-term motor learning. The broader implications of these findings are considered.
Keywords: cortex, fMRI, genetics, genotype, plasticity
Introduction
Brain-derived neurotrophic factor (BDNF) is the most abundant neurotrophin in the brain and is highly expressed throughout the central nervous system (CNS). This growth factor influences a wide range of brain events related to plasticity and repair (Cotman and Berchtold 2002). A single nucleotide polymorphism in the human BDNF gene at codon 66 (val66met) is present in one or both alleles in approximately 30% of people in the United States (Shimizu et al. 2004). The current study examined the effects of this BDNF polymorphism in relation to human brain motor system function, short-term plasticity, and motor learning.
The first goal of the current study was to define the effects that the val66met polymorphism has on brain motor system function, via functional magnetic resonance imaging (fMRI). Two competing hypotheses were tested using a brief (4-min) probe of the motor system. On the one hand, few effects might be expected as the val66met polymorphism affects activity-dependent, rather than constitutive, BDNF secretion (Egan et al. 2003), and more than 4 min are required for BDNF to undergo activity-dependent release and to then exert its effects. On the other hand, significant polymorphism effects might be seen with this probe as anatomical studies have found hippocampal and cortical atrophy in association with this polymorphism (Pezawas et al. 2004; Szeszko et al. 2005; Bueller et al. 2006; Ho et al. 2006; Frodl et al. 2007; Toro et al. 2009), and this suggests that polymorphism effects can be cumulative over time and thus might become apparent with even a brief probe of motor system function.
A second goal of this study was to examine the effect that this BDNF polymorphism has on human brain short-term plasticity, using both fMRI and behavioral endpoints. Functional neuroimaging studies have examined this issue in primary motor cortex, where the val66met polymorphism was found to be associated with deficient activity-dependent cortical plasticity over 30 min of motor training (Kleim et al. 2006) and with reduced after-effects of several repetitive transcranial magnetic stimulation (TMS) perturbations (Cheeran et al. 2008). However, after 30 min of activity, activity-dependent BDNF level increases are seen in multiple brain regions, beyond primary motor cortex (Ploughman et al. 2005), and so evidence for a polymorphism-related effect would be expected diffusely. The fMRI has established value for measuring the plasticity of representational maps throughout the brain across a short period of motor activity (Karni et al. 1995; Morgen et al. 2004; Floyer-Lea and Matthews 2005) and for measuring effects of genetic polymorphisms (Bookheimer et al. 2000; Egan et al. 2003; Hariri et al. 2003), and so was used in the current study for examining these issues. The reduced activity-dependent BDNF release associated with the val66met polymorphism might be expected to be associated with impaired neuronal processes involved in short-term plasticity, measured via changes in the fMRI signal. In addition, the reduction in activity-dependent BDNF release associated with the val66met polymorphism may be associated with decreased short-term motor learning, a hypothesis that was tested with a driving-based motor learning task.
To address the first goal, a cohort of healthy subjects underwent fMRI scanning during movement of the right index finger. Results were compared among subjects with, versus those without, a single copy of the BDNF val66met polymorphism. To address the second goal, these subjects then underwent 25 min of right index finger abduction/adduction training, after which fMRI was repeated, and the change in brain function across the 2 scans examined. This provided insight into the effects of the val66met polymorphism on short-term plasticity. To address the behavioral aim, short-term learning was assessed in a separate cohort of subjects, examined during a 15-min driving-based motor learning task. A repeat examination 4 days later provided information on polymorphism effects on retention.
Materials and Methods
Subjects
Healthy subjects provided written informed consent using procedures approved by the Institutional Review Board. Entry criteria were age 18–30 years, right-handed, no neurological or psychiatric diagnoses, and no contraindication to MRI (Kleim et al. 2007). Note that the current 2 subject cohorts have no overlap with enrollees in our prior studies of the BDNF polymorphism (Kleim et al. 2006). After behavioral testing (see below), a 10-cc blood sample was obtained from the nondominant arm of each subject for genotyping. From this pool, 25 subjects (10 with Val/Met genotype and 15 with Val/Val genotype; note that this contained the maximum number of recruited subjects with the Val/Met BDNF genotype) were invited to continue in Experiment 1, with all agreeing to do so. A separate cohort of 29 subjects (22 with Val/Val genotype and 7 with Val/Met genotype) meeting the first 3 entry criteria participated in Experiment 2.
In sum, Experiment 1 examined motor system function by performing fMRI of right index finger movement, then short-term plasticity by having subjects immediately complete 25 min of right first dorsal interosseus (FDI) muscle training followed by repeat fMRI. Experiment 2 examined short-term motor learning, long-term motor learning, and retention using a simulated driving-based motor learning task evaluated twice over a 4-day period.
Genotyping
Each subject's blood sample was genotyped for the BDNF val66met polymorphism. Genomic DNA was extracted from leukocytes by standard DNA extraction procedure. Polymerase chain reaction (PCR) amplifications of the 274-bp fragment were set up with the following forward and reverse primers (5′-aaagaagcaaacatccgaggacaag-3′; 5′-attcctccagcagaaagagaagagg-3′). Reactions were performed in a 50 ul volume containing 50 ng of total genomic DNA as template, 0.2 mM each deoxynucleotide triphosphate, 0.5 um each of the forward and reverse primers, and 1.5 U of Taq polymerase (Roche, Mannheim, Germany) in its 1× supplied buffer from the manufacturer. PCR conditions were 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by cooling to 4 °C. The initial denaturation was at 95 °C for 5 min, and the final elongation was at 72 °C for 5 min. PCR products were tested by gel electrophoresis on a 1% agarose gel stained with ethidium bromide.
Genotype screening was performed with denaturing high-performance liquid chromatography analysis on Transgenomic WAVE system (Transgenomic, Omaha, NE). PCR products were first denatured at 95 °C for 10 min and then slowly cooled to 65 °C at a rate of 1 °C/min. Following this 5-min incubation at 65 °C, PCR products were cooled to 4 °C at a rate of 1 °C every 5 s to form heteroduplexes. Temperature for successful detection of heteroduplexes was calculated using the Wavemaker software package (Transgenomic) and was also experimentally determined at 60.8 °C. First, we detected all heterozygous variants Val/Met after heteroduplex formation. In order to detect homozygous Met/Met variants, PCR products were mixed with a Val/Val control DNA to form heteroduplexes and run another time through the Wave system.
Heterozygous Val/Met and homozygous Met/Met were controlled by sequencing. PCR products were purified by ExoSAP-IT (Amersham, Piscataway, NJ) and directly sequenced using the PRISM Ready Reaction Sequencing Kit (PE Applied Biosystems, Foster City, CA) on an automatic sequencer (ABI 3130; PE Applied Biosystems). Sequence data were analyzed using Sequencher (version 4.0.5, Genecode Corp., Ann Arbor, MI) software.
Behavioral Assessments
A medical history was obtained from each subject, as well as determination of handedness (Oldfield 1971). Motor behavior at baseline was characterized using 3 standard motor assessments (Strauss et al. 2006). First, maximum right hand index finger tapping speed was assessed using a mechanical counter attached to a solid wooden board. Subjects were asked to tap as rapidly as possible for 10 s and then rest for 15 s. After every third trial, subjects rested for 2 min. Trials were administered until 5 consecutive trials rendered scores within a 5-step range, up to a maximum of 10 total trials. Second, time to complete the 9-hole pegboard was measured, as a test of dexterity. Subjects placed pegs with the right hand, in any order, into the holes until all were filled and then removed each peg individually. Two trials separated by a 1-min break were performed. Third, the maximum force of strength of lateral pinch grip was measured using a standard gauge. Maximum force exerted over 3 successive trials, with a 1-min break between, was recorded.
Experiment 1
MRI Preparation
In a room near to the MRI scanner, subjects were briefly familiarized with the fMRI scanning procedures. Subjects lay flat, with right forearm pronated and stabilized in a splint that was attached with Velcro to a small button box. The button box was positioned on the stomach such that the right elbow was at 45°. The right index finger had free movement and rested comfortably on top of the rightmost button.
Subjects then spent approximately 5 min rehearsing the activities to be performed during fMRI scanning and between scanning sessions. First, subjects used video goggles to learn the 2 commands that would be presented during fMRI data acquisition, REST and MOVE. The first video command was a nonflashing red circle below the word REST. Subjects were instructed to relax whenever this image appeared on screen. The second command was a 1-Hz flashing green circle below the word MOVE. Subjects were instructed to abduct the right index finger from the right to the left button (a 15-deg movement) and back again each second, as directed by the flashing circle.
Next, subjects learned the 2 commands that would be presented during the 25 min of finger training that would occur between the 2 fMRI scans, FAST and HARD. One command was a nonflashing green circle beneath the word FAST. Subjects were instructed to ab-/adduct the right index finger back and forth as fast as possible for the duration of any period when this image appeared on the screen. The other command was of a nonflashing green circle, visible for 5 s and absent for 1 s, beneath the word HARD. Subjects were asked to abduct the right index finger to the left button and then press hard and hold the button for as long as the green circle was visible. Subjects were instructed to return the right index finger to a relaxed resting position on the right button once the circle disappeared.
Note that actual finger movements during this rehearsal session were brief, lasting no more than 20 s on average. Upon establishing that subjects understood and could complete each of these commands in response to these video cues, subjects were then escorted to the fMRI scanning facility.
Baseline fMRI Scan
During MRI scanning, subjects wore protective headphones and video goggles and were positioned as during the preparation session, with left arm at side and right forearm splinted and pronated, elbow flexed, and splint attached to the button box atop the stomach.
All MRI scans were conducted on a 1.5-Tesla Phillips scanner. Scanning sessions began with a whole-brain, high-resolution, volumetric T1-weighted anatomical scan (1 mm3 isotropic voxels). The baseline fMRI scan was then acquired using a blood oxygen level–dependent T2*-weighted gradient-echo echoplanar imaging sequence, with time repetition = 2.5 s, time echo = 40 ms, 25 axial slices, and slice thickness 4 mm with 1 mm interslice gap. This fMRI scan had subjects alternate 30 s of 1 Hz right FDI abduction across 15 deg (the “MOVE” video command), with 30 s of rest (the “REST” video command), for 4 min. Subjects were visually monitored, and proper task performance was confirmed by investigators throughout the scan.
Motor Training between the 2 fMRI Scans
With the subject still in the scanner, including head in the coil, 25 min of finger training was then immediately performed, that is, directly following the baseline fMRI scan. No MRI data were acquired during this training interval. The training was directed by video commands presented through the goggles. The content of the 25 min of finger training was precisely that used in a prior TMS study of short-term, experience-dependent plasticity (Kleim et al. 2006) and that had been briefly rehearsed by each subject during the MRI preparation session. The first activity was the “FAST” video cue, which involved right index finger rapid ab- and adduction movements as fast as possible for 30 s, alternating with 30 s rest; this was repeated 5 times followed by a 2-min break. Next was the “HARD” video cue, which had subjects press with the abducted right index finger hard, at 1 Hz, for 5 of 6 s, across a 3-min period; this was followed by a 2-min break. Subjects then repeated a second round of these 2 tasks, for a total of 25 min of training. Subjects were continually monitored for proper task performance in all cases.
Posttraining fMRI Scan
Immediately after the 25 min of finger training, the 4 min fMRI scan contrasting right FDI movement with rest was then repeated, using parameters identical to those employed at baseline (see “Baseline fMRI Scan,” above).
Data Analysis
Two fMRI activation maps were made for each subject, one for the baseline fMRI scan and one for the posttraining fMRI scan, using the following protocol. Note that these first level analyses were performed blinded to genotype. The fMRI data were analyzed using Statistical Parametric Mapping software (SPM5). The first 2 volumes from each functional scan were removed due to tissue nonsaturation. Remaining volumes were realigned to the first collected volume. Each subject's anatomical and fMRI data were coregistered and then spatially normalized into MNI stereotactic space. The fMRI data were then spatially smoothed (8 mm FWHM) and high-pass filtered. Statistical analyses were carried out using a general linear model and a standard hemodynamic response function. For each subject, an activation map was created by contrasting images obtained at rest with images obtained during right FDI movement. This was done twice, once for the baseline fMRI scan and once for the posttraining fMRI scan. A difference map was then constructed for each subject, representing the subtraction of baseline from the posttraining activation map.
These activation maps were analyzed in 2 ways, first with “whole-brain analyses” to explore effects across the entire cerebrum and second with “regional analyses” to probe 5 specific motor regions of interest (ROIs).
In the whole-brain analyses, group maps were generated. Thus, for each of the 2 genotype groups, a baseline fMRI group map was generated and separately a posttraining fMRI group map was generated. These analyses used random effects methods, with significance set at P < 0.001, uncorrected for multiple comparisons, with secondary analysis at P < 0.05 performed where indicated. First, a 1-sample t-test was used to characterize baseline activation, first across all subjects, then across each of the 2 genotype groups separately. This provided the total volume of significant brain activation as well as the volume and location of each separate cluster of significant activation. The 2 subject groups were then directly contrasted at baseline, using a 2-sample t-test. Next, a paired t-test was used to characterize the change across training (posttraining vs. baseline), across all subjects.
Differences between the genotype groups across the training period were probed by examining the time X group interaction, which was tested using a flexible factorial repeated-measures analysis of variance (ANOVA) model, at P = 0.01. In order to gain insight into the contribution that each genotype group made to the observed time X group effects, results from the flexible factorial repeated-measures ANOVA model were further probed using the parameter estimates (beta values) for the most significant voxel within each cluster. Each group's beta values were examined separately, using a 1-sample t-test (P < 0.05, JMP, SAS, Cary, NC), to determine whether the mean value was significantly different from zero.
Two forms of regional analyses were included to probe results within individual subject's activation maps. The first regional analyses measured the volume of activation within each of 5 motor cortical ROIs: hand area of left and right primary sensorimotor cortex, left and right premotor cortex, and a midline supplementary motor area (SMA). Each of these was constructed as a 12-mm sphere centered at coordinates derived from prior motor activation studies (see http://hendrix.imm.dtu.dk/services/jerne/ninf/voi.html). For each subject's baseline activation map as well as their difference map, the volume of significant (P < 0.001) activation was measured in each of these 5 regions, using the SPM5 small volume correction method. A second regional analysis assessed the magnitude of activation, measured as the mean task-related fMRI signal change. This was calculated in each of these 5 motor ROIs, also on both the baseline maps and the difference maps. For each regional analysis, results were compared across the 2 genotype groups, with significance set at P = 0.01 to correct for multiple comparisons.
For subject demographic and motor behavior data, continuous data were analyzed using 2-tailed t-tests and ANOVA. Categorical variables were analyzed using chi-square testing.
Experiment 2
Driving-Based Motor Learning Task
A task previously developed (Marchal Crespo and Reinkensmeyer 2008) by members of our group to evaluate implicit motor learning was used to explore genotype effects in the current study. Subjects were seated in front of a computer screen with a steering wheel (Logitech MOMO) attached to the desk. All subjects were instructed to place 2 hands on the wheel at “10 and 2” and keep them there for the duration of the task. Subjects were told to use the steering wheel to guide a vehicle on the screen through a curving track on the floor. The track had a black line down its center, and subjects were told to use the steering wheel to stay centered over the black line, with a computer recording deviation from the black line as extent of error. The track used a simulated environment via V-Realm Builder 2.0 software.
The vehicle was programmed to not change direction instantaneously, providing a level of demand that supported a need for motor learning. Subjects therefore had to begin turning the steering wheel before the track changed in order to minimize tracking errors. At the completion of each circuit, subjects were given a 10-s rest before the next circuit began. On the first day, subjects completed the same circuit 15 times, each taking approximately 60 s. Subjects returned approximately 4 days later, when the driving circuit was repeated. The mean tracking error, defined as the mean of the absolute value between the black line and the actual steered path, was calculated for each trial and expressed as root mean squared (RMS), with a cap at 2.0.
Data Analysis
For subject demographic and motor behavior data, continuous data were analyzed using 2-tailed t-tests and ANOVA. Categorical variables were analyzed using chi-square testing.
For analysis of errors on the driving-based motor learning task, repeated-measures ANOVA was performed on mean tracking error (RMS) values over the first visit's 15 driving laps, examining the main effects of time and of genotype group, as well as their interaction. Change from the end of the first visit to the second visit was analyzed using a paired t-test. “Short-term learning” was operationally defined as driving error over the course of 15 laps (ca. 15 min), as assessed using repeated-measures ANOVA described above. “Long-term learning” was operationally defined as driving error on the second visit (day 5, i.e., lap 16) compared with driving error on the first lap of day 1 (i.e., lap 1). “Retention” was operationally defined as driving error on the second visit (day 5, i.e., lap 16) compared with driving error on the last lap of day 1 (i.e., lap 15). Secondarily, retention was examined simply as driving error present on lap 16.
Results
Experiment 1
Subjects
Of the 25 subjects who completed the study, one was removed from analyses due to excessive head motion during fMRI data acquisition. The remaining 24 subjects were composed of 9 subjects with the Val/Met genotype (one copy of the val66met polymorphism) and 15 subjects with the Val/Val genotype (polymorphism absent). Demographic and behavioral measures (Table 1) did not differ between the 2 genotype groups. All subjects correctly completed the finger abduction/adduction training that was performed between fMRI scans.
Table 1.
Demographics and baseline behavior
| Experiment 1: fMRI |
Experiment 2: motor learning |
|||||
| Val/Val | Val/Met | P | Val/Val | Val/Met | P | |
| n | 15 | 9 | 22 | 7 | ||
| Age (years) | 24.1 ± 0.7 | 23.6 ± 1.0 | 0.62 | 24.0 ± 0.7 | 23.6 ± 1.2 | 0.73 |
| Gender (male/female) | 9/6 | 5/4 | 0.83 | 16/6 | 2/5 | 0.07 |
| Race | 0.24 | 0.13 | ||||
| Asian | 2 | 3 | 1 | 1 | ||
| Caucasian | 9 | 4 | 20 | 5 | ||
| Hispanic | 3 | 1 | 1 | 1 | ||
| Black | 1 | 1 | 0 | 0 | ||
| Handedness | 1.7 ± 0.3 | 1.8 ± 0.1 | 0.79 | 1.4 ± 0.22 | 1.3 ± 0.8 | 0.80 |
| Pegboard (s) | 17.9 ± 0.6 | 17.8 ± 1.0 | 0.93 | 17.3 ± 0.4 | 17.3 ± 0.7 | 0.97 |
| Finger tapping rate (Hz) | 47.6 ± 2.7 | 51.1 ± 3.0 | 0.42 | 51.4 ± 9.7 | 42.1 ± 7.9 | 0.03* |
| Force of pinch grip (N) | 197.2 ± 13.0 | 193.3 ± 12.9 | 0.85 | 183.45 ± 42.2 | 147.2 ± 46.0 | 0.06 |
Note: Values are mean ± standard error of the mean. Pegboard scores are time to complete the 9-hole pegboard test. Handedness scores indicate subjects were strongly right handed (+2 = right handed, 0 = ambidextrous, −2 = left handed). Significant (P < 0.05) differences are marked with an asterisk (*).
Baseline fMRI Scan
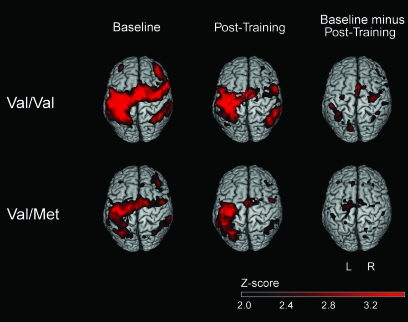
At baseline, among all 24 subjects, activation occurred in a network that included left primary sensorimotor cortex, bilateral premotor cortex, SMA, and bilateral cerebellum. However, separate analysis of the baseline fMRI map for each of the genotype groups found that activation volume was larger in Val/Val than Val/Met subjects, by several measures. Whole-brain analysis (P < 0.001) found that the total volume of significant brain activation in Val/Val subjects (178 cc) was larger than in Val/Met subjects (63 cc). Results of the group map cluster analyses at baseline are presented in Table 2. Note that, for each of the sensorimotor regions where both genotype groups showed activation, the volume of activation was consistently larger in Val/Val subjects (Table 2, Fig. 1). In addition to these regions that showed activation in both groups, there were regions that showed activation in only 1 of the 2 groups. In most cases, however, those clusters activated solely by Val/Met subjects were smaller in volume than those clusters activated solely by Val/Val subjects (Table 2). When baseline fMRI maps for the 2 groups were directly compared via a 2-sample t-test, there were 4 brain regions where the 2 groups showed significant differences in volume of activation, though only at threshold of P < 0.05. Three of these 4 were due to larger activation among Val/Val subjects, including right and left sensorimotor cortex as well as a region spanning bilateral dorsal cerebellum. One of these 4 was due to a larger activation among Val/Met subjects, in left cerebellum, more ventrally.
Table 2.
Regions of brain activation at baseline
| Val/Val |
Val/Met |
|||||
| Brain region | Vol | Mean z | x, y, z | Vol | Mean z | x, y, z |
| Left primary sensorimotor cortex, premotor cortex, SMA | 13 134 | 6.34 | −30, −12, 68 | 5095 | 4.24 | −8, −16, 64 |
| Left cerebellum | 1701 | 5.76 | −30, −62, −28 | 487 | 4.6 | −34, −54, −38 |
| Right cerebellum | 3645 | 6 | 24, −50, −30 | 694 | 4.12 | 22, −56, −32 |
| Right inferior parietal lobule | 905 | 4.3 | 56, −42, 30 | 391 | 4.11 | 64, −36, 30 |
| Left thalamus, ventral posteromedial nucleus | 96 | 4.02 | −14, −20, 6 | |||
| Right thalamus, ventrolateral nucleus | 695 | 5.64 | 14, −14, 4 | |||
| Left striatum | 448 | 5.5 | −26, −2, 6 | |||
| Right striatum | 311 | 4.34 | 20, 4, 4 | |||
| Right dorsolateral prefrontal cortex | 862 | 4.35 | 38, 40, 30 | |||
| Left inferior parietal lobule | 556 | 4.23 | −34, −68, 54 | |||
| Right motor cortex | 117 | 3.79 | 50, −4, 50 | |||
| Left cingulate motor area | 43 | 3.59 | −8, −24, 50 | |||
| Left superior parietal lobule | 118 | 3.28 | −28, −48, 62 | |||
| Left midbrain | 117 | 3.54 | −8, −14, −18 | |||
Note: Vol = volume of cluster, reported in voxels (8 mm3 each); mean z = mean z-score for the peak voxel within each cluster; x, y, z are reported in MNI coordinates. Results are the regions of significant activation in each group, in the whole-brain analysis, from separate 1-sample t-tests performed on each genotype group at P < 0.001, uncorrected for multiple comparisons, at baseline. In addition to above, note that the Val/Val group also showed significant activation in left superior temporal gyrus, right middle temporal gyrus, and the left occipital lobe, whereas the Val/Met group also showed significant activation in bilateral superior temporal gyri.
Figure 1.
The fMRI group activation maps, contrasting right FDI movement versus rest. At baseline, Val/Val subjects have larger motor system activation as compared with Val/Met subjects. Across 25 min of training, both groups showed reduced activation volume, as expected, however, the time × group analysis disclosed differences between groups.
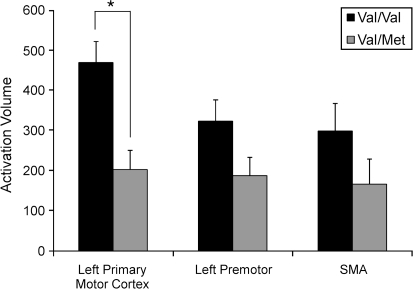
Regional analysis was consistent with the above, with Val/Val subjects having significantly larger activation volume at baseline as compared with Val/Met in the hand area of left primary sensorimotor cortex (P = 0.01; Fig. 2), with the differences present in other motor regions not reaching significance. Note that regional task-related fMRI signal change did not differ across the 2 subject groups in any of the 5 motor ROIs.
Figure 2.
Activation volume during the baseline fMRI scan is provided for 3 of the motor ROIs. Volume is reported in voxels (mean ± standard error of the mean). Val/Val subjects showed significantly larger activation within left primary motor cortex hand area (*P = 0.01), as well as a trend toward larger activation within left premotor cortex (P = 0.092) and SMA (P = 0.099).
Change across the Training Period
A paired t-test among all 24 subjects found that, across the 25-min period of finger training, brain activation showed a significant volume reduction within a region that spanned from left inferior parietal lobule through primary sensorimotor cortex to SMA and a significant volume increase within right medial primary sensorimotor cortex and bilateral posterior cingulate.
When this change over time was examined as a function of genotype (time × group interaction map), whole-brain analysis disclosed significant differences between the 2 groups within several brain regions (Table 3). Review of the parameter estimates (beta values) for each region provided insight as to how each group contributed to these observed differences (Table 3). These differences between the 2 groups in change over time were due to 2 processes: greater activation volume “increases” by Val/Val subjects, relative to Val/Met subjects, along the edge of the main activation clusters, and greater activation volume “decreases” by Val/Met subjects, relative to Val/Val subjects. For example, in bilateral sensorimotor cortex, along the ventromedial edge of the sensorimotor cortex activation cluster, subjects in the Val/Val group showed a focus with significantly greater increase in activation volume over time, as compared with subjects in the Val/Met group. In the left superior temporal gyrus, along the posterior edge, subjects in the Val/Met group showed a focus with significantly greater decrease in activation volume over time, as compared with subjects in the Val/Val group. In left medial frontal cortex, both patterns coexisted.
Table 3.
Brain areas where change over time differed as a function of genotype
| Brain region | Which genotype group accounted for finding | Vol | Mean z | x, y, z |
| Right sensorimotor cortex (medial edge) | Val/Val expansion | 269 | 3.37 | 26, −28, 48 |
| Left sensorimotor cortex (medial edge) | Val/Val expansion | 210 | 3.25 | −20, −36, 46 |
| Left medial frontal cortex | Val/Val expansion and Val/Met reduction | 182 | 3.61 | −8, 36, 46 |
| Left superior temporal gyrus anteriorly | Val/Val expansion and Val/Met reduction | 180 | 3.37 | −54, −4, −12 |
| Left superior temporal gyrus posteriorly | Val/Met reduction | 209 | 3.48 | −46, −60, 6 |
Note: Table 3 lists the brain areas in which the change in activation volume over time differed between the 2 genotype groups, based on the time × group interaction map. Review of the parameter estimates (beta values) within each of these brain regions provided insight as to which genotype groups accounted for the observed differences. Vol = volume of cluster, reported in voxels (8 mm3 each); mean z = mean z-score for the peak voxel within each cluster; x, y, z are reported in MNI coordinates.
Regional analyses did not reveal any significant differences between the 2 genotype groups in change over time in activation volume within any of the 5 motor ROIs. Furthermore, when change in activation volume was reexamined as “percent” change, which helps correct for observed differences between groups at baseline, again no significant difference between groups was found. Also, there was no difference in task-related fMRI signal change within any of the 5 motor ROIs across the training period.
Experiment 2
Subjects
A separate cohort of 29 subjects (age 23.9 ± 0.6 years) was enrolled for motor learning testing on the driving-based motor learning task. Of these, 22 were Val/Val and 7 were Val/Met. The 2 groups were well matched, with no differences in distribution of age, gender, or race (Table 1), though Val/Met subjects had a slightly slower rate of finger tapping. Note too that there were no significant differences between the subjects in Experiment 1 and the subjects in Experiment 2 in genotype distribution or in any of the measures in Table 1.
Driving-Based Motor Learning Task
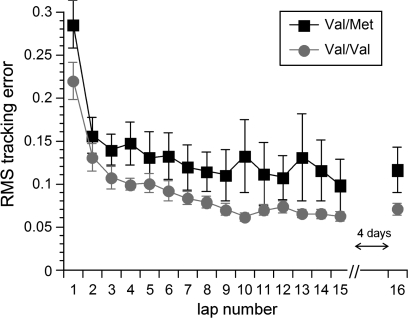
At baseline (lap 1), differences between the 2 genotype groups did not reach significance (P = 0.12). Short-term motor learning was present across all subjects over the 15 laps from the day 1 visit as the main effect of time was significant (F14,14 = 7.7, P = 0.0002; see Fig. 3). The main analysis of interest was that short-term learning was poorer in Val/Met subjects: driving error over the 15 laps of visit 1 differed according to BDNF genotype as the main effect of group was also significant (F1,27 = 4.5, P < 0.05). Note that although short-term error was greater in Val/Met subjects, both groups did nonetheless show significant short-term learning, as when each group was analyzed separately, the main effect of time over the 15 laps from the day 1 visit was significant for both Val/Val (F1,21 = 142.2, P < 0.0001) and Val/Met (F1,6 = 23.2, P < 0.003) subjects. Note too that driving error did not vary over time as a function of BDNF genotype as the time × group interaction term did not reach significance (F14,14 = 2.0, P = 0.11). In other words, both groups started at a similar level and both groups showed significant short-term learning, but this short-term learning was less in Val/Met subjects as they made more errors while learning.
Figure 3.
Driving error (reported as RMS tracking error) is presented for each lap of the driving-based motor learning task (mean ± standard error of the mean). Regarding short-term learning, over the first 15 laps, this driving error differed according to BDNF genotype (P < 0.05), with greater error among Val/Met subjects. Regarding retention, Val/Val subjects showed better retention of motor learning after 4 days, as within-subject change in driving error between the last lap of day 1 (“lap # 15”) and the lap performed on day 5 (“lap #16”) increased in Val/Met (P = 0.05), but not in Val/Val (P > 0.25), subjects.
Both groups also showed significant long-term learning as the lap driven on day 5 (lap #16) had significantly less error than the first lap of day 1 (lap #1), P < 0.0005 for each group separately, with no difference between groups (P > 0.5). However, 2 measures suggested that retention was poorer among Val/Met subjects. First, within-subject change in driving error between the last lap of day 1 (lap #15) and the lap performed on day 5 (lap #16) increased in Val/Met (P = 0.05), but not in Val/Val (P > 0.25), subjects. Secondarily, Val/Met subjects made significantly greater error than Val/Val subjects on lap 16 (P = 0.026). Thus, both groups showed long-term learning, but retention was weaker in Val/Met subjects.
Discussion
This study aimed to define the impact that the val66met BDNF polymorphism has on human brain motor system function, short-term plasticity, and motor learning. Results identified larger baseline activation volumes (including within bilateral sensorimotor cortex) in Val/Val subjects, who lacked this polymorphism, as compared with Val/Met subjects, who had one copy of this polymorphism. BDNF genotype was also associated with differences in short-term plasticity across 25 min of finger training, with Val/Val subjects showing sites of relatively greater activation expansion (in areas such as bilateral sensorimotor cortex) and Val/Met subjects showing areas of activation reduction. On a driving-based motor learning task, Val/Val subjects showed less error during short-term learning and greater retention over 4 days, relative to Val/Met subjects. Together, the current findings suggest that this polymorphism affects human brain motor system function and short-term motor system plasticity and is associated with greater error in short-term learning plus poorer retention.
Serial fMRI scans across a brief training period were used to study the reduction of activity-dependent BDNF release that is associated with the val66met polymorphism (Egan et al. 2003). Across all subjects, 25 min of right finger abduction/adduction training changed brain activation volume in a manner typical of that described in prior fMRI studies of short-term activity (Karni et al. 1995; Morgen et al. 2004; Floyer-Lea and Matthews 2005), such as reduced activation volume within sensorimotor cortex contralateral to movement. However, when change over time was examined as a function of BDNF genotype, differences were apparent. Val/Val subjects showed several foci where over time, activation volume expanded relative to Val/Met subjects (Table 3). In addition, Val/Met subjects showed several foci where over time, activation volume was reduced relative to Val/Val subjects. In other foci, these 2 processes were coexistent within the same brain regions (Table 3). These genotype-related differences over time might reflect polymorphism effects on those components of short-term cortical plasticity that are known to be influenced by BDNF, such as neuronal recruitment (Prakash et al. 1996; Monfils et al. 2005) or synaptic strengthening via long-term potentiation (Patterson et al. 1996; Mu and Poo 2006) or long-term depression (Ikegaya et al. 2002). Furthermore, note that several of the brain regions where Val/Val showed activation volume expansion over time relative to Val/Met (Table 3), such as left ventromedial sensorimotor cortex, were along the edge of the main activation sites associated with motor task performance (Table 2), an observation that might provide insight into the mechanism of polymorphism effects. Liepert et al. (2006) have found that changes in representational maps over time are influenced by inhibitory tone in the surrounding cortical regions. Therefore, some of the genotype-related differences over time might also reflect polymorphism effects on BDNF-related modulation of cortical inhibition (Wardle and Poo 2003; Hong et al. 2008) and excitability (Desai et al. 1999).
The current findings across a period of short-term training are consistent with prior studies that also suggested that the val66met polymorphism modifies short-term plasticity in humans. Egan et al. (2003) found the polymorphism to be associated with abnormal modulation of hippocampal function during a memory task. Kleim et al. (2006) found the polymorphism to be associated with dampening of motor cortex map plasticity across 30 min of motor practice. Cheeran et al. (2008) found the polymorphism to be associated with abnormal motor cortex plasticity in response to various forms of repetitive TMS perturbation, for example, subjects with the polymorphism showed reduced or absent after-effects of both inhibitory and excitatory repetitive TMS protocols. This convergence of findings to some extent mitigates concerns that the current short-term plasticity results merely reflect baseline differences among groups, such as in map size. Also reducing concerns regarding impact of baseline differences is that the areas where the 2 groups differed over time (Table 3) were not the same as the areas where the 2 groups differed at baseline. Together, the findings suggest that the reduction in activity-dependent BDNF release that is associated with the val66met polymorphism alters neuronal processes in a manner that produces differences (Table 3) in brain response to short-term training.
At baseline, prior to training, the overall pattern of motor system activation was similar across the 2 genotype groups, but the activation volume in Val/Met subjects was smaller as compared with Val/Val subjects, within several brain regions including the hand area of sensorimotor cortex contralateral to movement (Figs 1 and 2). The finding of polymorphism-related differences in motor system organization during a 4-min behavioral probe might seem unexpected given that this polymorphism affects only activity-dependent BDNF release and approximately 6–30 min (Hartmann et al. 2001; Poo 2001; Balkowiec and Katz 2002; Zhang and Poo 2002; Tanaka et al. 2008) are necessary for BDNF to be released and to exert its effects on cellular function. Though mechanisms of long-term plasticity were not directly evaluated in the current study, the observed differences in motor system organization at baseline might reflect a chronic or cumulative polymorphism effect, akin to the hippocampal and cortical atrophy described in carriers of this polymorphism (Pezawas et al. 2004; Szeszko et al. 2005; Bueller et al. 2006; Ho et al. 2006; Frodl et al. 2007). In this regard, note that the direction of between-group differences across short-term training (Val/Val only showed greater expansions, Val/Met only showed greater reductions; Table 3) was generally preserved when examining between-group differences at baseline (generally, Val/Val showed larger activation volumes). The baseline fMRI scan might thus be considered to be a reflection of polymorphism-influenced experience across the lifetime preceding study enrollment, possibly including development. The reason why genotype-related differences in the baseline fMRI scan were present in activation volume but not % signal change is unclear but might reflect the nature of the polymorphism effects on neuronal recruitment (Prakash et al. 1996; Monfils et al. 2005). Note that the current fMRI study found differences in hand sensorimotor cortex organization at baseline in relation to the val66met polymorphism, but our prior TMS study (Kleim et al. 2006) did not. This divergence in findings might reflect the different processes that are measured by TMS (cortical excitability, at rest) versus fMRI (afferent and efferent neuronal activity, during movement, as reflected through neurovascular coupling).
The behavioral data might be useful for interpreting these fMRI findings. The Val/Met subjects had smaller motor cortex map volumes at baseline, relative to Val/Val subjects. Smaller motor cortex activation volume can be a gain, for example, reflecting greater efficiency of motor cortex resource (Jancke et al. 2000). Smaller motor cortex map volume can also be a loss, for example, representing reduced motor cortex resource availability (Zemke et al. 2003; Cramer et al. 2006). Val/Met subjects showed poorer short-term learning and retention on the driving test (Fig. 3), observations that support the latter interpretation. This suggests that the impaired activity-dependent BDNF release associated with the val66met polymorphism is associated with reduced motor system resource availability, perhaps due to immediate effects of the polymorphism and perhaps due to a lifetime of exposure. The driving-based motor learning task probed motor, attentional, memory, and visuospatial systems, and so these conclusions might pertain broadly across the brain. Reports that the polymorphism is associated with behavioral impairments in systems such as memory (Egan et al. 2003) support this. However, when considering the overall significance of the val66met polymorphism, “reduced plasticity” might also provide “greater stability,” at least for some aspects of brain function. Greater stability might be advantageous in selected contexts, such as chronic degenerative diseases. Consistent with this view, the val66met polymorphism appears to have a beneficial effect on cognitive status in certain disease settings, such as Parkinson's disease (Foltynie et al. 2008), Huntington's disease (Alberch et al. 2005), systemic lupus erythematosus (Oroszi et al. 2006), and multiple sclerosis (Zivadinov et al. 2007). Therefore, the interpreting the significance of this common polymorphism might depend on context, and whether plasticity or stability is prioritized.
The current study has several limitations. Polymorphism effects can interact with age (Nemoto et al. 2006), but only a narrow range of ages was enrolled in the current study. Additionally, the cognitive and behavioral effects associated with the val66met polymorphism have been shown to be more robust in Caucasians (Bath 2006; Hashimoto 2008), which were the majority of current enrollees, suggesting the need to examine study aims in other ethnic groups. Further studies are needed to measure polymorphism effects on longer term forms of brain plasticity. Future studies might also measure attention, mood, anxiety, and other behavioral features that might be affected by the val66met polymorphism (Chen et al. 2008; Rybakowski 2008) in order to understand the contribution of these factors to the observed motor system effects.
The current results suggest that the val66met polymorphism impacts short-term motor system plasticity and short-term learning. These findings might have clinical implications given the role that BDNF has in CNS repair (Ferrer et al. 2001; Matzilevich et al. 2002; Uchida et al. 2003). For example, one recent study (Siironen et al. 2007) found that a subject with the val66met polymorphism had poorer outcomes on the Glascow Outcome Scale after subarachnoid hemorrhage as compared with subjects lacking this polymorphism, and a preliminary report suggests similar findings after ischemic stroke (Cramer et al. 2009). However, repair occurs over weeks or more. Further studies are therefore needed to measure effects of the val66met polymorphism in longer term settings and in the setting of disease.
Funding
U.C. Irvine General Clinical Research Centers Program of the National Center for Research Resources, National Institutes of Health (M01 RR000827-29); and NIH (R01 NS058755).
Acknowledgments
Conflict of Interest: None declared.
References
- Alberch J, Lopez M, Badenas C, Carrasco JL, Mila M, Munoz E, Canals JM. Association between BDNF val66met polymorphism and age at onset in Huntington disease. Neurology. 2005;65:964–965. doi: 10.1212/01.wnl.0000175977.57661.b1. [DOI] [PubMed] [Google Scholar]
- Balkowiec A, Katz DM. Cellular mechanisms regulating activity-dependent release of native brain-derived neurotrophic factor from hippocampal neurons. J Neurosci. 2002;22:10399–10407. doi: 10.1523/JNEUROSCI.22-23-10399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath KG, Lee FS. Variant BDNF (Val66Met) impact on brain structure and function. Cogn Affect Behav Neurosci. 2006;6:79–85. doi: 10.3758/cabn.6.1.79. [DOI] [PubMed] [Google Scholar]
- Bookheimer S, Strojwas M, Cohen M, Saunders A, Pericak-Vance M, Mazziotta J, Small G. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, Zubieta JK. BDNF val66met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Cheeran B, Talelli P, Mori F, Koch G, Suppa A, Edwards M, Houlden H, Bhatia K, Greenwood R, Rothwell JC. A common polymorphism in the brain-derived neurotrophic factor gene (BDNF) modulates human cortical plasticity and the response to rTMS. J Physiol. 2008;586:5717–5725. doi: 10.1113/jphysiol.2008.159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Bath K, McEwen B, Hempstead B, Lee F. Impact of genetic variant BDNF (val66met) on brain structure and function. Novartis Found Symp. 2008;289:180–188. doi: 10.1002/9780470751251.ch14. discussion 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC. Exercise: a behavioral intervention to enhance brain health and plasticity. Trends Neurosci. 2002;25:295–301. doi: 10.1016/s0166-2236(02)02143-4. [DOI] [PubMed] [Google Scholar]
- Cramer S, Warren M, Enney L, Sanaee N, Hancock S, Procaccio V. International Stroke Conference. San Diego (CA): 2009. BDNF polymorphism and clinical outcome in the GAIN trials; p. 128. [Google Scholar]
- Cramer SC, Shah R, Juranek J, Crafton KR, Le V. Activity in the peri-infarct rim in relation to recovery from stroke. Stroke. 2006;37:111–115. doi: 10.1161/01.STR.0000195135.70379.1f. [DOI] [PubMed] [Google Scholar]
- Desai NS, Rutherford LC, Turrigiano GG. BDNF regulates the intrinsic excitability of cortical neurons. Learn Mem. 1999;6:284–291. [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Krupinski J, Goutan E, Marti E, Ambrosio S, Arenas E. Brain-derived neurotrophic factor reduces cortical cell death by ischemia after middle cerebral artery occlusion in the rat. Acta Neuropathol. 2001;101:229–238. doi: 10.1007/s004010000268. [DOI] [PubMed] [Google Scholar]
- Floyer-Lea A, Matthews PM. Distinguishable brain activation networks for short- and long-term motor skill learning. J Neurophysiol. 2005;94:512–518. doi: 10.1152/jn.00717.2004. [DOI] [PubMed] [Google Scholar]
- Foltynie T, Cheeran B, Williams-Gray CH, Edwards MJ, Schneider SA, Weinberger D, Rothwell JC, Barker RA, Bhatia KP. BDNF val66met influences time to onset of levodopa-induced dyskinesia in Parkinson's disease. J Neurol Neurosurg Psychiatry. 2009;80:141–144. doi: 10.1136/jnnp.2008.154294. [DOI] [PubMed] [Google Scholar]
- Frodl T, Schule C, Schmitt G, Born C, Baghai T, Zill P, Bottlender R, Rupprecht R, Bondy B, Reiser M, et al. Association of the brain-derived neurotrophic factor val66met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–416. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M, Heumann R, Lessmann V. Synaptic secretion of BDNF after high-frequency stimulation of glutamatergic synapses. EMBO J. 2001;20:5887–5897. doi: 10.1093/emboj/20.21.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Moriguchi Y, Yamashita F, Mori T, Nemoto K, Okada T, Hori H, Noguchi H, Kunugi H, Ohnishi T. Dose-dependent effect of the Val66Met polymorphism of the brain-derived neurotrophic factor gene on memory-related hippocampal activity. Neurosci Res. 2008;61:360–367. doi: 10.1016/j.neures.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Ho BC, Milev P, O'Leary DS, Librant A, Andreasen NC, Wassink TH. Cognitive and magnetic resonance imaging brain morphometric correlates of brain-derived neurotrophic factor val66met gene polymorphism in patients with schizophrenia and healthy volunteers. Arch Gen Psychiatry. 2006;63:731–740. doi: 10.1001/archpsyc.63.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of BDNF transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegaya Y, Ishizaka Y, Matsuki N. BDNF attenuates hippocampal LTD via activation of phospholipase C: implications for a vertical shift in the frequency-response curve of synaptic plasticity. Eur J Neurosci. 2002;16:145–148. doi: 10.1046/j.1460-9568.2002.02051.x. [DOI] [PubMed] [Google Scholar]
- Jancke L, Shah N, Peters M. Cortical activations in primary and secondary motor areas for complex bimanual movements in professional pianists. Brain Res. 2000;10:177–183. doi: 10.1016/s0926-6410(00)00028-8. [DOI] [PubMed] [Google Scholar]
- Karni A, Meyer G, Jezzard P, Adams MM, Turner R, Ungerleider LG. Functional MRI evidence for adult motor cortex plasticity during motor skill learning. Nature. 1995;377:155–158. doi: 10.1038/377155a0. [DOI] [PubMed] [Google Scholar]
- Kleim JA, Chan S, Pringle E, Schallert K, Procaccio V, Jimenez R, Cramer SC. BDNF val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nat Neurosci. 2006;9:735–737. doi: 10.1038/nn1699. [DOI] [PubMed] [Google Scholar]
- Kleim JA, Kleim ED, Cramer SC. Systematic assessment of training-induced changes in corticospinal output to hand using frameless stereotaxic transcranial magnetic stimulation. Nat Protoc. 2007;2:1675–1684. doi: 10.1038/nprot.2007.206. [DOI] [PubMed] [Google Scholar]
- Liepert J, Haevernick K, Weiller C, Barzel A. The surround inhibition determines therapy-induced cortical reorganization. Neuroimage. 2006;32:1216–1220. doi: 10.1016/j.neuroimage.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Marchal Crespo L, Reinkensmeyer DJ. Haptic guidance can enhance motor learning of a steering task. J Mot Behav. 2008;40:545–556. doi: 10.3200/JMBR.40.6.545-557. [DOI] [PubMed] [Google Scholar]
- Matzilevich DA, Rall JM, Moore AN, Grill RJ, Dash PK. High-density microarray analysis of hippocampal gene expression following experimental brain injury. J Neurosci Res. 2002;67:646–663. doi: 10.1002/jnr.10157. [DOI] [PubMed] [Google Scholar]
- Monfils MH, Plautz EJ, Kleim JA. In search of the motor engram: motor map plasticity as a mechanism for encoding motor experience. Neuroscientist. 2005;11:471–483. doi: 10.1177/1073858405278015. [DOI] [PubMed] [Google Scholar]
- Morgen K, Kadom N, Sawaki L, Tessitore A, Ohayon J, Frank J, McFarland H, Martin R, Cohen L. Kinematic specificity of cortical reorganization associated with motor training. Neuroimage. 2004;21:1182–1187. doi: 10.1016/j.neuroimage.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Mu Y, Poo MM. Spike timing-dependent LTP/LTD mediates visual experience-dependent plasticity in a developing retinotectal system. Neuron. 2006;50:115–125. doi: 10.1016/j.neuron.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Nemoto K, Ohnishi T, Mori T, Moriguchi Y, Hashimoto R, Asada T, Kunugi H. The val66met polymorphism of the brain-derived neurotrophic factor gene affects age-related brain morphology. Neurosci Lett. 2006;397:25–29. doi: 10.1016/j.neulet.2005.11.067. [DOI] [PubMed] [Google Scholar]
- Oldfield R. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9:97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Oroszi G, Lapteva L, Davis E, Yarboro CH, Weickert T, Roebuck-Spencer T, Bleiberg J, Rosenstein D, Pao M, Lipsky PE, et al. The Met66 allele of the functional val66met polymorphism in the brain-derived neurotrophic factor gene confers protection against neurocognitive dysfunction in systemic lupus erythematosus. Ann Rheum Dis. 2006;65:1330–1335. doi: 10.1136/ard.2006.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploughman M, Granter-Button S, Chernenko G, Tucker BA, Mearow KM, Corbett D. Endurance exercise regimens induce differential effects on brain-derived neurotrophic factor, synapsin-I and insulin-like growth factor I after focal ischemia. Neuroscience. 2005;136:991–1001. doi: 10.1016/j.neuroscience.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Prakash N, Cohen-Cory S, Frostig RD. RAPID and opposite effects of BDNF and NGF on the functional organization of the adult cortex in vivo. Nature. 1996;381:702–706. doi: 10.1038/381702a0. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK. BDNF gene: functional val66met polymorphism in mood disorders and schizophrenia. Pharmacogenomics. 2008;9:1589–1593. doi: 10.2217/14622416.9.11.1589. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Iyo M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: the possibility to explain ethnic mental traits. Am J Med Genet B Neuropsychiatr Genet. 2004;126:122–123. doi: 10.1002/ajmg.b.20118. [DOI] [PubMed] [Google Scholar]
- Siironen J, Juvela S, Kanarek K, Vilkki J, Hernesniemi J, Lappalainen J. The Met allele of the BDNF val66met polymorphism predicts poor outcome among survivors of aneurysmal subarachnoid hemorrhage. Stroke. 2007;38:2858–2860. doi: 10.1161/STROKEAHA.107.485441. [DOI] [PubMed] [Google Scholar]
- Strauss E, Sherman E, Spreen O. A compendium of neuropsychological tests: administration, norms, and commentary. New York: Oxford University Press; 2006. [Google Scholar]
- Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, Sevy S, Ashtari M, Napolitano B, Bilder RM, Kane JM, et al. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol Psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Horiike Y, Matsuzaki M, Miyazaki T, Ellis-Davies GC, Kasai H. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Chupin M, Garnero L, Leonard G, Perron M, Pike B, Pitiot A, Richer L, Veillette S, Pausova Z, et al. Brain volumes and val66met polymorphism of the BDNF gene: local or global effects? Brain Struct Funct. 2009 doi: 10.1007/s00429-009-0203-y. [DOI] [PubMed] [Google Scholar]
- Uchida K, Baba H, Maezawa Y, Furukawa S, Omiya M, Kokubo Y, Kubota C, Nakajima H. Increased expression of neurotrophins and their receptors in the mechanically compressed spinal cord of the spinal hyperostotic mouse (twy/twy) Acta Neuropathol. 2003;106:29–36. doi: 10.1007/s00401-003-0691-4. [DOI] [PubMed] [Google Scholar]
- Wardle RA, Poo MM. Brain-derived neurotrophic factor modulation of GABAergic synapses by postsynaptic regulation of chloride transport. J Neurosci. 2003;23:8722–8732. doi: 10.1523/JNEUROSCI.23-25-08722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemke A, Heagerty P, Lee C, Cramer S. Motor cortex organization after stroke is related to side of stroke and level of recovery. Stroke. 2003;34:E23–E28. doi: 10.1161/01.STR.0000065827.35634.5E. [DOI] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Zivadinov R, Weinstock-Guttman B, Benedict R, Tamano-Blanco M, Hussein S, Abdelrahman N, Durfee J, Ramanathan M. Preservation of gray matter volume in multiple sclerosis patients with the Met allele of the rs6265 (val66met) SNP of brain-derived neurotrophic factor. Hum Mol Genet. 2007;16:2659–2668. doi: 10.1093/hmg/ddm189. [DOI] [PubMed] [Google Scholar]